Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(15):4302-4315. doi:10.7150/jca.124247 This issue Cite
Research Paper
Chemopreventive Targeting of Oncogenic Stemness: EGCG-Mediated Suppression of the HIF-1α-PPARγ-Cancer Stem Cells Transcriptional Signature in 3D Glioblastoma Spheroids
1. Laboratoire d'Oncologie Moléculaire, Département de Chimie, Université du Québec à Montréal, Montreal, QC, Canada, H3C 3P8.
2. Lady Davis Institute for Medical Research, SMBD-Jewish General Hospital, and Department of Surgery, McGill University, Montreal, QC, Canada, H3T 1E2.
Received 2025-8-26; Accepted 2025-10-15; Published 2025-10-20
Abstract
Background: Preventing the emergence and persistence of cancer stemness represents a promising strategy to reduce tumor aggressiveness and therapeutic failure. Cancer stem cells (CSCs), which contribute significantly to therapy resistance, recurrence, and metastasis, are sustained in part by metabolic reprogramming that enhances survival and self-renewal under stress conditions.
Methods: To model the hypoxic core of solid tumors, three-dimensional (3D) glioblastoma (GBM) spheroid cultures were generated using human U87, U118, U138, and U251 cell lines and compared to their respective two-dimensional (2D) monolayer cultures. Total RNA was extracted, and gene expression was analyzed via RT-qPCR and targeted gene arrays. Transient gene silencing was performed using specific siRNAs, while pharmacological intervention involved treatment with (-)-Epigallocatechin-3-gallate (EGCG), a bioactive phytochemical derived from green tea. Adipogenesis was evaluated using Oil Red O staining.
Results: Compared to conventional 2D cultures, 3D spheroids exhibited elevated expression of hypoxia-inducible factor-1 alpha (HIF-1α) and upregulation of peroxisome proliferator-activated receptor gamma (PPARγ), identified through adipogenesis array screening. Adipogenic activity persisted in the 3D spheroid model, and EGCG treatment effectively suppressed the upregulation of HIF-1α and PPARG transcripts. This led to a significant downregulation of adipogenic genes (CEBPD, FOXO1, BMP2, BMP7) and CSC-associated markers (CD44, PROM1, ABCB5, ABCG2), accompanied by reduced spheroid growth.
Conclusions: These findings underscore EGCG's chemopreventive potential in disrupting early HIF-1α-mediated molecular pathways that reinforce GBM stemness. By targeting hypoxia-driven metabolic reprogramming, EGCG offers a dietary-based approach to modulate the CSC niche and potentially delay or prevent GBM progression. Moreover, the use of 3D spheroid models highlights their relevance in preclinical chemoprevention research, bridging the gap between simplistic 2D cultures and the complex biology of solid tumors.
Keywords: metabolic reprograming, cancer stem cells, 3D spheroids, HIF, glioblastoma, adipogenesis, EGCG, PPARG
Introduction
Cancer stemness is a central driver of tumor resilience and treatment resistance, making it one of the most pressing therapeutic targets in oncology. Cancer stem cells (CSCs), a specialized subpopulation within tumors, possess the capacity for self-renewal, tumor initiation, evasion of conventional therapies, and promotion of recurrence and metastasis [1]. These properties enable CSCs to regenerate tumors even after aggressive treatment, contributing to clinical relapse and poor patient outcomes [2, 3]. Consequently, dismantling the molecular mechanisms that support CSCs survival, often involving metabolic plasticity, signaling pathways, and adaptive resistance, is increasingly prioritized in therapeutic strategies aimed at eradicating the root drivers of malignancy rather than merely reducing tumor burden [4]. Recent approaches include combining stemness-targeted therapies with chemotherapy or immunotherapy, although achieving therapeutic efficacy while minimizing toxicity remains a significant challenge [5, 6]. Overall, targeting cancer stemness holds the potential to redefine therapeutic success, from transient remission to durable disease control and, ultimately, long-term cancer prevention.
