Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(7):1145-1152. doi:10.7150/jca.18026 This issue Cite
Research Paper
Isolation and Identification of miRNAs in exosomes derived from serum of colon cancer patients
Department of Oncology, Beijing Friendship Hospital, Capital Medical University, Beijing 100050, China
Received 2016-10-21; Accepted 2017-3-6; Published 2017-4-9
Abstract
Objective: To isolate exosomes from the serum of colon cancer patients and identify RNAs in the small vesicles.
Methods: ExoQuick-TC™ Exosome Precipitation Solution was used for exosome isolation and the shapes of exosomes were observed under a transmission electron microscope. Mass spectrometry was used to identify the classification of miRNAs encapsulated in exosomes, and the expression levels of miRNA-21,-133a, and -181b in exosomes were detected by RT-PCR.
Results: Exosomes isolated from serum of colon cancer patients were circular-or oval-shaped and vary in size with a diameter of 40-100 nm. Mass spectrometry shows that the main RNAs in exosomes are small RNAs; the levels of these small RNAs in exosomes are significantly higher compared with fresh serum. There is still a tiny amount of small RNAs in exosome-free serum, but the amounts are significantly lower than that in exosomes. No more RNAs were detected in the repeated freezing and thawing serum, but there were still some RNAs detectable in the exosomes extracted from these serums. MiRNA-21, -133a and -181b can be detected in exosomes, and the level of miRNA-21 is associated with early diagnosis of colon cancer.
Conclusion: This study proves that commercial kits for exosome separation are more convenient and time-saving and that mass spectrometry is capable of identifying the miRNAs in exosomes. Compared with direct extraction of miRNAs from the serum, the method of isolating exosomes from the serum firstly and then extracting miRNAs from the exosomes can enhance the stability and integrity of their inner miRNAs. Also, we demonstrate that the exosomal miRNA-21 expression is associated with the early diagnosis of colon cancer.
Keywords: miRNAs, exosomes
Introduction
Exosomes are adsorbed vesicles with a lipid bilayer and about 50-100 nm in diameter, 1.13-1.19g/ml in density, and may precipitate at 100,000 × g centrifugation [1], with a classic “cup” or “dish” morphology. Exosomes can be released from secretory multivesicular bodies when fused to plasma membranes, and can also be formed through the breakaway of intracellular endosomes from cytoplasmic membranes [2]. Exosomes were initially considered as garbage-bag-wrapped obsolete plasma membranes or molecular membrane fragments. However, with the discovery of B lymphocyte antigen-presenting exosomes in mid-1990s, they were found to be closely linked to the function of immune system [3]. Thereafter, the research of exosomes was focused on the immune response, especially the physiological interaction between immune system and tumor cells [4-9].
Exosomes derived from tumor cells are rich in proteins and nucleic acids from parental cells, and thus can be seen as a source of tumor antigens to induce anti-tumor immune response. However, they can also inhibit the immune system and form a suitable niche for tumor cell growth, promoting tumor growth, metastasis, and drug resistance. Especially, tumor-derived exosomes contain tumor-specific microRNAs (miRNAs), making exosomes an ideal marker for tumor diagnosis. It was proposed that exosome-miRNAs can be used as diagnostic and prognostic markers for lung cancer [10], glioblastoma [11], esophageal squamous cell carcinoma [12], prostate cancer [13], breast cancer [14], kidney cancer [15], leukemia [16], colon cancer [17], ovarian cancer [18,19], pancreatic cancer [20] and other types of tumors. Similarly, signal pathways will change in the process of cell malignancy, while miRNAs released from exosomes are changed at the same time, indicating that alteration of signaling pathways in the cell malignant process are mediated by specific output of miRNAs [21]. Therefore, analysis of exosomes is mostly likely to assist us in determining the malignancy and development of a particular cancer [22, 23].
