Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(2):510-521. doi:10.7150/jca.26578 This issue Cite
Research Paper
Lycopene upregulates ZO-1 and downregulates claudin-1 through autophagy inhibition in the human cutaneous squamous cell carcinoma cell line COLO-16
1. Jiangsu Key Laboratory of Molecular Biology for Skin Diseases and STIs, Institute of Dermatology, Chinese Academy of Medical Sciences & Peking Union Medical College, Nanjing, 210042, China
2. Dermatology and Venereology Department, Tianjin Medical University General Hospital, Tianjin, China.
# these authors contribute to this work equally.
Received 2018-4-9; Accepted 2018-12-6; Published 2019-1-1
Abstract
Lycopene, a kind of carotenoid, has been reported to have an inhibitory function on tumor cell migration. However, the potential role of lycopene in the treatment of cutaneous squamous cell carcinoma (cSCC) remains unclear. Therefore, we assessed the biological effects of lycopene in the human cSCC cell line COLO-16, human epidermal keratinocytes (HEKs) and the immortalized human keratinocyte cell line HaCaT. We found that lycopene inhibited the cell proliferation and migration of COLO-16 cells but not normal keratinocytes. In addition, lycopene upregulated the protein levels of ZO-1 in COLO-16 and HaCaT cells but not in HEKs. In contrast, lycopene upregulated the protein level of claudin-1 in HEKs but downregulated claudin-1 in COLO-16 cells. Lycopene led to a decrease in autophagic flux in COLO-16 cells in a mechanistic target of rapamycin complex 1 (MTORC1)-dependent manner. Importantly, autophagy inhibition contributed to the lycopene-induced regulation on ZO-1 and claudin-1 in COLO-16 cells. Moreover, JNK inhibitor (SP600125) and MEK inhibitor (U0126) treatment abolished the increase in phosphorylated MTOR and ribosomal protein S6 as well as the increase in ZO-1 and the decrease in claudin-1 in lycopene-treated COLO-16 cells. Gene silencing of JNK and ERK also prohibited ZO-1 upregulation and claudin-1 downregulation. In conclusion, lycopene upregulates ZO-1 expression and downregulates claudin-1 expression through the activation of ERK, JNK and MTORC1 as well as the inhibition of autophagy in human cSCC cells. Our findings demonstrate that autophagy plays a key role in lycopene-mediated pharmacological effects. This study indicates that lycopene might be a useful chemopreventive agent against cSCC.
Keywords: lycopene, cutaneous squamous cell carcinoma, keratinocyte, autophagy, tight junction
Introduction
Lycopene, a kind of carotenoid, is present in various fruits, especially tomatoes. Carotenoids are known to accumulate in the skin and play a protective role [1]. Among the dietary carotenoids, lycopene possesses the best antioxidative activity, and it has been reported to have capabilities in the prevention of various diseases, including cancers and atherosclerosis [2, 3]. Lycopene exerts an inhibitory function on tumor cell migration [4, 5], and suppression MAPK and PI3K/Akt pathways has been supposed to play a role [6]. Increasing evidence suggests a photoprotective effect of lycopene on human skin [7], and recently, the inhibitory effect of lycopene on squamous cell carcinoma proliferation and invasion was evaluated [8]. Cutaneous squamous cell carcinoma (cSCC) is the most common nonmelanoma skin cancer [9]. An increasing annual cSCC incidence has emerged as a public health problem, and it has been reported to have increased by 2 to 8% since the 1960s [10, 11]. However, the potential role of lycopene in the treatment of cSCC remains unclear.
Cell migration is an important process in morphogenesis, regeneration and tumor progression [12], and it is a common characteristic of cancer cells. It has been reported that the weakening of cell junctions is followed by the onset of cell migration [12]. Broken cell-cell junctions lead to migration promotion [13]. At least 40 different tight junctions (TJs) have been identified to function as barriers and fences [14]. TJs have been divided into integral membrane TJ proteins and plaque proteins. The former group includes claudins, which contribute to intercellular homophilic and heterophilic adhesions; the plaque proteins include zonula occludens (ZO)-1, ZO-2, ZO-3, which serve as scaffold proteins [15]. Among the ZO family, ZO-1 was the first to be identified, and its role has clarified more thoroughly than those of ZO-2 and ZO-3. ZO-1 has been reported to be involved in the regulation of cell migration through the alteration of ZO-1 subcellular location [16] or the binding of ZO-1 to MRCKβ, a cytoskeletal dynamics regulatory protein kinase [17]. In addition, ZO-2 inhibition increases the migrative capacities of tumor cells [18]. Among the claudins, claudin-1 is primarily expressed in keratinocytes [19]. Claudin-1 has been reported to act as a promoter in tumor cell migration [20, 21]. Interestingly, silencing of both ZO-1 and ZO-2 has been found to impair the polymerization of claudins [22].
Autophagy is a self-degradative process that has been identified to be involved in diverse cellular processes [23]. The mechanistic target of rapamycin complex 1 (MTORC1), whose repression usually results in autophagy induction, is a key regulator of the autophagy machinery [24]. Some studies have shown that the impact of autophagy on cell migration depends on cell type. For instance, the enhancement of autophagy promotes cell migration in human endometrial stromal cells [25, 26]. Although autophagy is involved in various cellular functions, it remains to be clarified whether autophagy modulates TJ expression. Although lycopene has been reported to induce autophagy in myocardioblasts [27], its regulatory effect on autophagy remains unclear.
Here, we assessed the regulatory effect of lycopene on cell viability, proliferation and migration and detected the molecules involved in the regulation of cell cycle progression, cell junctions and MAPK pathways. We especially focused on autophagy and the autophagic pathway involving MTORC1 in the human cSCC cell line COLO-16, human epidermal keratinocytes (HEKs) and the immortalized human keratinocyte cell line HaCaT. Most importantly, we investigated whether autophagy is involved in lycopene-induced TJ protein regulation.
Materials and Methods
Cell culture
HaCaT and COLO-16 cells (human squamous cell carcinoma cell line [28]) were maintained in the Chinese Academy of Medical Sciences & Peking Union Medical College (as described previously [29]). As previously described[30, 31], HaCaT and COLO-16 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) containing 10% fetal bovine serum (Gibco, Invitrogen, Thermo Fisher, Carlsbad, CA, USA). Primary human epidermal keratinocytes were cultured in Keratinocyte-SFM (Gibco, Invitrogen, Thermo Fisher) as previously described [31]. Cells were incubated at 37°C under 5% CO2.
Reagents and antibodies
The compounds used in this study included lycopene (sc-205738, Santa Cruz Biotechnology, Dallas, TX, USA) dissolved in tetrahydrofuran (0.1% was used in the non-treatment cells as solvent control), rapamycin, trehalose, E64d, pepstatin, acridine orange (AO), and dimethyl sulfoxide (DMSO) (all from Sigma-Aldrich, St. Louis, MO, USA). Other compounds included torin 1 (Tocris, Bristol, UK) and U0126 and SP600125 (both from Cell Signaling Technology, Danvers, MA, USA). All primary and secondary antibodies were purchased from Cell Signaling Technology.
