3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(15):4616-4625. doi:10.7150/jca.56014 This issue Cite
Research Paper
RNF43 and PWWP2B inhibit cancer cell proliferation and are predictive or prognostic biomarker for FDA-approved drugs in patients with advanced gastric cancer
1. Hallym Translational Research Institute, Hallym University Sacred Heart Hospital, Anyang, 14066, Republic of Korea.
2. Department of Internal Medicine, Hallym University Medical Center, Hallym University College of Medicine, Anyang-si, Gyeonggi-do 14068, Republic of Korea.
3. Department of Surgery, Hallym University Medical Center, Hallym University Kangnam Sacred Hospital, Singil-ro Yeongdeungpo-gu, Seoul, 07441, Republic of Korea.
4. Department of Pathology, Hallym University Medical Center, Hallym University College of Medicine, Anyang-si, Gyeonggi-do 14068, Republic of Korea.
5. Department of Bio-medical Gerontology, Ilsong Institute of Life Sciences, Hallym University, Anyang, Gyeonggi-do, Republic of Korea.
*The authors contributed equally to this work.
Received 2020-11-17; Accepted 2021-5-19; Published 2021-6-1
Abstract

Background: Abnormal regulation of genes has been closely related to gastric cancer. The characterization of gastric cancer has necessitated the development of new therapeutics as well as the identification of prognostic markers to predict the response to novel drugs. In our study, we used RNA sequencing analyses to show that on gastric cancer tissues to identification of gastric cancer prognostic markers. We specifically chose to study RNF43 because it inhibits gastric cancer-related Wnt/β-catenin signaling by interacting with Wnt receptors. PWWP2B was chosen because it is a gene which is downregulated in gastric cancer.
Methods: Utilizing RNA sequencing analysis, we evaluated the mRNA expression profile in gastric cancer patients. Also, we used HAP1 cells which is a human near-haploid cell line derived from the male chronic myelogenous leukemia cell line KBM-7. These cell line has one copy of each gene, ensuring the edited allele will not be masked by additional alleles. We investigated the screening of 1,449 FDA-approved drugs in HAP1, HAP1 RNF43 KO and HAP1 PWWP2B KO cells. RNA sequencing data reveals that RNF43 and PWWP2B expression were down-regulated in recurrence gastric cancer patients. Next, we investigated the anti-cancer effects of selected drugs in RNF43 and PWWP2B down-regulated MKN45 gastric cancer cells and xenograft model.
Results: Among these FDA-approved drugs, three drugs (docetaxel trihydrate, pelitinib and uprosertib) showed strong inhibitory effects in RNF43 KO cells and PWWP2B KO cells. In MKN45 xenograft model, tumor volumes were significantly reduced in the docetaxel trihydrate, uprosertib or pelitinib-treated group. Our data demonstrated that RNF43 and PWWP2B are a biomarker that predict recurrence of gastric cancer.
Conclusions: Our findings suggest that docetaxel trihydrate, uprosertib and pelitinib could be used as novel therapeutic agents for the prevention and treatment of gastric cancer with a decrease in RNF43 and PWWP2B expression.
Keywords: gastric cancer, PWWP2B, RNF43, docetaxel trihydrate, pelitinib, uprosertib
Introduction
Gastric cancer (GC) is the fifth most common cancer and third leading cause of cancer deaths worldwide, especially in Eastern Europe, South America, and Eastern Asia (mainly China, Japan, and Korea) [1-4]. However, gastric cancer incidence has decreased markedly in Asian countries in recent years [5]. In South Korea, despite a decline in incidence, it is the second most common cancer [6]. In addition, unsatisfactory treatment outcomes are caused by differences of the molecular basis and intrinsic biological factors. The interplay between poor prognoses related genes such as c-MET, H. pylori, the gastric epithelium, and microbiota is among the most critical factors determining the fate of treatment outcome [7-9]. In an effort to overcome this problem, and to develop and identify new drug candidates, determining tumor characteristics and treatment parameters is important in Eastern Asia. Furthermore, the majority of cancers are loss-of-function events that defy standard inhibitor-based drug screening strategies. One strategy for overcoming the loss of function is to perform high-throughput screening using target gene knockout (KO) cell lines.
RNF43 is encoded by Wnt target genes, and the loss of expression of this E3 ligase has been predicted to result in hyper-responsiveness to endogenous Wnt signals [10]. RNF43 is a tumor suppressor gene in mucinous ovarian cancers, mucinous pancreatic precancerous cysts, and gastric cancer [11-15]. Loss-of-function mutations in RNF43 promotes tumor cell proliferation and result in neoplastic transformation [10]. Recent studies revealed that RNF43 suppresses proliferation and induces apoptosis in gastric carcinoma cells [11, 16]. The PWWP domain is an essential component of DNMT3B that promotes tumorigenesis and contributes to aberrant DNA methylation in carcinogenesis [17].
In this study, we applied an RNA sequencing (RNA-seq) approach to identify RNF43 (i.e., verify a known marker) and PWWP2B (i.e., explore a novel marker) genes differentially expressed in gastric cancer and adjacent normal tissues from 34 patients. The HAP1 cell line was used to determine how the loss-of-function of RNF43 or PWWP2B correlate with gastric cancer. HAP1 cell line has one copy of each gene, ensuring the edited allele will not be masked by additional alleles. To identify Food and Drug Administration (FDA)-approved drugs that selectively target cancer cells with inactivated RNF43 and PWWP2B genes, we performed a high-throughput screening of 1,449 drugs in HAP1, HAP1 RNF43 KO, and HAP1 PWWP2B KO cells. However, HAP1 cell lines were originally derived from human hematopoetic cells. Therefore, anti-cancer effects of selected drugs were re-tested in RNF43 and PWWP2B down-regulated MKN45 gastric cancer xenograft model. This study has the potential to identify additional genes and drugs involved in GC that may contribute to human GC disease.