It has been proved that exosome-secreted miRNAs have numerous biofunctions. The transport of miRNAs can be mediated by exosomes and this transport mode may be the mechanism of genetic exchange between cells [24]. Human EB virus (EBV)-derived miRNAs can be transported through exosomes and regulate the target genes in the recipient virus [25]. Moreover, it is demonstrated that in the supernatant of colon cancer cell lines, miRNAs can be protected by encapsulation in exosomes [26]. It is also indicated that the stability and integrity of miRNAs can be enhanced after encapsulation in exosome, and can produce proteins to resist digestion of RNA enzyme and other environmental damages [27,28]. Therefore, high-throughput analysis of signals from tumor-derived exosomes in different forms and stages may contain a huge amount of information that can be used for early diagnosis and tumor stage-specific interventions. The stability, cell type specificity and easy access from body fluids make exosomes potential valuable factors for cancer diagnosis and targeted treatment.
In this study, we use an ExoQuick-TC™ Exosome Precipitation Solution kit for exosome extraction, and further analyze the shape and size of exosomes as well as specific miRNAs in them. We expect to provide an experimental basis for research about the associations of colon-cancer-derived exosomes with cancer diagnosis, staging, and drug efficacy and drug resistance.
Materials and methods
Serum, reagents and equipment
Twenty colon cancer patients (13 males, 7 females) confirmed by histology or cytology were randomly chosen from patients hospitalized in Department of Oncology, Beijing Friendship Hospital, Capital Medical University between 2013-1-1 and 2015-5-1, together with 20 serum from healthy medical examination control subjects. The study complied with the Declaration of Helsinki. All subjects gave written informed consent.
A ExoQuick-TC™ Exosome Precipitation Solution kit (System Biosciences Corporation, USA), a total serum RNAs (including miRNAs) extraction miRNeasy Serum/Plasma kit (QIAGEN Company, Germany), as well as a miRNA reverse transcription TagMan MicroRNA Reverse Transcription kit and Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) kits (Life Technologies Corporation, USA) were used here.
Other instruments included an ultrafleXtreme MALDI-TOF/TOF mass spectrometer(Bruker Daltonics, Germany), a H7650 transmission electron microscope (HITACHI Company, Japan); a PCR instrument (Applied Biosystems Gene Co., Ltd., USS), an UV gel image analysis system (Bio-rad Company, America),a water bath (Shanghai Jing Hong Experimental Equipment Co., China), an electronic balance (Sartorius Company, Japan), high-low-speed desktop centrifuge and rotor (Heraeus Company, Germany), palm centrifuges (Jiangsu Haimen QiLinbeier Instruments Company, China), a RNA/DNA analyzer (Pharmacia Company, USA), and a pH gauge (Sartorius Corporation, Japan).
Isolation of serum
From each subject, whole blood (2ml) was exsanguinated and placed in a refrigerator at 4ºC for 1 hour, followed by centrifugation at 3000 rpm for 10 min by a low-speed centrifuge. The supernatant was serum, which was placed in a refrigerator at -80 ºC after separation.
Extraction of exosomes
ExoQuick-TCTM Exosome Precipitation Solution kit was used for exosome extraction following the instructions: Collect serum 250μl and centrifuge at 3000 × g for 15 min to remove cells and cell debris. Transfer supernatant to a sterile vessel and add 63μl ExoQuick Exosome Precipitation Solution to it. Then refrigerate overnight. The next day centrifuge ExoQuick/serum mixture at 1500 × g for 30 min. After centrifugation, the exosomes may appear as a beige or white pellet at the bottom of the vessel. Aspirate supernatant. Spin down residual ExoQuick solution by centrifugation at 1500 × g for 5 min. Remove all traces of fluid by aspiration, taking great care not to disturb the precipitated exosomes in pellet. Resuspend exosome pellet in 1/10 to 1/100 of original volume using sterile Phosphate Buffered Saline (PBS) and aliquot in cryogenic vials and store at -80°C until ready for use.