Cytotoxicity measurement
To determine the cytotoxicity of lycopene, a Cell Counting Kit-8 (CCK-8) (Beyotime Biotechnology, Haimen, Jiangsu, China) was used according to the manufacturer's instructions. Cells were seeded onto 96-well plates and then treated with lycopene for 24 hours. Next, CCK-8 reagent was added and incubated for 2 hours. The optical density was measured at a wavelength of 450 nm.
Cell proliferation assay
A bromodeoxyuridine (BrdU) cell proliferation ELISA kit (number 11647229001, Roche Applied Science, Basel, Switzerland) was used according to the manufacturer's instructions. In the Brdu assay, cells were seeded onto 96-well plates with or without lycopene incubation and cultured for 24 hours. Then BrdU was added, and the cells were incubated for 3 hours. Next, BrdU incorporation was determined by ELISA according to the manufacturer's instructions. The optical density was measured at 370 nm and 492 nm wavelengths. The incorporation of BrdU was calculated as follows: absorbance at 370 nm - absorbance at 492 nm.
Colony formation Assay
Cells were treated with or without lycopene for 24 hours. Then, the cells were seeded onto 60 mm culture dishes (5 × 103 cells per dish) and cultured for 10 days at 37 ℃. The cells were washed with PBS and fixed in methanol for 5 minutes. Then, the cells were stained with Giemsa stain for 10 minutes to visualize the colonies. A colony cluster containing more than 20 cells was defined as positive. Images were acquired using a Canon EOS 100D digital camera.
Cell migration assay
An OrisTM Cell Migration Assay kit (collagen I coated) was used according to the manufacturer's instructions (CMACC5.101, Platypus Technologies LLC, Madison, WI, USA). Cells were seeded at a density of 5 × 104 cells per well and cultured for 24 hours. The well stoppers were removed, and the culture medium with or without lycopene was replaced, followed by incubation for 24 hours. Micrographs were captured under an inverted microscope. The cell migration data were calculated by the following formula: (100% - areas without cell migration/area isolated by stopper) × 100.
Western blotting
Cells were lysed in RIPA lysis buffer (Beyotime Biotechnology) containing phosphatase and protease inhibitors (Roche Applied Science). The protein concentration was determined with a BCA assay (Beyotime Biotechnology). The proteins were loaded onto 4-15% precast polyacrylamide gels (Bio-Rad Laboratories, Hercules, CA, USA) and then transferred onto PVDF membranes (Bio-Rad Laboratories). The PVDF membranes were blocked with 3% BSA and incubated with the indicated primary and secondary antibodies. The protein bands were visualized using a chemiluminescence kit (Bio-Rad Laboratories). The density of the protein bands was quantified using Quantity One software. GAPDH served as the loading control.
AO staining assay
Autophagosomes in the cytoplasm, which are acidic vesicular organelles, were marked as red fluorescence by AO staining, while nuclei and cytoplasm were marked by deep and light green fluorescence, respectively. After washing with PBS, cells were incubated with 5 μg/mL AO for 10 minutes and then imaged by confocal microscopy (AO G: λex = 488 nm and λem = 515 nm; AO R: λex= 546 nm and λem = 620 nm). The red and green fluorescence intensities in the cells were determined with Quantity One software.
GFP-LC3B puncta analysis
LC3B-GFP transgene in the cells was generated using a Premo Autophagy Sensor LC3B-GFP BacMam 2.0 system (P36235, Invitrogen, Thermal Fisher) according to the manufacturer's instructions. After transfection for 12 hours, lycopene or E64d and pepstatin were added, and the cells were incubated for 24 hours. Cells were visualized by laser scanning confocal microscopy (GFP scanning: λex = 530 nm and λem = 500 nm). The number of LC3B-GFP puncta in the transfected cells was determined using ImageJ software (http://imagej.nih.gov/ij/).
RNA interference
COLO-16 cells were transfected with JNK or ERK siRNAs at a concentration of 100 nm when the confluence of cells was 30%-50% using 5 μL of Lipofectamine 2000 (Invitrogen, Thermal Fisher) according to the manufacturer's instructions. Cells transfected with scrambled siRNA served as the negative controls (NCs). At 48 hours after transfection, the cells were treated with or without 10 μM lycopene for 24 hours. The JNK siRNA (sc-39101) and scrambled siRNA for JNK (sc-37007) were both obtained from Santa Cruz. The ERK siRNA (GenePharma, Shanghai, China) sequences were 5'-GUG CUC UGC UUA UGA UAA UTT-3' (sense) and 5'-AUU AUC AUA AGC AGA GCA CTT-3' (antisense). The scrambled siRNA sequences for ERK (GenePharma) were 5'-UUC UCC GAA CGU GUC ACG UTT-3' (sense) and 5'-ACG UGA CAC GUU CGG AGA ATT-3' (antisense).
Statistical Analysis
The independent experiments were performed at different times. The results for statistical analysis were obtained from at least three independent experiments. The data were analysed by Student's t-test or univariate ANOVA. SPSS Statistics software (Version 17.0) was used for statistical analysis. Statistical significance was defined as a probability value of less than 0.05.
Results
Lycopene inhibits cell proliferation and migration of the cSCC cell line but not keratinocytes
First, we measured the effect of lycopene on cell viability using CCK-8 and BrdU incorporation assays. Treatment with various concentrations of lycopene (0, 5, 10, 15, 20 and 25 μM) for 24 hours did not impair the cell viability of COLO-16 cells, HEKs or HaCaT cells in the CCK-8 assay (Fig. 1a). Treatment with 5 and 10 μM lycopene increased the viability of HEKs. Next, we confirmed that 10 μM lycopene treatment for 24 hours did not impair DNA synthesis with BrdU incorporation assay (Fig. 1b), which indicated that lycopene does not lead to cell damage. Interestingly, we found that 10 μM lycopene treatment for 24 hours decreased cell colony formation in COLO-16 cells but not in HEKs or HaCaT cells, suggesting inhibition of cell proliferation (Fig. 1c).
Cells were treated with 0, 5, 10, 15, 20 or 25 μM lycopene for 24 hours, and cell toxicity assays were performed using a CCK-8 kit (a). Cells were treated with or without 10 μM lycopene for 24 hours and analysed for DNA synthesis (b), colony formation (c) and cell migration (d). The protein levels of PARP, caspase-3, cyclin B1, cyclin D1, CDK4, phospho-Histone H3, ZO-1, ZO-2, afadin and claudin-1 were determined by western blotting (f-j). The data are shown as the means ± SD of three independent experiments, and representative figures are shown. *: p < 0.05.