Materials and Methods
Study subjects and gastric tissue specimen collection
Gastric cancer and adjacent normal tissues obtained from 34 patients with total or subtotal gastrectomy who underwent initial surgery at Hallym University Sacred Heart Hospital from March 2014 to July 2015, were selected as the discovery cohort for RNA-seq. Cases that died within 30 days after surgery were excluded. All cases were prospectively followed up for at least 3 year. Table 1 summarizes the discovery sets. This study was approved by the Ethics Committee of Hallym University Sacred Heart Hospital (2015-I078). Written informed consent was obtained from all of the participants.
RNA-seq and differentially expressed gene (DEG) analyses
Gastric cancer and adjacent normal tissues from 34 patients were subjected to RNA-seq. Total RNA was extracted with TRIzol Reagent. Beads containing oligo (dT) were used to isolate poly(A) mRNA from total RNA. mRNA was fragmented, and first-step cDNA was synthesized with random hexamer primers using PCR. Second-step cDNA was synthesized using dNTPs, RNase H, and DNA polymerase I. Next, short double-stranded cDNA fragments were ligated to Illumina sequencing adaptors. DNA fragments were gel-purified and amplified by polymerase chain reaction (PCR). The amplified library was sequenced on an Illumina HiSeq 2500 sequencing machine. The raw reads were saved in the FASTQ format, and the dirty raw reads were removed before analyzing the data. Reads that could be uniquely mapped to a gene were used to calculate gene expression levels, which were measured based on the number of reads per kilobase of transcript per million mapped reads. We identified differentially expressed genes (DEGs) between paired tumor and normal samples and considered P ≤0.001 as significant.
Cell lines and cell culture
The male and female human cell lines used in this study. HAP1, HAP1 RNF43 KO, and HAP1 PWWP2B KO cells (derived from 40year male chronic myelogenous leukemia) were obtained from Horizon. HAP1 KO cell lines were generated using CRISPR-Cas9 gene editing technology in Horizon Genomics (Vienna Austria) as single clones. SNU620 (derived from 59 year female gastric adenocarcinoma), MKN28 (derived from 37 year male gastric tubular adenocarcinoma), MKN45 (derived from 62year female gastric adenocarcinoma), and Kato III (derived from 57year male Signet ring cell gastric adenocarcinoma) cells were obtained from the Korean Cell Line Bank (KCLB, http://cellbank.snu.ac.kr). The cells were grown and maintained under conditions of 100% humidity and 5% CO2 at 37°C in Iscove's modified Dulbecco's medium (IMDM) or RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% streptomycin and penicillin (Invitrogen Life Technologies, Rockville, MD, USA). The cells were plated onto tissue culture flasks (T-75 cm2) at a density of 1 × 107/mL in hormonally defined IMDM or RPMI 1640 medium. The medium was changed every 3 days until the cells reached 80-90% confluence, at which point they were used in the experiments.
Morphological evaluation
Microscopical studies of the HAP1, HAP1 RNF43 KO, and HAP1 PWWP2B KO cells were carried out with a Microscope. The magnification was 400 fold.
Cell size analysis
HAP1, HAP1 RNF43 KO, and HAP1 PWWP2B KO cells seeded onto 6-well plates at a density 5 × 104 cells per mL. Cell size was determined using a CytoFLEX flow cytometer (Beckman Coulter, Brea, CA, USA). The use of the flow cytometry parameters forward (FSC) and sideward (SSC) scatter of the cells give an indication on gene KO effects through the relative cell size.
Quantitative real-time PCR analysis
RNA was isolated from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instruments, and quantified on a NanoDrop ND-1000 device (Thermo Scientific, Wilmington, DE, USA). Complementary DNA (cDNA) was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Quantitative real-time (qRT) PCR was performed using Power SYBR Green PCR Master Mix on a LightCycler 96 instrument (Roche Applied Science, Indianapolis, IN, USA). The transcript levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used for sample normalization. Primer sequences were as follows: RNF43 (FW 5'-CTG TCA CTG GCT AGC AAG G-3'; RW 5'-AGC TTC TCA GCG TCA TTA CC-3'), PWWP2B (RT2 qPCR Primer Assay, Qiagen, Inc., Valencia, CA, USA), and GAPDH (FW 5'-GAG TCA ACG GAT TTG GTC G-3'; RW 5'-TGG AAT CAT ATT GGA ACA TGT AAA C-3').
Cell proliferation assay
The proliferation of HAP1, HAP1 RNF43 KO, and HAP1 PWWP2B KO cells was assessed using the MTT assay. Cells were cultured in 96-well plates (2 × 104/mL) for 0, 24, 48, and 72 h. MTT solution (5 mg/mL) was added at the end of incubation, which was continued for 4 h. The reaction was terminated by adding a detergent reagent (0.01 M HCl). The absorbance was read at 570 nm using a microplate reader (Synergy 2 Multi-Mode Microplate Reader; BioTek, Winooski, VT, USA).
Colony forming assay
HAP1, HAP1 RNF43 KO, and HAP1 PWWP2B KO cells were diluted and seeded at a density of approximately 5,000 cells per well in 6-cm dish. After incubation for 7 days, colony formation and growth were visualized with crystal violet staining. After the wells were photographed, the dye was solubilized with methanol and the optical density was measured at 570 nm using a microplate reader.
Cell migration assay
HAP1, HAP1 RNF43 KO, and HAP1 PWWP2B KO cells were diluted and seeded at a density of approximately 1 × 105 cells per well in 6-well plates or 24-well plates. After incubation for 1 day, a straight scratch was made on the cells using a P200 pipette tip. The cells were then washed with PBS and further cultured with or without docetaxel trihydrate, pelitinib, and uprosertib in IMDM. After incubation for 0, 48, and 72 h, the gap width of the scratch re-population was photographed and then compared with the initial gap size at 0 h.