Morphological analysis of exosomes
After separation, exosomes were fixed in a 0.01 M phosphate buffer solution (PBS, containing 4% paraformaldehyde, PH 7.4) at 4ºC overnight. On the next day, after being washed with PBS, the exosomes were fixed in 1% OsO4 for 30 min. After being rinsed with distilled water, the exosome precipitate was dehydrated in concentration gradient alcohol, stained by 1% uranyl acetate for 30 min, and then embedded in TaAb 812. After polymerization at 60 ºC overnight, the precipitate was sliced, and the ultrathin sections were observed under a transmission electron microscope.
Extraction of RNA in exosomes
Use the miRNeasy Serum/Plasma Kit for exosome-RNA extraction. Add 1ml QIAzolLysis Reagent to 200ul exosome. Mix by vortexing or pipetting up and down. Place the tube containing the lysate on the benchtop at room temperature (15-25°C) for 5 min. Add 200ul chloroform to the starting sample to the tube containing the lysate and cap it securely. Vortex or shake vigorously for 15 s. Place the tube containing the lysate on the benchtop at room temperature (15-25°C) for 2-3 min. Centrifuge for 15 min at 12,000 × g at 4°C. Transfer the upper aqueous phase to a new collection tube. Avoid transfer of any interphase material. 900ul 100% ethanol and mix thoroughly by pipetting up and down several times. Pipet up to 700μl of the sample, including any precipitate that may have formed, into an RNeasy MinElute spin column in a 2 ml collection tube. Close the lid gently and centrifuge at ≥8000 × g (≥10,000 rpm) for 15 s at room temperature (15-25°C). Discard the flow-through. Repeat previous step using the remainder of the sample. Discard the flow through. Add 700μl Buffer RWT to the RNeasy MinElute spin column. Close the lid gently and centrifuge for 15 s at ≥8000 × g (≥10,000 rpm) to wash the column. Discard the flow-through. Pipet 500μl Buffer RPE onto the RNeasy MinElute spin column. Close the lid gently and centrifuge for 15 s at ≥8000 × g (≥10,000 rpm) to wash the column. Discard the flow-through. Pipet 500μl of 80% ethanol onto the RNeasy MinElute spin column. Close the lid gently and centrifuge for 2 min at ≥8000 × g (≥10,000 rpm) to wash the spin column membrane. Discard the collection tube with the flow-through. Place the RNeasy MinElute spin column into a new 2 ml collection tube. Open the lid of the spin column, and centrifuge at full speed for 5 min to dry the membrane. Discard the collection tube with the flow-through. Place the RNeasy MinElute spin column in a new 1.5 ml collection tube. Add 14μl RNase-free water directly to the center of the spin column membrane. Close the lid gently, and centrifuge for 1 min at full speed to elute the RNA.
Molecular weight and species of total RNAs identification in exosomes
Using a mass spectrometer, we drawn 1ulof 3-Hydroxy picolinicacid (3-HPA) as substrate spot on the panel of the target sample, which dried naturally. Then 1μlof a sample solution was spotted as the substrate, which dried naturally. After that, using a Smart beamTM laser, we employed a Ground steel sample target to obtain a spectrum of molecular weight and species of the total RNAs.
Detection of miR-21, miR-131a and miR-181b in exosomes by RT-PCR
First, apply exosome-miRNAs as template, using TagMan MicroRNA Reverse Transcription Kit to reverse miRNA to cDNA. The systems are as follows: dNTP mix 0.15ml, Multiscribe RT enzyme (50U/ul) 1.0ul, 10×RT Buffer 1.5ul, RNase Inhibitor (20U/ul) 0.19ul, Unclease free water 4.16ul. Polymerase Chain Reaction(PCR) conditions were: 16 °C 30 min, 42 °C 30 min, 85 °C 5min to reverse miRNA to cDNA. Then apply cDNA as template, and the total systems are: TaqMan Universal Master Mix II 10.0ul, TaqMan Assay 1.0ul, cDNA+RNase-free water 9.0ul. Quantitative PCR conditions were: pre-denaturation at 95 °C, 10 min followed by 95°C 15 s, 60°C 60 s, totally 40 cycles to detect the expression of miR-21,131a and 181b in serum and exosomes of colon cancer patient. Amplification products stored at -80 °C. Use small nuclear RNA U6 (U6 snRNA) as internal reference, applying the formula: F = 2-△△t to calculate the fold-change in the expression of the above miRNAs. △Ct= CtmiRNA-CtU6, △△Ct=△Ct colon cancer exosome-miRNA-△Ct normal human exosoem-miRNA, in which F represents the expression fold-change of target miRNAs in exosomes of colon cancer patients compared to in exosomes of normal controls. For normal controls, △△Ct=0, F=1.