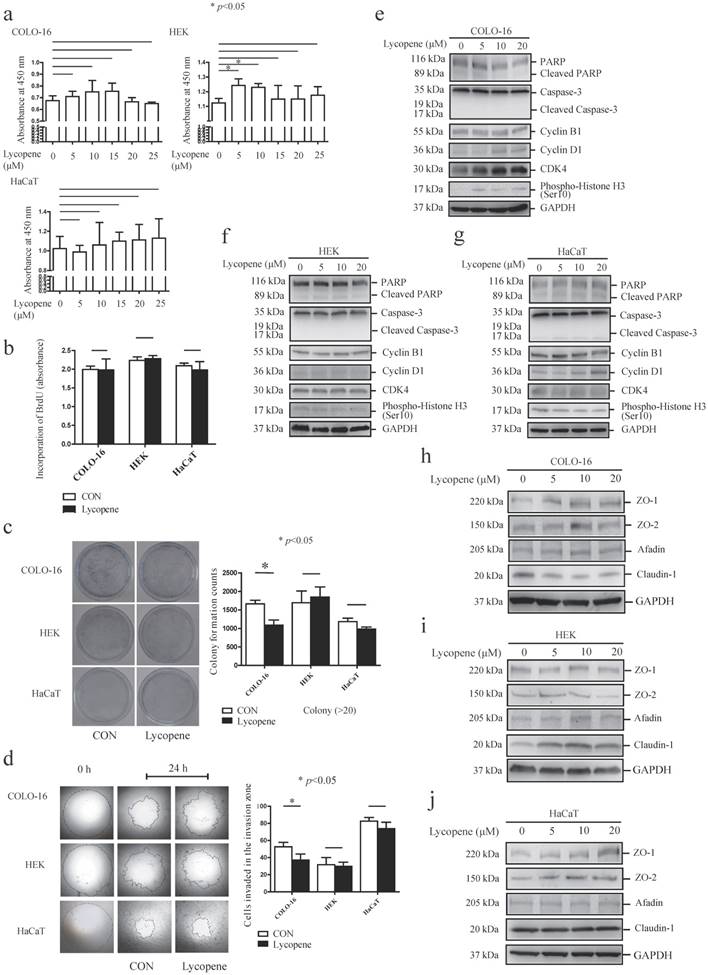
Importantly, transwell migration studies showed that 10 μM lycopene treatment for 24 hours inhibited cell migration only in COLO-16 cells (Fig. 1d). These data demonstrated that the inhibitory effect on cell proliferation and migration is stronger in keratinocyte-derived cancer cells compared to normal keratinocytes.
Lycopene did not induce apoptosis of keratinocytes, but upregulated the cell cycle regulatory proteins Cyclin D1 and CDK4 in COLO-16 cells
We determined the effects of lycopene on basal cell processes such as apoptosis and cell cycle progression in the above three cell types. An effector of apoptosis, caspase-3 is responsible for the cleavage of many proteins, and it was cleaved into 17 and 19 kDa fragments when apoptosis occurs [32]. Poly(ADP-ribose) polymerase (PARP) is a target of active caspase-3, and its cleavage is another marker of apoptosis process[33]. First, we found that 5, 10 and 20 μM lycopene treatment did not lead to the cleavage of PARP or caspase-3 in the three cell types assessed (Fig. 1e-g). Next, we detected the expression of several key cell cycle molecules, cyclin B1, cyclin D1, cyclin-dependent kinase 4 (CDK4) and histone H3. Overexpression of cyclin B1, cyclin D1 and CDK4 has been found in various cancers [34-36]. Cyclin D1 can facilitate cell cycle progression via forming an activating complex with cyclin-dependent kinase 4/6 (CDK4/6)[34]. Cyclin B1 is an important regulator of the G2/M phase [37]. The phosphorylation of histone H3 ser10 is the key event of chromosome condensation and cell cycle progression [38]. We found that 5, 10 and 20 μM lycopene treatment upregulated expression levels of cyclin D1 and CDK4 in COLO-16 cells (Fig. 1e). However, upregulation of CDK4 was not observed in HEKs and HaCaT cells, and upregulation of cyclin D1 was only observed in HaCaT cells treated with 20 μM lycopene (Fig.1f-g).
Lycopene differentially regulates TJ protein expression
Considering the close interplay between TJ proteins and cell migration, we next investigated whether lycopene treatment regulates the expression of TJs in COLO-16 cells, HEKs and HaCaT cells. We found that lycopene upregulated the protein levels of ZO-1 in COLO-16 cells (10 μM lycopene produced the strongest effect) and HaCaT cells (20 μM lycopene produced the strongest effect) but not in HEKs. In contrast, lycopene upregulated the protein level of claudin-1 in HEKs but not in COLO-16 or HaCaT cells. Importantly, lycopene downregulated the expression of claudin-1 in COLO-16 cells. ZO-2 and afadin, an adherens junction protein, were not affected by lycopene in any of the three types of cells assessed (Fig. 1h-j). These data indicate that lycopene treatment differentially regulates TJ protein expression.
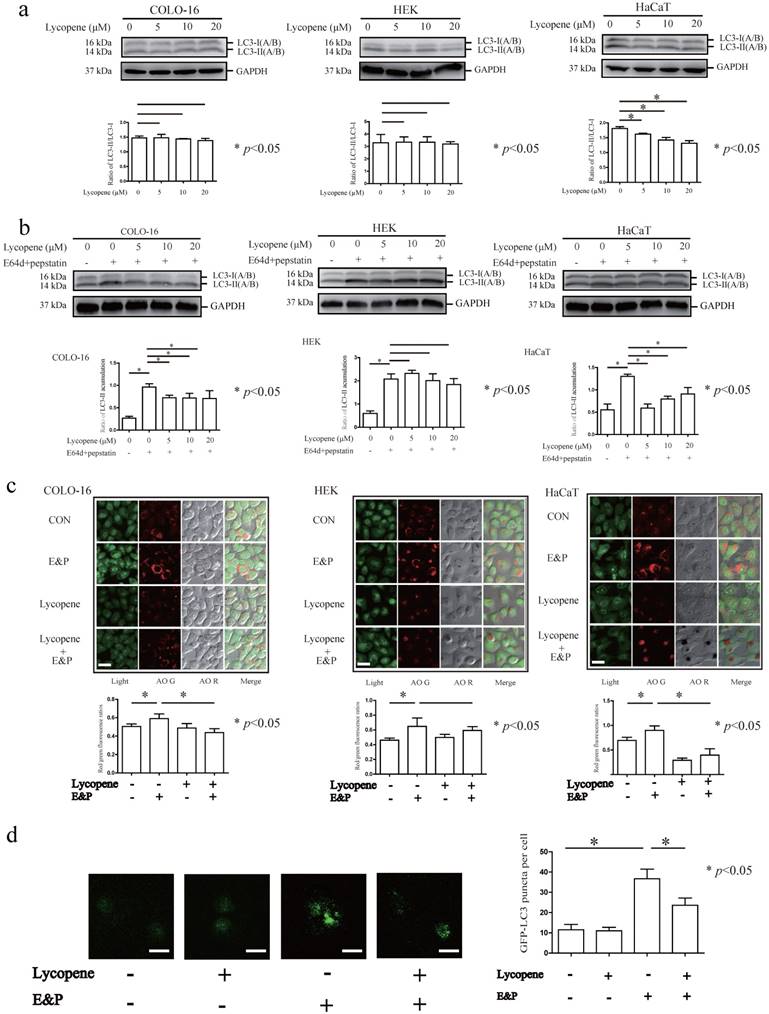
Lycopene decreases autophagy flux in COLO-16 cells
Microtubule-associated protein 1 light chain 3 (LC3) is the most commonly used autophagy marker. The cytosolic form of LC3 (LC3-I) is converted to the lipidated form (LC3-II) when autophagy is induced [39]. However, newborn LC3-II is degraded after autophagolysosome formation. Therefore, the autophagy flux can be determined in the presence of lysosomal inhibitors that block LC3-II degradation [39]. The conversion from LC3-I to LC3-II was decreased in HaCaT cells treated with 5, 10 and 20 μM lycopene for 24 hours (Fig. 2a). In this study, LC3-II accumulation was observed after treatment with the lysosomal inhibitors E64d and pepstatin (E&P) for 24 hours, indicating the basal autophagic flux in the three cell types assessed (Fig. 2b). Furthermore, we observed that LC3-II levels (LC3-II/loading control) were decreased in the 5, 10 and 20 μM lycopene treated COLO-16 and HaCaT cells in the presence of E&P compared with the cells treated with E&P alone. AO staining is a complementary method to monitor autophagy through the visualization of autophagic vacuoles. The red/green fluorescence ratios of COLO-16 and HaCaT cells, but not HEKs, were decreased in 10 μM lycopene-treated cells in the presence of E&P compared with the cells treated with E&P alone (Fig. 2c). These data suggest that lycopene treatment leads to a decrease in autophagic flux in COLO-16 and HaCaT cells but not in HEKs. The inhibition of autophagy flux in 10 μM lycopene-treated COLO-16 cells was confirmed by GFP-LC3B puncta analysis, which monitored the GFP-tagged LC3 aggregation on the autophagosome membrane (Fig. 2d).