High-throughput drug screening
An FDA-approved compound library of 1,448 drugs was purchased from Selleck Chemicals. c-MET inhibitor INC280 was supplied from Novartis (Basel, Switzerland). HAP1, HAP1 RNF43 KO, and HAP1 PWWP2B KO cells were seeded at a density of 2,000 or 3,000 cells per well in 384-well, clear-bottom culture plates with 20 µL IMDM or RPMI 1640 medium containing 10% FBS for 24 h. Then, 10 μM of FDA-approved drug was added to the wells, and the cells were incubated for an additional 48 h. Control cells were not exposed to drugs. On the day of the proliferation assay, the medium was removed, 20 μL of fresh medium was added to each well of the 384-well plates, followed by 5 μL of MTS solution (Cell Titer 96 Aqueous One Solution Cell Proliferation Assay Kit; Promega, Madison, WI, USA), and the plates were incubated at 37 °C for 4 h in a humidified environment with 5% CO2. The absorbance was read at 490 nm using a PerkinElmer (Waltham, MA, USA) EnVision luminescence microplate reader. Data were validated using the Zʹ factor analysis. The percentage inhibition was expressed as [viability level of test samples/viability level of control)] × 100.
Confirmatory growth inhibition assays
The half maximal inhibitory concentrations of the selected drugs of HAP1, HAP1 RNF43 KO, and HAP1 PWWP2B KO cells were measured using the MTS assay for selected drugs at concentrations of 40, 20, 10, 5, 2.5, 1.25, 0.625, and 0.3125 µM for 48 h. On the day of the proliferation assay, medium was removed, and 100 μL of fresh medium was added to each well of 96-well plates, followed by 20 μL of MTS solution, and the plates were incubated at 37 °C for 1 h in a humidified environment with 5% CO2. The absorbance was read at 490 nm using a microplate reader (Synergy 2 Multi-Mode Microplate Readers; BioTek, Winooski, VT, USA). The IC50 values were determined after fitting growth inhibition curves to dose-response curves using GraphPad Prism software (GraphPad Software Inc., CA, USA).
Association of RNF43 and PWWP2B expression with clinicopathological characteristics in 34 gastric cancer patients
| Characteristics | RNF43 low/PWWP2B low (%) | RNF43 high1/PWWP2B low (%) |
|---|---|---|
| Age | 69.0 (44-87) | 62.5 (61-64) |
| Sex | ||
| Male | 17(50.0) | 2(5.9) |
| Female | 15(44.1) | 0 |
| Tumor location | ||
| Pylorus | 1 (2.9) | 0 |
| Antrum | 20 (58.8) | 2 (5.9) |
| Body | 6 (17.7) | 0 |
| Pylorus ~ Antrum | 1 (2.9) | 0 |
| Antrum ~ Body | 4 (11.8) | 0 |
| Lauren's classification | ||
| Diffuse | 15 (44.1) | 0 |
| Intestinal | 6 (17.7) | 1 (2.9) |
| Mixed | 11 (32.4) | 1 (2.9) |
| Cancer stage (TNM class) | ||
| I | 1 (2.9) | 0 |
| II | 9 (26.5) | 0 |
| III | 22 (64.7) | 0 |
| IV | 0 | 2 (5.9) |
| Recurrence | ||
| Yes | 11 (32.3) | 0 |
| No | 21 (61.8) | 2 (5.9) |
1High, fold change >2.0.
Apoptosis analysis
HAP1, HAP1 RNF43 KO, HAP1 PWWP2B KO, and GC (SNU620 cells: high-RNF43, low-PWWP2B; Kato III cells: middle-RNF43 and middle-PWWPB; MKN28: low-RNF43 and high-PWWP2B; and MKN45 cells: low-RNF43 and low-PWWP2B) cells seeded onto 6-well plates at a density 5 × 104 cells per mL were treated with the respective IC50 values of docetaxel trihydrate, pelitinib, and uprosertib (Table 2). Cell death was determined using the Annexin V-APC/Propidium Iodide (PI) Apoptosis Detection Kit (Thermo Fisher Scientific, Rockford, IL, USA) on a CytoFLEX flow cytometer (Beckman Coulter, Brea, CA, USA). The percentages of intact and apoptotic cells were calculated using CytExpert software (Beckman Coulter).
In vivo tumor growth inhibition studies
All the experiments and animal handling procedures in this study were approved by the Animal Experimental Ethics Committee of the Asan Medical Center, Seoul, Korea. Six-week-old male BALB/c-nu/nu mice (Joongang Laboratory Animal Inc., Seoul, Korea) were housed in cages, and maintained at 23 ºC with a 12-h light/dark cycle under specific pathogen free conditions. Each mouse was inoculated subcutaneously (s.c.) into the right flank with either 1 × 107 cells/mouse of RNF43 and PWWP2B down-regulated human gastric cancer cell line MKN45. When the average s.c. tumor volume reached 100 mm3 (day 0), the mice were randomly divided in the following treatment groups (5 mice per group): vehicle control, docetaxel (positive control, 5 mg/kg/weekly intraperitoneally (i.p.)), pelitinib (10 mg/kg/day orally), and uprosertib (10 mg/kg/day orally). Tumor size was measured twice every week with caliper (calculated volume = shortest diameter2 x longest diameter/2). Body weight and tumor size were recorded twice every week. After three weeks, the mice were sacrificed.
Statistical analysis
The data were statistically analyzed using Prism 5 (GraphPad Software Inc.). All values are presented as the mean ± standard deviation. Statistical significance was determined using one-way ANOV (Bonferroni's Multiple Comparison Test) or Fisher's exact test. A P value < 0.05 indicated statistical significance.
Results
Baseline characteristics
A total of 34 subjects were enrolled in this study to gain insight into the molecular pathogenesis of gastric cancer in a Korean population. We searched for genetic alterations using RNA-seq in gastric cancer samples and their matched adjacent normal tissues. By comparing the transcriptome sequences of the cancer tissues with their matched normal tissues, we identified differentially expressed genes, including RNF43 and PWWP2B (Figure S1). The associations of RNF43 and PWWP2B expression with clinicopathological characteristics are shown in Table 1. The subjects included 19 males (56%) and 15 females (44%), with a median age of 68.6 years (range: 44-87 years). At the 3-year follow-up 11 of 34 patients (32%) had gastric cancer relapse (11 of 11 [100%] in the low RNF43 and low PWWP2B expression group). Low expression of RNF43 and PWWP2B (100%) was significantly associated with recurrence (Table 1).