Statistical analysis
SPSS 19.0 statistical software was applied on the experimental data analysis. We select Wilcoxon rank sum test for two independent and paired samples to take the test, and select Mann-Whitney U test for variable analysis. P<0.05 was considered as statistically significant difference.
Results
Morphology of exosomes
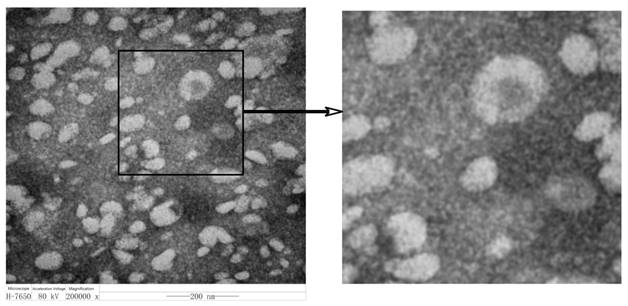
Different from the exosome extraction methods reported before [29-31], here we used commercial kits (this method has been rarely applied before) for exosome extraction. After extraction, colon-cancer-derived exosomes were diluted for observation under a transmission electron microscope. The results show exosomes are round- or oval-shaped and vary in size with a diameter about 40-100 nm (Figure 1). Results show that the kit-isolated exosomes are fully consistent with the morphology and sizes reported before, which prove that the commercial kits for exosome separation are reliable, time-saving and more convenient with higher purity and higher yield.
Results of RNAs identification in exosomes
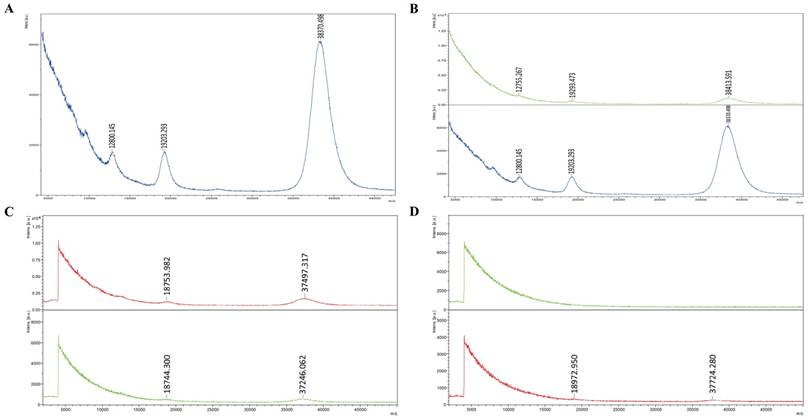
Mass spectrometry shows three peaks of RNAs in exosomes, with a molecular weight of 7000-8000, 15000-18000, 37000-38000 Daltons, respectively. Since the weight of single nucleotide is 300 Daltons, it is calculated that the RNA lengths in exosomes are about 23, 50 and 128 bases. Those RNAs all are small RNAs, according to species of RNAs, from which it is inferred that they are single-stranded miRNAs, self-folding formed complementary false double chain of miRNAs, and the subunit 5s of mRNA, containing no or little 18s or 28s subunit (Figure 2A). Clearly, the peak and thus the content of small-molecule RNAs in exosomes are significantly higher compared with those in fresh serum, suggesting exosomes can package and enrich small -molecule RNAs in them, making it a higher content and more detectable (Figure 2B). There are still a little of small-molecule RNAs in no-exosome serum, but significantly lower than the content of RNAs in exosomes (Figure 2C). No RNA was detected in repeated freezing and thawing serum, but in the repeated freezing and thawing exosomes extracted from serum, there are still certain levels of RNAs (Figure 2D).