Autophagy inhibition contributes to the regulatory effect of lycopene on ZO-1 and claudin-1
MTORC1, whose repression usually results in autophagy induction, is a key regulator of the autophagy machinery [24]. We observed an increase in the phosphorylation of MTOR at ser2448 and its substrates [40-42], including p70 S6 kinase at thr389, ribosomal protein S6 at ser235/236 and 4E-BP1 at ser65, in COLO-16 cells (Fig. 3a). These findings demonstrated that the MTORC1 pathway is involved in lycopene-induced autophagy inhibition in COLO-16 cells. Notably, we observed an increase in ribosomal protein S6 phosphorylation in HEKs and HaCaT cells treated with lycopene (Fig. 3a), although the changes in other components in the MTORC1 pathway were not as significant as in COLO-16 cells.
Cells were treated with 0, 5, 10 or 20 μM lycopene (a) with or without 10 μg/mL E64d and 10 μg/mL pepstatin (b) for 24 hours. Western blotting analysis was performed using a primary antibody against LC3A/B. GAPDH served as a loading control. The LC3-II / LC3-I and LC3-II / GAPDH ratios were calculated. Cells were treated with or without 10 μM lycopene in the presence or absence of 10 μg/mL E64d and 10 μg/mL pepstatin for 24 hours (c and d). Cells stained by AO (c) or transfected with GFP-LC3B (d) were visualized using a laser scanning confocal microscope. The means of the red/green fluorescence ratios (c) or the GFP-LC3 puncta (d) for individual cells were determined for statistical analysis. The data are shown as the means ± SD of three independent experiments, and representative figures are shown. Bars = 5 μm. *: p < 0.05.
Cells were treated with 0, 5, 10 or 20 μM lycopene (a), and then the protein levels and phosphorylation of MTOR, p70 S6 kinase, ribosomal protein S6 and 4E-BP1 were determined by western blotting. Cells were treated with or without 10 μM lycopene in the presence or absence of 80 nM rapamycin (b and c), 1 μM torin 1 (d) or 100 mM trehalose (f) for 24 hours, and the protein levels and phosphorylation of MTOR (b, d and f) and ribosomal protein S6 (b), as well as the protein levels of ZO-1, claudin-1 (c, d and f) and LC3 (d and f), were determined by western blotting. In addition, COLO-16 cells were treated with or without 10 μM lycopene in the presence or absence of 10 μg/mL E64d and 10 μg/mL pepstatin or 80 nM rapamycin (e), and the levels of LC3 protein were determined by western blotting. GAPDH served as a loading control. The LC3-II / LC3-I or LC3-II / GAPDH ratios were calculated. The data are shown as the means ± SD of three independent experiments, and representative figures are shown. *: p < 0.05.
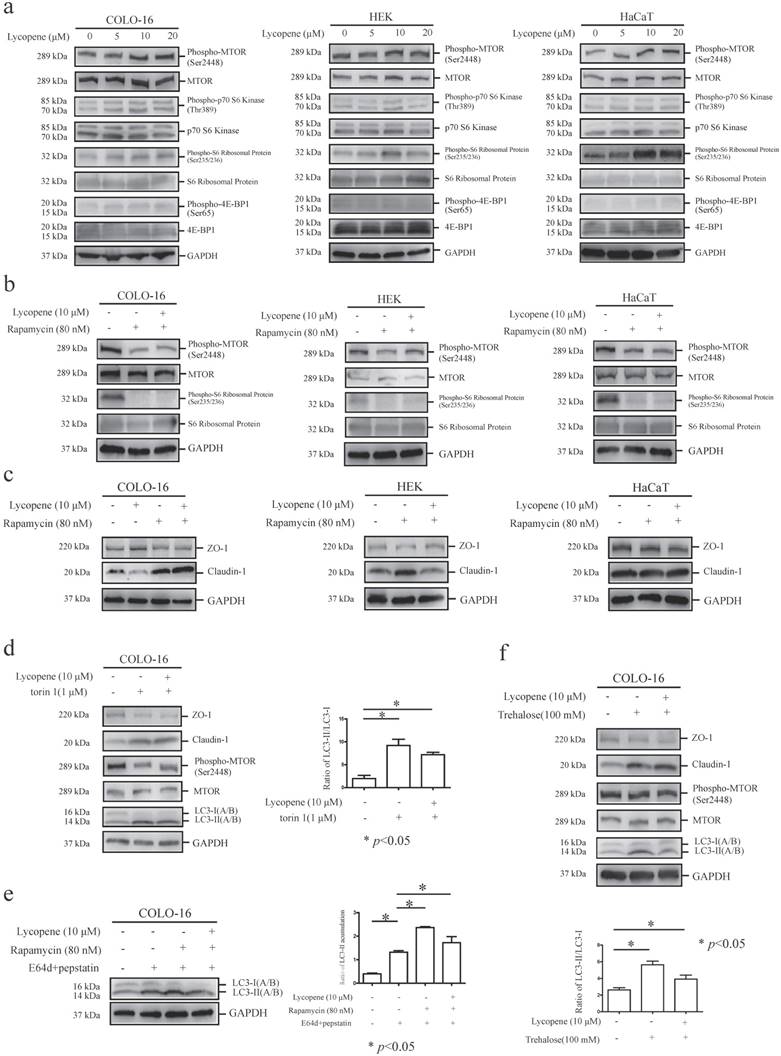
Rapamycin is an MTOR inhibitor and MTOR-dependent autophagy inducer [39]. Here, cells were co-incubated with lycopene and 80 nM rapamycin for 24 hours, and we found that rapamycin treatment decreased the phosphorylation of MTOR and ribosomal protein S6 in all three cell types assessed, indicating sensitivity to rapamycin. Importantly, the lycopene-induced increases in MTOR and ribosomal protein S6 phosphorylation were abolished in the presence of rapamycin in COLO-16 cells, suggesting that rapamycin treatment inhibited lycopene-induced MTORC1 activation (Fig. 3b).