Characterization of RNF43 KO and PWWP2B KO cells. A Representative images (left column) and cell size (middle column) of RNF43KO and PWWP2B cells (x400). B The MTT cell proliferation assay was performed after 0, 24, 48, and 72 h. Data represent the mean value of three experiments performed in triplicate. C The effects of RNF43 and PWWP2B KO on cell proliferation were evaluated by crystal violet staining. D The wound-healing assay was used to assess the effects of RNF43 and PWWP2B KO on the migration ability of HAP1 cells. Both RNF43 KO and PWWP2B KO cells showed increased migration ability compared with control cell lines. WT, wild type; KO, knockout; *** P < 0.001 compared with the WT.
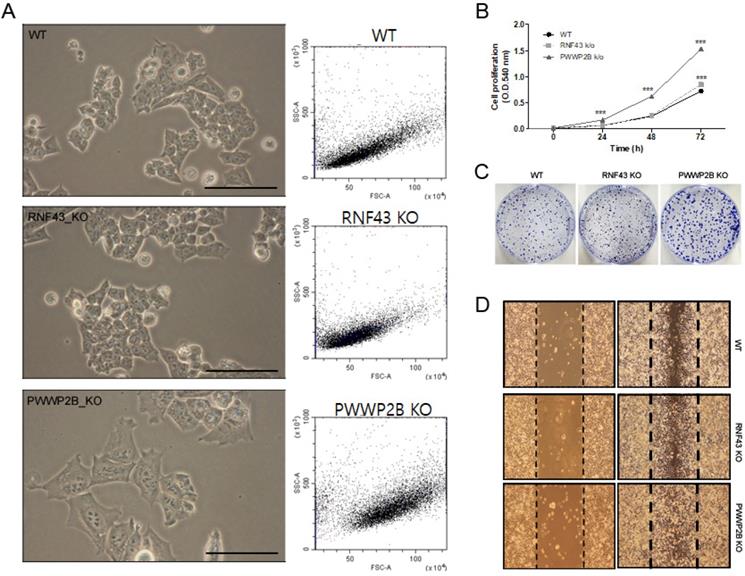
Cell proliferation and migration regulation by RNF43 and PWWP2B
In light microscopy we first investigated the cell size of RNF43 KO or PWWP2B KO compared to wild type (WT) cells (Figure 1A). The KO of RNF43 and PWWP2B show only visible influence on the cell size of PWWP2B KO cells. The morphological parameters of the FACS analysis shows only altered cell size in PWWP2B KO cells (Figure 1A). The effects of RNF43 KO and PWWP2B KO on cell proliferation, we investigated cell proliferation using MTT and colony formation assays (Figure 1B and 1C). RNF43 KO and PWWP2B KO increased the proliferation and colony formation of HAP1 cells. Especially, PWWP2B KO cell line formed colonies much larger than WT and RNF43 KO cells, suggesting that the PWWP2B KO might endow gastric cancer cells with accelerated proliferative capability. Then we investigated cell migration using the wound-healing assay. RNF43 KO and PWWP2B KO associated with increased migration in HAP1 cells (Figure 1D). These results suggest that the loss of RNF43 and PWWP2B may modulate cell migration.
FDA-approved drug library screen
We to try select effective drugs in recurrence gastric cancer patients with low RNF43 and low PWWP2B expression, by using HAP1 cell line. Those cells have one copy of each gene, ensuring the edited allele will not be masked by additional alleles. To evaluate the effect of the 1,449 FDA-approved drugs on HAP1, HAP1 RNF43 KO, and HAP1 PWWP2B KO cell viability, cells were treated with 10 µM of FDA-approved drugs for 48 h, and the inhibitory effect of the drugs was evaluated using the MTS assay. After 48 h, the cell viability of the drug-treated cells was lower than that of the untreated control cells, with 9 drugs resulting in cell viability less than 40% in HAP1 cells. The cells were treated with different concentrations of each drug for 48 h, and the optimal dose was determined by evaluating cell viability using MTS assays. Treatment with the nine drugs (aprepitant, docetaxel trihydrate, ethinyl estradiol, griseofulvin, INC280, pelitinib, pimobendan, tepotinib, and uprosertib) decreased cell viability in a dose-dependent manner (n = 3). The IC50 values of the nine drugs were determined using non-linear regression analysis (Table 2). Among these drugs, docetaxel trihydrate, pelitinib, and uprosertib showed the best inhibition rates.
IC50 of selected drugs in HAP1, HAP1 RNF43 KO and HAP1 PWWP2B KO cells
| Drug | HAP-1 | PWWP2B | RNF43 |
|---|---|---|---|
| IC50 (uM) | |||
| Aprepitant | 17.25 | 14.89 | 16.25 |
| Docetaxel Trihydrate (nM) | 19.83 | 19.31 | 17.19 |
| Ethinyl Estradiol | 18.85 | 15.88 | 16.42 |
| Griseofulvin | 16.00 | 13.46 | 15.56 |
| INC280 | 39.13 | 37.56 | 28.08 |
| Pelitinib | 3.14 | 2.03 | 0.71 |
| Pimobendan | 13.23 | 11.81 | 12.38 |
| Tepotinib | 23.17 | 21.38 | 19.07 |
| Uprosertib | 4.93 | 3.02 | 2.99 |
Effects of three drugs on cell migration
To determine the inhibitory effects of docetaxel trihydrate, pelitinib, and uprosertib on HAP1, HAP1 RNF43 KO, and HAP1 PWWP2B KO cells, cell migration was examined by performing a wound healing assay with the respective IC50 values of the three drugs (Figure 2A-C). The wound gaps in the cells treated with each of the three drugs were significantly wider than those of the untreated groups at 48 and 72 h. IC50 values of these drugs, pelitinib and uprosertib showed the best inhibitory effect.