In this study, we prove that the exosomes mainly contain small-molecular RNAs by using mass spectrometry. Different from other methods that directly extract miRNAs from the serum, we first isolated exosomes from the serum and then extracted miRNAs from the exosomes. This method has higher yield and higher purity, which prove that exosomes can enhance the stability and integrity of their inner miRNAs and prevent them from being destroyed by RNA enzymes and other environmental factors and are favorable for further research.
The expression of miR-21, -131a and -181b in exosomes derived from serum of patients with colorectal cancer
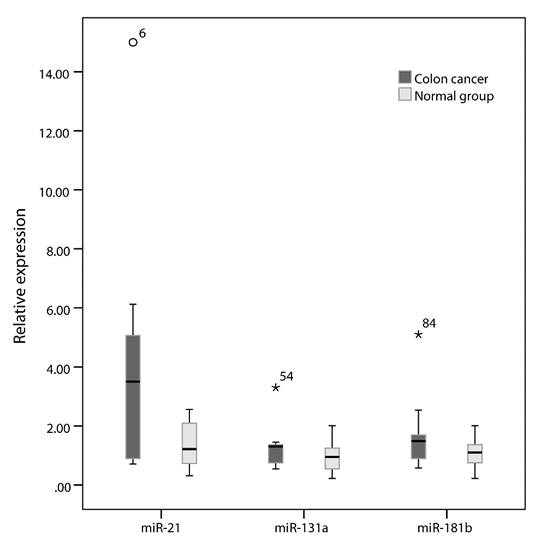
The expression levels of targeted miRNAs are represented as median and interquartile range [(P, P)]. Results show that the relative expressions of miR-21, -133a and -181b in the exosomes from serum of colon cancer patients are 3.974 (0.897, 5.145), 1.312(0.732, 1.469) and 1.486 (0.890, 1.692), respectively. The expression of miR-21 is significantly higher compared with exosomes from normal healthy serum (P=0.007) (Figure 3).
Among the six newly-diagnosed and untreated patients from all 20 cases, the relative expression of miR-21in exosomes is 5.398 (0.986, 5.857), which is significantly higher with a relative expression fold change>5 times (P<0.001).
Here we prove for the first time that in the serum exosomes isolated from colon cancer patients, the miRNA-21 expression is associated with early diagnosis of colon cancer.
Morphology of exosomes
RNAs expression in serum and exosomes derived from colon cancer patients. A: Mass spectrometry show the peaks of RNAs in exosomes are small RNAs. B: The content of small-molecule RNAs in exosomes is significantly higher than in fresh serum (The figure above shows the peak of small RNAs in fresh serum of colon cancer patients; the figure below shows the peak of small RNAs in exosomes derived from the same serum). C: There are still a little of small-molecule RNAs in no-exosome serum, but significantly lower than the content of RNAs in exosomes (The figure above shows the content of small RNAs in exosomes; the figure below shows the content of small RNAs in exosome-free serum). D: No RNA was detected in repeated freezing and thawing serum, but in the exosomes extracted from repeated freezing and thawing serum, there are still certain levels of RNAs. (The figure above shows the content of small RNAs in repeated freezing and thawing serum; the figure below shows the content of small RNAs in exosome extracted from repeated freezing and thawing serum.)
Box plots of miR-21, miR-131a and miR-181b expression in each group
Discussion
Exosomes were found in early 1980s, but were initially regarded as garbage-bag-wrapped abandoned plasma membranes or membrane molecular fragments, so they did not attract the attention of researchers. However, recent studies show that exosomes are closely associated with the function of the immune system [3]. In the 2010s, scientists discovered that miRNA and mRNA can be loaded as "goods” in exosomes [24], which indicate the possession of bioactivity. Since then, research of exosomes gradually attracts researchers. The present study is focused on the miRNA isolation from colon-cancer-derived exosomes, aiming to lay a foundation for further application in clinic.