Next, we found that rapamycin treatment prohibited the lycopene-induced upregulation of ZO-1 and downregulation of claudin-1 in COLO-16 cells (Fig. 3c), indicating that the MTORC1 pathway is involved in lycopene-related TJ regulation. We validated this result using another MTOR inhibitor, torin 1 [43] (Fig. 3d). Interestingly, rapamycin treatment also abolished the lycopene-induced upregulation of claudin-1 in HEKs (Fig. 3c). Rapamycin and torin 1 have both been reported to induce autophagy via inhibiting the MTORC1 pathway in some cells [39, 43]. We found that rapamycin treatment attenuated the lycopene-induced decrease in LC3-II accumulation (Fig. 3e), and torin 1 also prohibited the decrease in conversion from LC3-I to LC3-II in COLO-16 cells (Fig. 3c). These data indicate that autophagy inhibition might contribute to the lycopene-induced regulation of ZO-1 and claudin-1. Therefore, we further confirmed the key role of autophagy regulation using the MTOR-independent autophagy inducer trehalose [30]. We found that trehalose treatment enhanced the conversion from LC3-I to LC3-II in an MTOR-independent mechanism and abolished the lycopene-induced upregulation of ZO-1 and downregulation of claudin-1 (Fig. 3f). These data demonstrate that lycopene-induced autophagy inhibition plays a crucial role in the lycopene-mediated regulation of TJ expression.
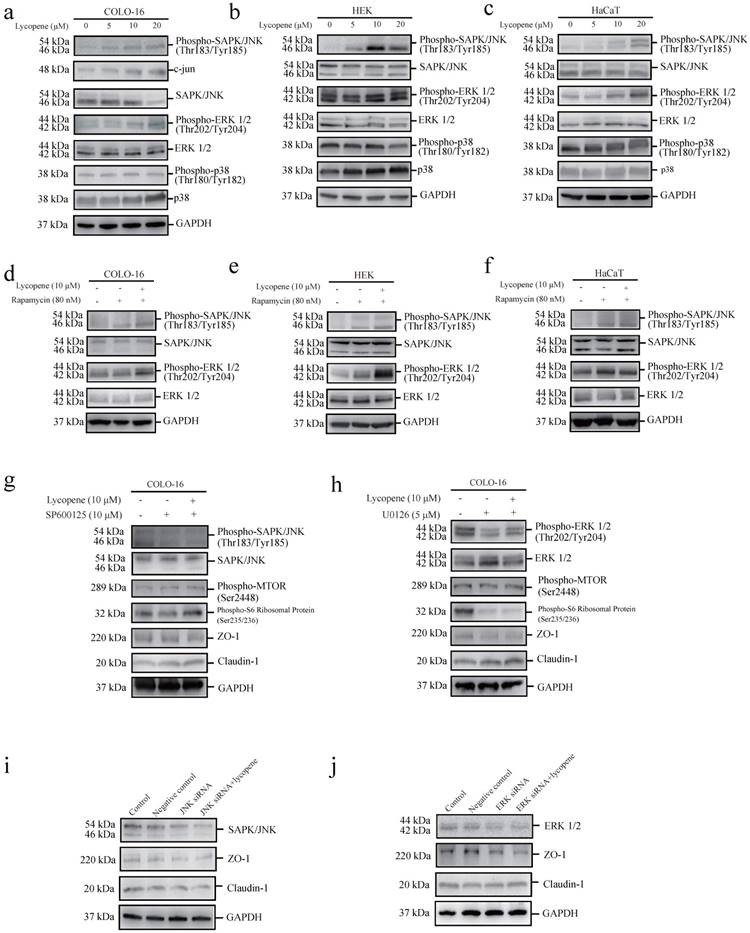
Lycopene activates MTORC1 signalling through the JNK and ERK pathways in COLO-cells
c-Jun N-terminal kinase (JNK) [44], signal-regulated kinase (ERK1/2) [45] and p38 MAPK [46] have been reported to be capable of positively regulating the MTOR pathway. We found that lycopene treatment led to increases in JNK and ERK phosphorylation, but not p38 MAPK phosphorylation, in the three cell types assessed (Fig. 4a-c). We confirmed JNK activation in lycopene-treated COLO-16 cells by measuring the protein level of c-jun, a substrate of the JNK pathway. We observed that lycopene treatment led to an increase in c-jun expression (Fig. 4a). Considering the close interplay between MTOR signalling and MAPK signalling (including JNK, ERK and p38 MAPK signalling), we explored whether the activation of JNK or ERK could be abolished by inhibiting the lycopene-induced activation of MTORC1 signalling using rapamycin treatment. We found that rapamycin treatment did not prohibit the increase in JNK and ERK phosphorylation in the three cell types assessed (Fig. 4d-f). Furthermore, we found that both a JNK inhibitor (SP600125) and a MEK inhibitor (U0126) abolished the lycopene-induced increase in phosphorylated MTOR and ribosomal protein S6 in COLO-16 cells (Fig. 4g, h). These findings demonstrate that lycopene treatment activates MTORC1 signalling through the JNK and ERK pathways in COLO-16 cells.
We further used the JNK inhibitor (SP600125) and the MEK inhibitor (U0126) to investigate whether the JNK and ERK pathways are involved in the lycopene induced regulation of ZO-1 and claudin-1. We found that both inhibitors abolished the lycopene-induced increase in ZO-1 expression and decrease in claudin-1 expression in COLO-16 cells (Fig. 4g, h), indicating that the activation of the ERK and JNK pathways contributes to the lycopene-induced changes in TJ proteins. In addition, gene silencing of JNK and ERK also prohibited the upregulation of ZO-1 and the downregulation of claudin-1 in COLO-16 cells (Fig. 4i, j).
Discussion
In this study, we demonstrate that lycopene treatment upregulates ZO-1 expression and downregulates claudin-1 expression through autophagy inhibition in human cSCC cells. Furthermore, we reveal that the activation of JNK and ERK contributes to the lycopene-induced activation of MTORC1, upregulation of ZO-1 and downregulation of claudin-1. These effects might be involved in the lycopene-induced inhibition of cell migration.
The exact regulatory effect of lycopene on autophagy and its underlying mechanism remain unclear. Chen et al [27] reported that lycopene increased the autophagy level in myocardioblasts. Buyuklu et al, found that lycopene treatment improved LC3B expression in contrast agents-induced nephropathy [47]. Here, our study verified that the antioxidant and anticarcinogenic agent lycopene is a novel autophagy inhibitor in keratinocytes and keratinocyte-derived cancer cells. However, lycopene-induced autophagy inhibition is mediated by an MTOR-dependent mechanism, which should be considered in further investigations, as Karayannopulou et al [48], observed a significantly elevated level of phosphorylated MTOR in cSCC tissues. The MTOR inhibitor rapamycin has been reported to oppose the carcinogenesis of epidermal cancer [49] and to exhibit an antitumoral effect in kidney transplant recipients with previous cSCC [50].