Effects of three drugs on cell apoptosis
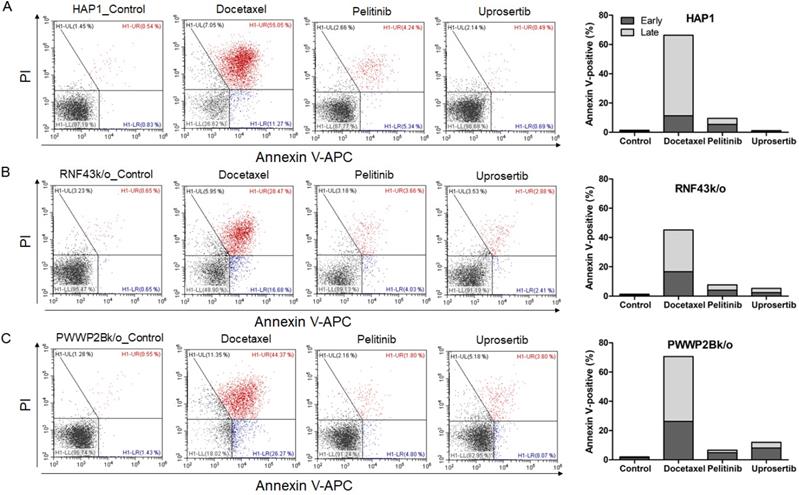
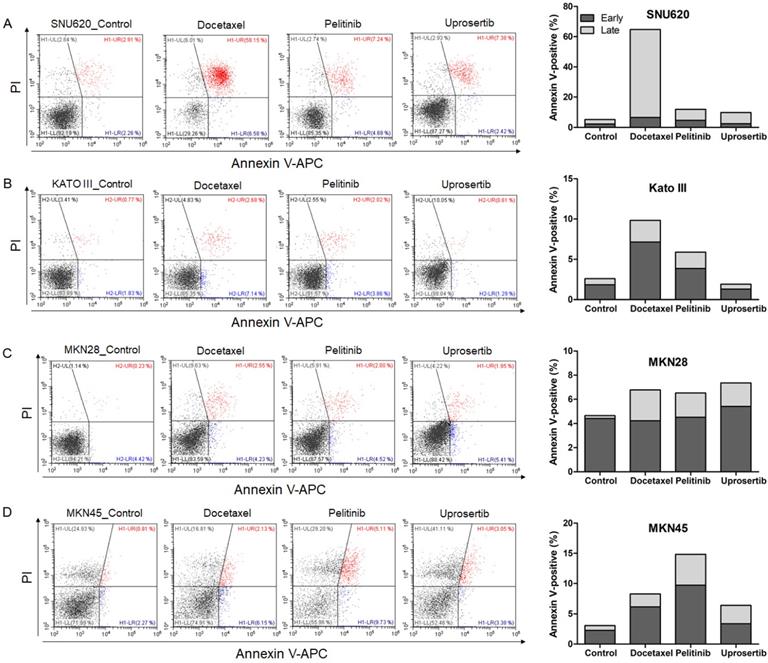
To evaluate the effects of docetaxel trihydrate, pelitinib, and uprosertib on cell death in HAP1, HAP1 RNF43 KO, HAP1 PWWP2B KO cells, apoptosis was examined by flow cytometry (Figure 3). However, these cell lines were leukemia cell line. Therefore, we evaluate the apoptosis effects on GC cells (SNU620 cells: high-RNF43, low-PWWP2B; Kato III cells: middle-RNF43 and middle-PWWPB; MKN28: low-RNF43 and high-PWWP2B; and MKN45 cells: low-RNF43 and low-PWWP2B) was examined (Figure 4). Cells were to assess early apoptosis and the rate of apoptosis in a cell population. Docetaxel trihydrate showed the significantly induced cell death rates in HAP1, RNF43 KO, PWWP2B KO, SNU620, and Kato III cells (Figure 3-4). Pelitinib showed the best cell death rates in MKN45 cells (Figure 4D). RNF43 and PWWP2B genes were weakly expressed in MKN45 cells compared with the other gastric cancer cell types (Figure S2). The percentage of apoptotic cells was 10%, 15%, and 6% after exposure to docetaxel trihydrate, pelitinib, and uprosertib, respectively, while that of control cells was only 3% (Figure 4). In contrast, these drugs were not effects MKN28 cells, which highly express PWWP2B (Figure 4 and Figure S2).
Anti-migration activities of docetaxel, pelitinib, and uprosertib in HAP1, HAP1 RNF43 KO, and HAP1 PWWP2B KO cells. The wound-healing assay of HAP1, HAP1 RNF43 KO, and HAP1 PWWP2B KO cells treated with the respective half maximal inhibitory concentration (IC50) values of docetaxel trihydrate, pelitinib or uprosertib. Docetaxel trihydrate-, pelitinib- and uprosertib-treated cells showed inhibited migration ability compared with control cell lines. Experiments were repeated three times. WT, wild type; Con, control; KO, knockout.

Apoptotic activities of docetaxel, pelitinib and uprosertib in HAP1, HAP1 RNF43 KO and HAP1 PWWP2B KO cells. Flow cytometric assay of A) HAP1, B) HAP1 RNF43 KO, and C) HAP1 PWWP2B KO cells treated with the respective half maximal inhibitory concentration (IC50) values of docetaxel trihydrate, pelitinib or uprosertib. Docetaxel trihydrate-, pelitinib- and uprosertib-treated cells showed induced apoptosis ability compared with control cell lines. Experiments were repeated three times. PI, propidium iodide; Con, control; KO, knockout.