At present, the commonly-used methods of exosome isolation include ultracentrifugation, sucrose density gradient combined with ultracentrifugation ultrafiltration, magnetic activated cell sorting (MACS), and a variety of commercial kits. However, no perfect method can ensure the yield, purity and bioactivity of exosomes at the same time. Ultracentrifugation is the most commonly-used one, which is based on different centrifugal speeds and layer-by-layer centrifugation, and ultimately gains exosomes smaller than 100 nm in diameter. This method guarantees the production of exosomes, which, however, are inevitably mixed with vesicles of other bodies and thus reduce the purity of extraction. The sucrose density gradient centrifugation aims to add a sample to the density gradient sucrose for centrifugal sedimentation or sedimentation equilibrium, and under a certain centrifugal force, assigns exosomes to a specific gradient centrifugation location. Although exosomes obtained by this method are purer, the preparatory work is very complicated and time-consuming. MACS aims to pre-coat exosomes specific antibodies in magnetic beads, incubate them with exosomes together, then wash and re-suspend after ultracentrifugation. This method has high specificity, easy-to-operate, and guarantees the complete form of exosomes, but the beads antibody may affect the bioactivity of exosomes and the stability of subsequent experiments. In the present study, we use an ExoQuickExosome Precipitation Solution kit for exosome extraction. The principle of this kit is that there are many holes which have consistent diameter as exosomes and will crosslink and wrap exosomes, and filter out exosomes like a sieve. This method is simple, easy, and the whole centrifugal process only takes 45 min. Besides the very high yield and purity of exosomes, this method does not affect the bioactivity of exosomes and the subsequent experiment. These characteristics all prove that this method is appropriate for exosome extraction.
As reported, miRNA-21 expression is associated with the diagnosis, staging, evolution and prognosis of tumors, and the expression detection may help to observe clinical curative effect and to judge reoccurrence and prognosis of tumors [32,33]. Moreover, miRNAs are ubiquitous in various body fluids, including serum, plasma, tears, milk, saliva, and pleural and peritoneal effusions. Thus, miRNAs can be sampled from various sources. However, miRNAs are usually unstable in plasma and serum, and are susceptible to degradation after repeated freeze-thaw, long-time placement at room temperature, or under harsh conditions (e.g. strong acid, strong base, DNA enzymes or RNA enzymes). These limitations inhibit the yield and purity of miRNA extraction. Nevertheless, high-quality serum miRNA extraction is the first step that decides whether serum miRNA is effective for diagnosis, and also directly decides the accuracy of diagnosis results. There are many methods for extraction of serum miRNAs [34, 35], including commercial kits, phenol-chloroform, and Trizol extraction. Most studies state that commercial kits are very effective and outperform the phenol-chloroform method and the Trizol method in terms of simple steps, no addition of reagent or centrifugation, time-saving and labor-saving, higher efficiency and higher yield. Thus, commercial kits are currently the most commonly-used method for miRNA extraction. However, commercial kits are more expensive at a cost of 10 dollars per reaction. Thus, the keys are how to reduce miRNA degradation as much as possible, improve the miRNA yield from each reaction, and to effectively eliminate impurities such as proteins. Thus, in this study, different from previous studies for direct miRNA extraction from serum, we introduced a new concept and substance - exosomes in our paper. Exosomes are able to enrich and encapsulated the miRNAs in the serum, so the exosomes are rich in miRNAs. Moreover, like "protective membranes", exosomes protect the miRNAs from being destroyed by RNA enzymes or environmental factors, and thereby stabilize miRNAs. In this study, we first extracted the exosomes that could enrich miRNA in the serum. Then a commercial kit was used to extract and isolate miRNA from the exosomes. These steps ensure the yield and stability