Cells were treated with 0, 5, 10 or 20 μM lycopene (a-c). Western blotting analysis was performed to determine the protein levels or phosphorylation of ERK, JNK and p38 MAPK. The protein level of c-jun was determined in COLO-16 cells (a). Cells were treated with or without 10 μM lycopene in the presence or absence of 80 nM rapamycin (d-f), 10 μM SP600125 (g) or 5 μM U0126 (h) for 24 hours. Western blotting analysis was performed to determine the protein levels of ZO-1, claudin-1, JNK and ERK, as well as the phosphorylation of JNK and ERK. In addition, the phosphorylation of MTOR and ribosomal protein S6 was determined in COLO-16 cells (g and h). (i and j): The COLO-16 cells in the control, negative control, RNAi JNK (or ERK) and RNAi JNK (or ERK) plus lycopene groups were treated with transfection agent (Lipofectamine 2000) alone, scrambled siRNA, JNK siRNA (or ERK siRNA) and JNK siRNA (or ERK siRNA) followed by 10 μM lycopene, respectively. The transfection agents were given in the same volume (5 μL) in the control, negative control, RNAi of JNK (or ERK), and RNAi of JNK (or ERK) plus lycopene groups. GAPDH served as a loading control. Representative figures are shown from three independent experiments.
Interestingly, lycopene exhibits an effect against cSCC cells, although it enhances MTOR activity. In addition to autophagy regulation, the MTOR pathway participates in various cellular processes, such as metabolism [51], cell proliferation [52], angiogenesis [53] and cell cycle progression [54]. It should be considered that lycopene-induced effects on extensive cell processes associated with MTOR signalling might lead to undesirable consequences of the use of lycopene in cancer treatments.
Claudin-1 binds to ZO-1 through its ZO-1 binding site, contributing to the formation of tight junction strands [55]. Although both ZO-1 and claudin-1 belong to the family of TJ proteins, their roles might be distinct in carcinogenesis and tumor development. For example, Morita et al found that ZO-1 and claudin-1 were strongly expressed in tumor cells with keratinization but that ZO-1 was decreased or unexpressed in unkeratinized tumor cells[56]. Therefore, lycopene-induced ZO-1 upregulation should be explored in further studies on cSCC treatment. Our study also showed that lycopene differentially regulates the expression of ZO-1 and claudin-1 along with its anti-tumor effects.
Lycopene inhibits platelet-derived growth factor (PDGF)-AA/-AB-induced migration in smooth muscle cells and fibroblasts [57]. In addition, lycopene exhibits an inhibitory effect on migration in prostate cancer cell lines [5] and SK-Hep1 human hepatoma cell lines [4]. Interestingly, lycopene inhibits PDGF-BB induced retinal pigment epithelial cell migration by suppressing ERK and p38 MAPK activation. Although we observed that lycopene could inhibit migration of cSCC cell line, and the ERK and JNK signalling were activated. We speculate that the molecular mechanisms of lycopene-induced effects involve histological specificity.
There are limited reports on how autophagy is involved in the regulation of TJ protein function. Yang et al [58], found that amino acid deprivation led to the impairment of barrier function including decreased expression of ZO-1 and claudin-1 and increased permeability, in a porcine intestinal epithelial cell model. Importantly, they found that enhancement of autophagy with rapamycin partially attenuated the abovementioned barrier dysfunction, and inhibition of autophagy with 3-methyladenine enhanced the dysfunction. Li et al [59] also reported that inducing autophagy with rapamycin or lithium carbonate reversed the decrease in ZO-1 in brain microvascular endothelial cells associated with ischemia-reperfusion injury. Intriguingly, our study demonstrates that lycopene upregulates ZO-1 expression and downregulates claudin-1 expression by inhibiting autophagy flux in the cSCC cell line. Although our results are not completely consistent with those of previous reports on the role of autophagy in TJ protein regulation, the findings strongly indicate that autophagy plays an important role in the function of TJ proteins. The underlying mechanism of how the autophagy machinery modulates TJ proteins needs to be elucidated through additional investigation.
In this study, we found that an effect of lycopene was present in cSCC cells but not in normal keratinocytes. In fact, such tumor-selective effects were also observed in some previous studies. For example, lycopene inhibited proliferation and downregulated acidic isoforms of cytokeratin 19 in breast cancer cells but not in normal breast cells [60]. However, the mechanisms leading to the differential regulation between cancer and normal cells has not been clarified. In our previous study, we found that lycopene inhibited the phosphorylation of epidermal growth factor receptor (EGFR) in COLO-16 cells but not in HaCaT cells and HEKs [61]. Lycopene has been reported to inhibit EGFR gene expression in prostate cancer cells [62]. EGFR has been reported to possibly be involved in the modulation of SCC growth and migration [63]. Thus, we speculate that the selective regulation of some key pathways, such as lycopene-mediated EGFR signalling might contribute to the different effects of lycopene on keratinocyte-derived cancer cells and normal keratinocytes.
In summary, our study reports a novel effect of lycopene on the expression of the TJ proteins ZO-1 and claudin-1 through the activation of ERK, JNK and MTORC1 as well as the inhibition of autophagy in cSCC cells. Our data demonstrate that autophagy plays a key role in lycopene-mediated pharmacological effects. These findings broaden the scope of investigations on the molecular mechanisms underlying the anti-tumor effects of natural agents. However, we could not determine whether the ERK and JNK pathways are the crucial mediators of the lycopene-induced effects, or whether changes in ZO-1 and claudin-1 lead to the lycopene-induced inhibition of cell migration. Our study indicates that lycopene might be a useful chemopreventive agent, and the potential value of lycopene is worthy of additional investigation in future studies.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (No. 81773342, 81673083, 81773338, 81703153), CAMS Innovation Fund for Medical Sciences (2017-I2M-1-017, 2016-I2M-1-005), PhD Programs Foundation of Ministry of Education of China (No. 20131106120046), the Open Foundation of State Key Laboratory of Pollution Control and Resource Reuse (PCRRF17032). Song Xu, Xu Chen and Wenbo Bu, are supported by the PUMC Youth Fund and Fundamental Research Funds for the Central Universities (3332015116, 3332014008, 3332015026, 2016RC320005, 2016ZX320014).