In vivo anti-tumor efficacy of three drugs in tumor xenografts
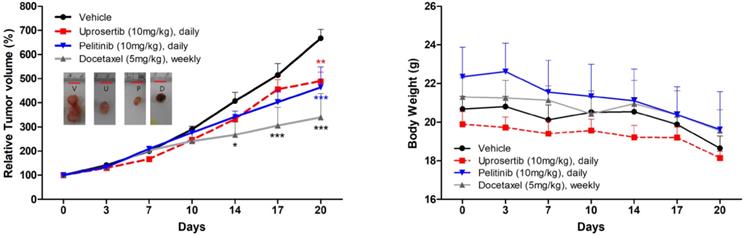
Prompted by the in vitro data supporting a potential anti-tumor activity of docetaxel trihydrate, pelitinib, and uprosertib, we examined the in vivo efficacy of 3 drugs on the growth of MKN45 xenograft models. As demonstrated in Figure 5, mice bearing s.c. MKN45 tumors were treated with docetaxel trihydrate (blocking tubulin), pelitinib (blocking EGFR), and uprosertib (blocking AKT). The docetaxel trihydrate, pelitinib, and uprosertib could significantly inhibit tumor growth at 3 weeks with the inhibition rate 49%, 31%, and 27% in KMN45 xenografts, respectively. The docetaxel trihydrate and pelitinib were well tolerated as demonstrated by the weight gain of treatment groups over the treatment period (Figure 5).
Discussion
Unsatisfactory treatment outcomes occur in Asian countries due to differences in the intrinsic biological factors and rate of diagnosis of gastric cancer between Western and Eastern countries, which represents a major impediment in this field. Therefore, in the present study, 1,449 FDA-approved drugs were screened according to the tumor characteristic status in Korea.
Aberrant regulation of Wnt/β-catenin signaling is observed in colon, ovarian, lung, prostate, liver, breast, and gastric cancers [16, 18-22]. Wnt/β-catenin signaling mediates the epithelial-to-mesenchymal transition in gastric cancer, a process whereby epithelial cells are converted into migratory and invasive cells [23, 24]. RNF43 encodes a transmembrane ubiquitin E3 ligase and is a tumor-suppressing gene that suppresses the Wnt/β-catenin signaling pathway, which is intimately involved in the etiopathogenesis of several cancers [11, 25, 26]. Using a gastric cancer cohort, we retrospectively evaluated the relationship between RNF43 and PWWP2B expression levels and clinical characteristics (Table 1). We found that gastric cancer recurrence patients showed downregulated RNF43 and PWWP2B (100%). In addition, we found RNF43 and PWWP2B mutations in 7 and 6 of 34 gastric cancer patients, respectively. In addition, 34 gastric cancer patients carried germline RNF43 mutations. The RNF43 and PWWP2B genes are potential biomarkers candidates for patients with advanced gastric cancer. As far as we know, the RNF43 gene is well known to be associated with gastric cancer, but the PWWP2B gene has not been found to be associated with gastric cancer. Therefore, we examined the action of RNF43 and PWWP2B in RNF43 KO and PWWP2B KO cell lines. Our results showed that the cellular capacity for proliferation and migration was markedly increased in RNF43 KO and PWWP2B KO cell lines compared with the wild type control. These data suggest that downregulation of RNF43 and PWWP2B might promote proliferation of gastric cancer cells and could be a condition for the conversion of normal gastric epithelial cells into cancerous cells.
Apoptotic activities of docetaxel, pelitinib, and uprosertib in gastric cancer cell lines established from metastasis to the peritoneal cavity (Kato III and SNU620 cells) and gastric carcinoma cell lines (MKN28 and MKN45). Flow cytometric assay of A) SNU620, B) Kato III, C) MKN28, and D) MKN45 cells treated with 12nM, 5uM, and 5uM concentration values of docetaxel trihydrate, pelitinib or uprosertib. Docetaxel trihydrate-, pelitinib- and uprosertib-treated cells showed induced apoptosis ability compared with control cell lines. Experiments were repeated three times. PI, propidium iodide; Con, control; KO, knockout.
Anti-tumor growth activities of docetaxel, pelitinib, and uprosertib in vivo. The effects of relative tumor volume and body weight of MKN45 xenograft mouse treatmented with docetaxel trihydrate (D; 5mg/kg/weekly, intraperitoneally), pelitinib (P; 10mg/kg/day, oral gavage) or uprosertib (U; 10mg/kg/day, oral gavage) at described concentrations. All error bars are SD. * P < 0.05, ** P < 0.01, and *** P < 0.001 compared with control (V; vehicle) group.
Used simultaneously, cell- and target-based screening procedures might be the optimal methods for promoting cancer drug discovery. They can be used to screen large numbers of drugs to determine their therapeutic potential, and correlations between target genes and clinical characteristics. Therefore, in this study, 1,449 FDA-approved drugs were screened to determine whether they could be used as therapeutic agents for the treatment of gastric cancer using growth inhibition assays of HAP1, HAP1 RNF43 KO, and HAP1 PWWP2B KO cells. Among the 1,449 FDA-approved drugs tested, nine (aprepitant, docetaxel trihydrate, ethinyl estradiol, griseofulvin, INC280, pelitinib, pimobendan, tepotinib, and uprosertib) showed high inhibitory activity; therefore, these drugs were selected for further study. Table 2 presents the IC50 values and effective doses of these drugs. Consistently, we have observed the anti-cancer effects using the docetaxel trihydrate, pelitinib, and uprosertib in concentrations as low as 17nmol/L-3 umol/L for in vitro and 5-10 mg/kg for in vivo studies. Docetaxel trihydrate, pelitinib, and uprosertib, which showed the highest inhibition and apoptotic rates of the tested drugs, have been shown to have therapeutic utility. For example, AKT signaling is responsible for development of resistance in cancer to various chemotherapeutics drugs [27]. The downstream protein GSK3ß is major AKT substrate. Docetaxel trihydrate is used widely to treat recurrent or gastric cancer [28]. Docetaxel treatment inhibit phosphorylation state of GSK3ß [29]. Pelitinib is used to treat lung cancer [30]. Pelitinib is also an inhibitor of epidermal growth factor receptor (EGFR), making it active against putative EGFR-dependent tumor types [31, 32]. In addition, Pelitinib inhibit EGF-induced activation of AKT and ERK1/2 in cancer cells [33]. Uprosertib is used in recurrent or persistent ovarian cancer, endometrial cancer, and melanoma [34]. Uprosertib is a broad AKT inhibitor used not only for the treatment of gastric cancer, but also for AKT-dependent cancers [35]. It is well-known that active AKT phosphorylates Gsk3β on serine 9, thereby inactivating it [36]. Gsk3β inhibits Wnt/β-catenin signaling by inhibiting AKT in gastric cancer.