of miRNA and firmly underlie the subsequent experiments. Then we further validated through mass spectromery and proved that the RNAs in the exosomes are mainly miRNAs, which is consistent with the study by Valadi [24]. The miRNAs in the exosomes are significantly richer than fresh serum. The serum without exosomes still contains a certain amount of miRNA, but the content is significantly lower than that in the exosomes. The serum after repeated freeze-thaw was not found with any miRNA, but the exosomes thereby isolated were still identified with a certain level of miRNAs. Afterwards, we used reverse transcriptase-polymerase chain reaction (RT-PCR) to detect three types of miRNAs in the exosomes. At present, the methods for miRNA detection include Northern Blot, microarray chip, in-situ hybridization, RT-PCR, rolling circle amplification, and the use of conjugated polymers. Though microarray analysis is able to analyze the miRNA expression profile, there are many novel chips and improved techniques, but the sensitivity and specificity of microarray analysis should be further improved. Various probe marking techniques, amplification methods and detection methods should be further integrated and improved. Analysis of in-situ miRNA hybridization shows that though the use of LNA probe improves the analytical sensitivity and reliability, the efficiency and sensitivity of hybridization remain to be improved. The sensitivity of new methods such as ISH should be further enhanced together with in-situ amplification techniques. Though the existing miRNA amplification methods have pros and cons, RT-PCR is the most widely-used amplification method, and there are many types of commercial kits. Here we used RT-PCR into miRNAs detection. Compared with other methods, RT-PCR is superior in terms of no marking, simple operations, low costs, and high sensitivity. RT-PCR is able to get results within short time, detect the expressions of multiple miRNAs, and even require less RNAs. Here RT-PCR was successfully used to identify the expressions of miR-21, -133a and -181b in the serum exosomes of colon cancer patients. We also prove that miR-21 is significantly higher expressed in exosomes derived from colon cancer patients, especially in patients at the first diagnosis or before any treatment. The miR-21 expression is about 5 folds than normal people. This difference is significantly larger than the direct miR-21 extraction from the serum and facilitates early diagnosis.
Our study is also limited by the relatively small number of cases of colon cancer patients; however, our study showed that Exosomes can enhance the stability and integrity of miRNAs and prevent miRNAs from RNA enzyme digestion and other environmental damage which may be applied to clinical drug development.
Acknowledgements
This study was supported by grants from the Beijing Talents Fund (No. 2014000021469G268, to Lei Zhao), Rising Star Program from Beijing Friendship Hospital, CMU (No. yyqdkt 2015-10, to Lei Zhao) and Beijing Municipal "215" High-level Health Person Foundation Project (No.2014-3-004, to Jing Yu).
Contributions
Z. L. and C. B. W. designed the study. The data were analyzed by Z. L., Y. J., W. J., L. H. H., and C. J. J.. The figures were prepared by Z. L. and L. H. H.. Z. L. wrote the paper. All authors reviewed the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zhao L, Liu W, Xiao J. et al. The role of exosomes and "exosomal shuttle microRNA" in tumorigenesis and drug resistance. Cancer Lett. 2015;356:339-46
2. Gould SJ, Raposo G. As we wait: Coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles. 2013;2:10.3402 /jev.v2i0.20389
3. Raposo G, Nijman HW, Stoorvogel W. et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161-72
4. Bobrie A, Thery C. Exosomes and communication between tumours and the immune system: Are all exosomes equal? Biochem Soc Trans. 2013;41:263-7
5. Clayton A, Mason MD. Exosomes in tumour immunity. Curr Oncol. 2009;16:46-9
6. Gutierrez-Vazquez C, Villarroya-Beltri C, Mittelbrunn M. et al. Transfer of extracellular vesicles during immune cell-cell interactions. Immunol Rev. 2013;251:125-42
7. Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373-83
8. Thery C. Exosomes: Secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15