Author Contributions
Xu Chen, Li Li, Suyun Bi and Zhihai Zhou wrote the main manuscript text. Li Li, Song Xu, Xu Chen, Mengli Zhang Suyun Bi and Wenbo Bu performed the experiments and statistical analysis and jointly prepared all figures in this work. Xu Chen, Heng Gu, Li Li and Min li jointly developed the study design. Xu Chen is corresponding author. All authors reviewed the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Scarmo S, Cartmel B, Lin H, Leffell DJ, Welch E, Bhosale P. et al. Significant correlations of dermal total carotenoids and dermal lycopene with their respective plasma levels in healthy adults. Archives of biochemistry and biophysics. 2010;504:34-9
2. Agarwal S, Rao AV. Tomato lycopene and its role in human health and chronic diseases. CMAJ: Canadian Medical Association journal = journal de l'Association medicale canadienne. 2000;163:739-44
3. Kavanaugh CJ, Trumbo PR, Ellwood KC. The U.S. Food and Drug Administration's evidence-based review for qualified health claims: tomatoes, lycopene, and cancer. Journal of the National Cancer Institute. 2007;99:1074-85
4. Hwang ES, Lee HJ. Inhibitory effects of lycopene on the adhesion, invasion, and migration of SK-Hep1 human hepatoma cells. Experimental biology and medicine. 2006;231:322-7
5. Elgass S, Cooper A, Chopra M. Lycopene treatment of prostate cancer cell lines inhibits adhesion and migration properties of the cells. International journal of medical sciences. 2014;11:948-54
6. Chan CM, Fang JY, Lin HH, Yang CY, Hung CF. Lycopene inhibits PDGF-BB-induced retinal pigment epithelial cell migration by suppression of PI3K/Akt and MAPK pathways. Biochemical and biophysical research communications. 2009;388:172-6
7. Grether-Beck S, Marini A, Jaenicke T, Stahl W, Krutmann J. Molecular evidence that oral supplementation with lycopene or lutein protects human skin against ultraviolet radiation: results from a double-blinded, placebo-controlled, crossover study. The British journal of dermatology. 2017;176:1231-40
8. Shimamoto S, Arai S, Enomoto T, Koji T, Morita O. [Oral health condition of the population in special elderly nursing home]. Shigaku = Odontology; journal of Nihon Dental College. 1989;77:1416-22
9. Dooley TP, Reddy SP, Wilborn TW, Davis RL. Biomarkers of human cutaneous squamous cell carcinoma from tissues and cell lines identified by DNA microarrays and qRT-PCR. Biochemical and biophysical research communications. 2003;306:1026-36
10. Glass AG, Hoover RN. The emerging epidemic of melanoma and squamous cell skin cancer. Jama. 1989;262:2097-100
11. Green A. Changing patterns in incidence of non-melanoma skin cancer. Epithelial cell biology. 1992;1:47-51
12. Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nature reviews Molecular cell biology. 2009;10:445-57
13. Zhuang Q, Liu C, Qu L, Shou C. Synuclein-gamma promotes migration of MCF7 breast cancer cells by activating extracellular-signal regulated kinase pathway and breaking cell-cell junctions. Molecular medicine reports. 2015;12:3795-800
14. Gonzalez-Mariscal L, Betanzos A, Nava P, Jaramillo BE. Tight junction proteins. Progress in biophysics and molecular biology. 2003;81:1-44
15. McNeil E, Capaldo CT, Macara IG. Zonula occludens-1 function in the assembly of tight junctions in Madin-Darby canine kidney epithelial cells. Molecular biology of the cell. 2006;17:1922-32
16. Nasu Y, Ido A, Tanoue S, Hashimoto S, Sasaki F, Kanmura S. et al. Hepatocyte growth factor stimulates the migration of gastric epithelial cells by altering the subcellular localization of the tight junction protein ZO-1. Journal of gastroenterology. 2013;48:193-202
17. Huo L, Wen W, Wang R, Kam C, Xia J, Feng W. et al. Cdc42-dependent formation of the ZO-1/MRCKbeta complex at the leading edge controls cell migration. The EMBO journal. 2011;30:665-78
18. Luczka E, Syne L, Nawrocki-Raby B, Kileztky C, Hunziker W, Birembaut P. et al. Regulation of membrane-type 1 matrix metalloproteinase expression by zonula occludens-2 in human lung cancer cells. Clinical & experimental metastasis. 2013;30:833-43
19. Brandner JM, Kief S, Grund C, Rendl M, Houdek P, Kuhn C. et al. Organization and formation of the tight junction system in human epidermis and cultured keratinocytes. European journal of cell biology. 2002;81:253-63
20. Bhat AA, Ahmad R, Uppada SB, Singh AB, Dhawan P. Claudin-1 promotes TNF-alpha-induced epithelial-mesenchymal transition and migration in colorectal adenocarcinoma cells. Experimental cell research. 2016;349:119-27
21. Zhou B, Blanchard A, Wang N, Ma X, Han J, Schroedter I. et al. Claudin 1 promotes migration and increases sensitivity to tamoxifen and anticancer drugs in luminal-like human breast cancer cells MCF7. Cancer investigation. 2015;33:429-39
22. Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M. et al. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 2006;126:741-54
23. Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F. et al. Molecular definitions of autophagy and related processes. The EMBO journal. 2017;36:1811-36
24. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology. 2011;13:132-41
25. Liu H, Zhang Z, Xiong W, Zhang L, Xiong Y, Li N. et al. Hypoxia-inducible factor-1alpha promotes endometrial stromal cells migration and invasion by upregulating autophagy in endometriosis. Reproduction. 2017;153:809-20
26. Ferraresi A, Phadngam S, Morani F, Galetto A, Alabiso O, Chiorino G. et al. Resveratrol inhibits IL-6-induced ovarian cancer cell migration through epigenetic up-regulation of autophagy. Molecular carcinogenesis. 2017;56:1164-81
27. Chen F, Sun ZW, Ye LF, Fu GS, Mou Y, Hu SJ. Lycopene protects against apoptosis in hypoxia/reoxygenationinduced H9C2 myocardioblast cells through increased autophagy. Molecular medicine reports. 2015;11:1358-65
28. Moore GE, Merrick SB, Woods LK, Arabasz NM. A human squamous cell carcinoma cell line. Cancer research. 1975;35:2684-8
29. Zhang M, Tao Y, Ma P, Wang D, He C, Cao Y. et al. Synthesis and characterization of a new retinoic acid ECPIRM as potential chemotherapeutic agent for human cutaneous squamous carcinoma. Anti-cancer agents in medicinal chemistry. 2015;15:1204-12
30. Chen X, Li M, Li L, Xu S, Huang D, Ju M. et al. Trehalose, sucrose and raffinose are novel activators of autophagy in human keratinocytes through an mTOR-independent pathway. Scientific reports. 2016;6:28423
31. Xu S, Li L, Li M, Zhang M, Ju M, Chen X. et al. Impact on Autophagy and Ultraviolet B Induced Responses of Treatment with the MTOR Inhibitors Rapamycin, Everolimus, Torin 1, and pp242 in Human Keratinocytes. Oxidative medicine and cellular longevity. 2017;2017:5930639