Conclusions
In summary, the results of this study indicate that RNF43 and PWWP2B are downregulation in gastric cancers compared with normal adjacent gastric mucosa. From a clinical aspect, low RNF43 or PWWP2 gene expression showed worse prognosis in overall survival than high gene expression, respectively (http://www.stomachcancerdb.org/). Furthermore, a correlation between low RNF43 and PWWP2B expression and tumor recurrence was seen. Importantly, based on our preclinical finding, docetaxel trihydrate, pelitinib, and uprosertib have significant anti-cancer effects in cancers with low RNF43 and PWWP2B expression (HAP1 RNF43 KO, HAP1 PWWP2B KO cells, MKN45 cells, and KMN45 xenografts). Therefore, further studies are needed to elucidate their mechanisms of action should to aid in the discovery of new therapeutic agents for the treatment of gastric cancer.
Abbreviations
RNA-seq: RNA sequencing; FDA: Food and Drug Administration; DEG: differentially expressed gene; SNP: single-nucleotide polymorphism; indels: insertions/deletions; IMDM: Iscove's modified Dulbecco's medium; FBS: fetal bovine serum; IHC: Immunohistochemical; IC50: half maximal inhibitory concentration.
Supplementary Material
Supplementary figures.
Acknowledgements
Ethics Statement
Ethical registry was obtained from the Institutional Review Board and Ethics Committee of Hallym University Sacred Heart Hospital prior to the beginning of the study under No. 2015-I078. Written, informed consent was obtained from all participants prior to their inclusion in this study.
Funding
This research was supported by the National R&D Program for Cancer Control, Ministry of Health and Welfare (HA17C0054), and Hallym University Research Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Peleteiro B, Severo M, La Vecchia C, Lunet N. Model-based patterns in stomach cancer mortality worldwide. Eur J Cancer Prev. 2014;23(6):524-531
2. Peleteiro B, Barros S, Castro C, Ferro A, Morais S, Lunet N. Worldwide burden of gastric cancer in 2010 attributable to high sodium intake in 1990 and predicted attributable burden for 2030 based on exposures in 2010. Brit J Nutr. 2016;116(4):728-733
3. Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, Negri E, La Vecchia C, Lunet N. Worldwide trends in gastric cancer mortality (1980-2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50(7):1330-1344
4. Yang L, Zheng R, Wang N, Yuan Y, Liu S, Li H, Zhang S, Zeng H, Chen W. Incidence and mortality of stomach cancer in China, 2014. Chinese journal of cancer research = Chung-kuo yen cheng yen chiu. 2018;30(3):291-298
5. Park MS, Yoon JY, Chung HS, Lee H, Park JC, Shin SK, Lee SK, Lee YC. Clinicopathologic Characteristics of Interval Gastric Cancer in Korea. Gut Liver. 2015;9(2):167-173
6. Jung KW, Won YJ, Kong HJ, Oh CM, Lee DH, Lee JS. Cancer Statistics in Korea: Incidence, Mortality, Survival, and Prevalence in 2011. Cancer Res Treat. 2014;46(2):109-123
7. Sohn SH, Lee YC. The genome-wide expression profile of gastric epithelial cells infected by naturally occurring cagA isogenic strains of Helicobacter pylori. Environmental toxicology and pharmacology. 2011;32(3):382-389
8. Sohn SH, Kim B, Sul HJ, Choi BY, Kim HS, Zang DY. Foretinib Inhibits Cancer Stemness and Gastric Cancer Cell Proliferation by Decreasing CD44 and c-MET Signaling. Onco Targets Ther. 2020;13:1027-1035
9. Sohn SH, Kim N, Jo HJ, Kim J, Park JH, Nam RH, Seok YJ, Kim YR, Lee DH. Analysis of Gastric Body Microbiota by Pyrosequencing: Possible Role of Bacteria Other Than Helicobacter pylori in the Gastric Carcinogenesis. Journal of cancer prevention. 2017;22(2):115-125
10. Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, van Es JH, Mohammed S, Heck AJ, Maurice MM. et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488(7413):665-669
11. Niu L, Qin HZ, Xi HQ, Wei B, Xia SY, Chen L. RNF43 Inhibits Cancer Cell Proliferation and Could be a Potential Prognostic Factor for Human Gastric Carcinoma. Cell Physiol Biochem. 2015;36(5):1835-1846
12. Ryland GL, Hunter SM, Doyle MA, Rowley SM, Christie M, Allan PE, Bowtell DD, Australian Ovarian Cancer Study G, Gorringe KL, Campbell IG. RNF43 is a tumour suppressor gene mutated in mucinous tumours of the ovary. J Pathol. 2013;229(3):469-476
13. Zou Y, Wang F, Liu FY, Huang MZ, Li W, Yuan XQ, Huang OP, He M. RNF43 mutations are recurrent in Chinese patients with mucinous ovarian carcinoma but absent in other subtypes of ovarian cancer. Gene. 2013;531(1):112-116
14. Jiang X, Hao HX, Growney JD, Woolfenden S, Bottiglio C, Ng N, Lu B, Hsieh MH, Bagdasarian L, Meyer R. et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A. 2013;110(31):12649-12654
15. Niu L, Qin HZ, Xi HQ, Wei B, Xia SY, Chen L. RNF43 Inhibits Cancer Cell Proliferation and Could be a Potential Prognostic Factor for Human Gastric Carcinoma. Cell Physiol Biochem. 2015;36(5):1835-1846