9. Jerez S, van Wijnen AJ, Galindo M. Proteomic analysis of exosomes and Exosome-Free conditioned media derived from human osteosarcoma cell lines reveal differential secretion of proteins related to biological functions and tumor progression. J Cell Biochem. 2017;118:351-60
10. Deng Z, Rong Y, Teng Y. et al. Exosomes miR-126a released from MDSC induced by DOX treatment promotes lung metastasis. Oncogene. 2017;36:639-51
11. Godlewski J, Krichevsky AM, Johnson MD. et al. Belonging to a network-microRNAs, extracellular vesicles, and the glioblastoma microenvironment. Neuro Oncol. 2015;17:652-62
12. Tanaka Y, Kamohara H, Kinoshita K. et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer-Am Cancer Soc. 2013;119:1159-67
13. Li J, Yang X, Guan H. et al. Exosome-derived microRNAs contribute to prostate cancer chemoresistance. Int J Oncol. 2016;49:838-46
14. Joyce DP, Kerin MJ, Dwyer RM. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int J Cancer. 2016;139:1443-8
15. Alvarez ML, Khosroheidari M, Kanchi RR. et al. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82:1024-32
16. Umezu T, Ohyashiki K, Kuroda M. et al. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene. 2013;32:2747-55
17. Zaharie F, Muresan MS, Petrushev B. et al. Exosome-Carried microRNA-375 inhibits cell progression and dissemination via bcl-2 blocking in colon cancer. J Gastrointestin Liver Dis. 2015;24:435-43
18. Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13-21
19. Au YC, Co NN, Tsuruga T. et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun. 2016;7:11150. doi: 10.1038/ncomms11150
20. Erb U, Zoller M. Progress and potential of exosome analysis for early pancreatic cancer detection. Expert Rev Mol Diagn. 2016;16:757-67
21. Barger JF, Rahman MA, Jackson D. et al. Extracellular miRNAs as biomarkers in cancer. Food Chem Toxicol. 2016;98:66-72
22. Dhondt B, Rousseau Q, De Wever O. et al. Function of extracellular vesicle-associated miRNAs in metastasis. Cell Tissue Res. 2016;365:621-41
23. Li H, Yang BB. Friend or foe: The role of microRNA in chemotherapy resistance. Acta Pharmacol Sin. 2013;34:870-9
24. Valadi H, Ekstrom K, Bossios A. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-9
25. Palma J, Yaddanapudi SC, Pigati L. et al. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012;40:9125-38
26. Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013;32:623-42
27. Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087-92
28. Kosaka N, Iguchi H, Yoshioka Y. et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442-52
29. Hildonen S, Skarpen E, Halvorsen TG. et al. Isolation and mass spectrometry analysis of urinary extraexosomal proteins. Sci Rep. 2016;6:36331. doi: 10.1038/srep36331
30. Quek C, Bellingham SA, Jung CH. et al. Defining the purity of exosomes required for diagnostic profiling of small RNA suitable for biomarker discovery. Rna Biol. 2016 doi: 10.1080/15476286.2016.1270005
31. Royo F, Diwan I, Tackett MR. et al. Comparative miRNA analysis of urine extracellular vesicles isolated through five different methods. Cancers (Basel). 2016;8:doi 10.3390/cancers8120112
32. Leite KR, Reis ST, Viana N. et al. Controlling RECK miR21 promotes tumor cell invasion and is related to biochemical recurrence in prostate cancer. J Cancer. 2015;6:292-301
33. Zhao Y, Wang Y, Yang Y. et al. MicroRNA-222 controls human pancreatic cancer cell line capan-2 proliferation by p57 targeting. J Cancer. 2015;6:1230-5
34. Roy S, Soh JH, Ying JY. A microarray platform for detecting disease-specific circulating miRNA in human serum. Biosens Bioelectron. 2016;75:238-46
35. Brunet-Vega A, Pericay C, Quilez ME. et al. Variability in microRNA recovery from plasma: Comparison of five commercial kits. Anal Biochem. 2015;488:28-35
Author contact
![]() Corresponding author: Dr. Bangwei Cao, Department of Oncology, Beijing Friendship Hospital, Capital Medical University, 95 Yong An Road, Xicheng District, Beijing 100050, China.TEL: +86-010-63139321. Fax: +86-010-63139321. E-mail address: oncologyedu.cn
Corresponding author: Dr. Bangwei Cao, Department of Oncology, Beijing Friendship Hospital, Capital Medical University, 95 Yong An Road, Xicheng District, Beijing 100050, China.TEL: +86-010-63139321. Fax: +86-010-63139321. E-mail address: oncologyedu.cn

 Global reach, higher impact
Global reach, higher impact