32. Scabini M, Stellari F, Cappella P, Rizzitano S, Texido G, Pesenti E. In vivo imaging of early stage apoptosis by measuring real-time caspase-3/7 activation. Apoptosis: an international journal on programmed cell death. 2011;16:198-207
33. Oliver FJ, de la Rubia G, Rolli V, Ruiz-Ruiz MC, de Murcia G, Murcia JM. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. The Journal of biological chemistry. 1998;273:33533-9
34. Alao JP. The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Molecular cancer. 2007;6:24
35. Shen M, Feng Y, Gao C, Tao D, Hu J, Reed E. et al. Detection of cyclin b1 expression in g(1)-phase cancer cell lines and cancer tissues by postsorting Western blot analysis. Cancer research. 2004;64:1607-10
36. Paternot S, Bockstaele L, Bisteau X, Kooken H, Coulonval K, Roger PP. Rb inactivation in cell cycle and cancer: the puzzle of highly regulated activating phosphorylation of CDK4 versus constitutively active CDK-activating kinase. Cell cycle. 2010;9:689-99
37. Jang SH, Kim AR, Park NH, Park JW, Han IS. DRG2 Regulates G2/M Progression via the Cyclin B1-Cdk1 Complex. Molecules and cells. 2016;39:699-704
38. Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends in genetics: TIG. 2004;20:214-20
39. Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1-222
40. Carayol N, Katsoulidis E, Sassano A, Altman JK, Druker BJ, Platanias LC. Suppression of programmed cell death 4 (PDCD4) protein expression by BCR-ABL-regulated engagement of the mTOR/p70 S6 kinase pathway. The Journal of biological chemistry. 2008;283:8601-10
41. Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T. et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes & development. 2005;19:2199-211
42. Duluc C, Moatassim-Billah S, Chalabi-Dchar M, Perraud A, Samain R, Breibach F. et al. Pharmacological targeting of the protein synthesis mTOR/4E-BP1 pathway in cancer-associated fibroblasts abrogates pancreatic tumour chemoresistance. EMBO molecular medicine. 2015;7:735-53
43. Ni HM, McGill MR, Chao X, Du K, Williams JA, Xie Y. et al. Removal of acetaminophen protein adducts by autophagy protects against acetaminophen-induced liver injury in mice. Journal of hepatology. 2016;65:354-62
44. March HN, Winton DJ. mTOR regulation by JNK: rescuing the starving intestinal cancer cell? Gastroenterology. 2011;140:1387-91
45. Zhang D, Han S, Wang S, Luo Y, Zhao L, Li J. cPKCgamma-mediated down-regulation of UCHL1 alleviates ischaemic neuronal injuries by decreasing autophagy via ERK-mTOR pathway. Journal of cellular and molecular medicine. 2017
46. Hernandez G, Lal H, Fidalgo M, Guerrero A, Zalvide J, Force T. et al. A novel cardioprotective p38-MAPK/mTOR pathway. Experimental cell research. 2011;317:2938-49
47. Buyuklu M, Kandemir FM, Ozkaraca M, Set T, Bakirci EM, Topal E. et al. Benefical effects of lycopene against contrast medium-induced oxidative stress, inflammation, autophagy, and apoptosis in rat kidney. Human & experimental toxicology. 2015;34:487-96
48. Karayannopoulou G, Euvrard S, Kanitakis J. Differential expression of p-mTOR in cutaneous basal and squamous cell carcinomas likely explains their different response to mTOR inhibitors in organ-transplant recipients. Anticancer research. 2013;33:3711-4
49. Sully K, Akinduro O, Philpott MP, Naeem AS, Harwood CA, Reeve VE. et al. The mTOR inhibitor rapamycin opposes carcinogenic changes to epidermal Akt1/PKBalpha isoform signaling. Oncogene. 2013;32:3254-62
50. Euvrard S, Morelon E, Rostaing L, Goffin E, Brocard A, Tromme I. et al. Sirolimus and secondary skin-cancer prevention in kidney transplantation. The New England journal of medicine. 2012;367:329-39
51. Yin F, Sharen G, Yuan F, Peng Y, Chen R, Zhou X. et al. TIP30 regulates lipid metabolism in hepatocellular carcinoma by regulating SREBP1 through the Akt/mTOR signaling pathway. Oncogenesis. 2017;6:e347
52. Shui X, Zhou C, Lin W, Yu Y, Feng Y, Kong J. Long non-coding RNA BCAR4 promotes chondrosarcoma cell proliferation and migration through activation of mTOR signaling pathway. Experimental biology and medicine. 2017;242:1044-50
53. Guru SK, Pathania AS, Kumar S, Ramesh D, Kumar M, Rana S. et al. Secalonic Acid-D Represses HIF1alpha/VEGF-Mediated Angiogenesis by Regulating the Akt/mTOR/p70S6K Signaling Cascade. Cancer research. 2015;75:2886-96
54. Pollard HJ, Willett M, Morley SJ. mTOR kinase-dependent, but raptor-independent regulation of downstream signaling is important for cell cycle exit and myogenic differentiation. Cell cycle. 2014;13:2517-25
55. Kobayashi J, Inai T, Shibata Y. Formation of tight junction strands by expression of claudin-1 mutants in their ZO-1 binding site in MDCK cells. Histochemistry and cell biology. 2002;117:29-39
56. Morita K, Tsukita S, Miyachi Y. Tight junction-associated proteins (occludin, ZO-1, claudin-1, claudin-4) in squamous cell carcinoma and Bowen's disease. The British journal of dermatology. 2004;151:328-34
57. Chen CP, Hung CF, Lee SC, Lo HM, Wu PH, Wu WB. Lycopene binding compromised PDGF-AA/-AB signaling and migration in smooth muscle cells and fibroblasts: prediction of the possible lycopene binding site within PDGF. Naunyn-Schmiedeberg's archives of pharmacology. 2010;381:401-14
58. Yang Y, Li W, Sun Y, Han F, Hu CA, Wu Z. Amino acid deprivation disrupts barrier function and induces protective autophagy in intestinal porcine epithelial cells. Amino acids. 2015;47:2177-84
59. Li H, Gao A, Feng D, Wang Y, Zhang L, Cui Y. et al. Evaluation of the protective potential of brain microvascular endothelial cell autophagy on blood-brain barrier integrity during experimental cerebral ischemia-reperfusion injury. Translational stroke research. 2014;5:618-26.60
60. Uppala PT, Dissmore T, Lau BH, Andacht T, Rajaram S. Selective inhibition of cell proliferation by lycopene in MCF-7 breast cancer cells in vitro: a proteomic analysis. Phytotherapy research. 2013;27:595-601
61. Bi SY, Li L, Xu S, Zhang ML, Gu H, Zhou ZH. et al. Regulatory effects of lycopene on the key signaling receptors in human cutaneous squamous cell carcinoma cell line COLO16. Chinese journal of dermatology. 2018;51:421-4
62. Rafi MM, Kanakasabai S, Reyes MD, Bright JJ. Lycopene modulates growth and survival associated genes in prostate cancer. Journal of nutritional biochemistry. 2013;24:1724-34
63. Zhang X, Wu L, Xiao T, Tang L, Jia X, Guo Y. et al. TRAF6 regulates EGF-induced cell transformation and cSCC malignant phenotype through CD147/EGFR. Oncogenesis. 2018;7:17
Author contact
![]() Corresponding author: Xu Chen: chenxcams.cn
Corresponding author: Xu Chen: chenxcams.cn

 Global reach, higher impact
Global reach, higher impact