16. Min BH, Hwang J, Kim NKD, Park G, Kang SY, Ahn S, Ahn S, Ha SY, Lee YK, Kushima R. et al. Dysregulated Wnt signalling and recurrent mutations of the tumour suppressor RNF43 in early gastric carcinogenesis. J Pathol. 2016;240(3):304-314
17. Lechner M, Boshoff C, Beck S. Cancer epigenome. Adv Genet. 2010;70:247-27
18. Wei CY, Zhang X, He S, Liu BL, Han HF, Sun XJ. MicroRNA-219-5p inhibits the proliferation, migration, and invasion of epithelial ovarian cancer cells by targeting the Twist/Wnt/beta-catenin signaling pathway. Gene. 2017;637:25-32
19. Liu L, Zhang YX, Cao WK. Highly expressed lncRNA LOC730101 promotes lung cancer cell growth through Wnt canonical pathway. Biochem Bioph Res Co. 2017;493(2):992-997
20. Murillo-Garzon V, Kypta R. WNT signalling in prostate cancer. Nat Rev Urol. 2017;14(11):683-696
21. Debebe A, Medina V, Chen CY, Mahajan IM, Jia C, Fu D, He L, Zeng N, Stiles BW, Chen CL. et al. Wnt/beta-catenin activation and macrophage induction during liver cancer development following steatosis. Oncogene. 2017;36(43):6020-6029
22. Cho SG. APC downregulated 1 inhibits breast cancer cell invasion by inhibiting the canonical WNT signaling pathway. Oncol Lett. 2017;14(4):4845-4852
23. Huang L, Wu RL, Xu AM. Epithelial-mesenchymal transition in gastric cancer. Am J Transl Res. 2015;7(11):2141-2158
24. Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 2006;66(17):8319-8326
25. de Lau W, Peng WC, Gros P, Clevers H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Gene Dev. 2014;28(4):305-316
26. Tsai JH, Liau JY, Yuan CT, Cheng ML, Yuan RH, Jeng YM. RNF43 mutation frequently occurs with GNAS mutation and mucin hypersecretion in intraductal papillary neoplasms of the bile duct. Histopathology. 2017;70(5):756-765
27. Janjigian YY, Azzoli CG, Krug LM, Pereira LK, Rizvi NA, Pietanza MC, Kris MG, Ginsberg MS, Pao W, Miller VA. et al. Phase I/II trial of cetuximab and erlotinib in patients with lung adenocarcinoma and acquired resistance to erlotinib. Clin Cancer Res. 2011;17(8):2521-2527
28. Cavanna L, Bodini FC, Stroppa EM, Banchini F, Michieletti E, Capelli P, Zangrandi A, Anselmi E. Advanced Gastric Cancer with Liver and Lymph Node Metastases Successfully Resected after Induction Chemotherapy with Docetaxel, Cisplatin and 5-Fluorouracil. Chemotherapy. 2014;60(4):224-227
29. Janjigian YY, Shah MA. Molecularly targeted therapies in advanced gastric cancer. Minerva Gastroenterol Dietol. 2011;57(1):75-88
30. To KKW, Poon DC, Wei YM, Wang F, Lin G, Fu LW. Pelitinib (EKB-569) targets the up-regulation of ABCB1 and ABCG2 induced by hyperthermia to eradicate lung cancer. Brit J Pharmacol. 2015;172(16):4089-4106
31. Hegedus C, Truta-Feles K, Antalffy G, Varady G, Nemet K, Ozvegy-Laczka C, Keri G, Orfi L, Szakacs G, Settleman J. et al. Interaction of the EGFR inhibitors gefitinib, vandetanib, pelitinib and neratinib with the ABCG2 multidrug transporter: implications for the emergence and reversal of cancer drug resistance. Biochem Pharmacol. 2012;84(3):260-267
32. Aravindan N, Aravindan S, Herman TS, Natarajan M. EGFR Tyrosine Kinase Inhibitor Pelitinib Regulates Radiation-Induced p65-Dependent Telomerase Activation in Squamous Cell Carcinoma. Radiation Research. 2013;179(3):304-312
33. Janjigian YY, Park BJ, Zakowski MF, Ladanyi M, Pao W, D'Angelo SP, Kris MG, Shen R, Zheng J, Azzoli CG. Impact on disease-free survival of adjuvant erlotinib or gefitinib in patients with resected lung adenocarcinomas that harbor EGFR mutations. J Thorac Oncol. 2011;6(3):569-575
34. Gungor H, Saleem A, Babar S, Dina R, El-Bahrawy MA, Curry E, Rama N, Chen M, Pickford E, Agarwal R. et al. Dose-Finding Quantitative 18F-FDG PET Imaging Study with the Oral Pan-AKT Inhibitor GSK2141795 in Patients with Gynecologic Malignancies. J Nucl Med. 2015;56(12):1828-1835
35. Dumble M, Crouthamel MC, Zhang SY, Schaber M, Levy D, Robell K, Liu Q, Figueroa DJ, Minthorn EA, Seefeld MA. et al. Discovery of novel AKT inhibitors with enhanced anti-tumor effects in combination with the MEK inhibitor. Plos One. 2014;9(6):e100880
36. Maurer U, Preiss F, Brauns-Schubert P, Schlicher L, Charvet C. GSK-3-at the crossroads of cell death and survival. J Cell Sci. 2014;127(7):1369-1378
Author contact
![]() Corresponding author: Dae Young Zang, Division of Hematology-Oncology, Department of Internal Medicine, Hallym University Medical Center, Hallym University College of Medicine, 22, Gwanpyeong-ro 170beon-gil, Dongan-gu, Anyang-si Gyeonggi-do 14086, Republic of Korea. Phone: +82-31-380-4167; Fax: +82-31-386-1528; E-mail: fhdzangor.kr; fhdzangcom.
Corresponding author: Dae Young Zang, Division of Hematology-Oncology, Department of Internal Medicine, Hallym University Medical Center, Hallym University College of Medicine, 22, Gwanpyeong-ro 170beon-gil, Dongan-gu, Anyang-si Gyeonggi-do 14086, Republic of Korea. Phone: +82-31-380-4167; Fax: +82-31-386-1528; E-mail: fhdzangor.kr; fhdzangcom.

 Global reach, higher impact
Global reach, higher impact