In solid malignancies such as glioblastoma (GBM), cancer stemness is increasingly recognized as a key contributor to therapeutic resistance and tumor recurrence. GBM stem cells (GSCs) exhibit remarkable phenotypic and metabolic plasticity, enabling dynamic transitions that promote survival under cytotoxic stress [7, 8]. Among these, mesenchymal GSCs demonstrate greater resistance to conventional therapies compared to proneural subtypes [9]. This functional heterogeneity is closely tied to differential metabolic dependencies, with distinct GSCs populations variably relying on glutamine, glucose, or lipid metabolism, factors that significantly influence treatment responsiveness [10-12]. Under therapeutic pressure, GBM tumors exploit this metabolic flexibility to evade immune surveillance and repopulate, driving disease relapse and poor clinical outcomes.
Recent studies highlight metabolic rewiring in GBM, particularly within hypoxic poorly vascularized tumor regions. In these niches, tumor cells upregulate genes involved in lipid biosynthesis, storage, and transport [13, 14]. Transcriptomic analyses reveal co-expression of adipogenic and stemness-associated markers in chemoresistant GBM populations [15], suggesting a functional link between these programs. Moreover, tumor cells can reprogram surrounding adipocytes or hijack adipogenic signaling pathways to support their growth and survival [16]. These findings underscore the therapeutic importance of targeting metabolic plasticity and stemness-related pathways in GBM to overcome resistance and improve long-term patient outcomes.
Emerging evidence suggests that cancer cells can co-opt adipogenic transcription factors, such as peroxisome proliferator-activated receptor gamma (PPARγ) and CCAAT/enhancer-binding protein alpha (C/EBPα), to facilitate metabolic adaptation within the nutrient-deprived, stress-inducing tumor microenvironment (TME) [17, 18]. This phenotypic mimicry enhances lipid uptake and utilization, promoting cellular resilience and survival. In rapidly proliferating solid tumors like GBM, hypoxic niches arise due to inadequate vascularization. Under these conditions, hypoxia-inducible factors (HIFs) are upregulated, triggering a broad transcriptional response that includes activation of adipogenesis-associated pathways [19, 20]. This metabolic reprogramming contributes to chemoresistance by altering drug permeability, reinforcing antioxidant defenses, and reshaping key components of the TME [19, 21]. To effectively study these adaptations and their therapeutic implications, experimental models that replicate hypoxia-induced metabolic and transcriptional dynamics, such as three-dimensional culture systems, are essential.
Three-dimensional (3D) spheroid cultures provide a physiologically relevant in vitro model that closely mimics the architecture, oxygen and nutrient gradients, and microenvironmental complexity of solid tumors [22]. Unlike traditional two-dimensional (2D) monolayer cultures, 3D spheroids exhibit structural heterogeneity, including proliferative outer layers, quiescent intermediate zones, and necrotic cores, reflecting the spatial dynamics of intratumoral regions [23]. The hypoxic core and associated nutrient gradients promote the expression of CSC-associated transcriptional markers such as SOX2, OCT4, PROM1 (CD133), and NANOG, enabling the study of CSC plasticity and behavior under tumor-like conditions [24]. Additionally, spheroids replicate diffusion limitations and cell-cell interactions that contribute to therapeutic resistance, making them valuable for investigating CSC-specific drug evasion strategies and screening agents that disrupt stemness-associated resilience pathways [25].
In this study, 3D GBM spheroids were employed as a physiologically relevant platform to assess the chemopreventive potential of (-)-epigallocatechin-3-gallate (EGCG), a bioactive compound derived from dietary green tea. By targeting the emergence of adipogenic features and CSC-associated transcriptional signatures, EGCG modulated the transcriptional landscape of GBM cells under conditions that better reflect the clinical disease phenotype. This diet-derived intervention highlights the potential for nutritional compounds to influence tumor plasticity and suppress the acquisition of stemness and metabolic reprogramming, hallmarks of therapeutic resistance and tumor recurrence. Importantly, such biologically grounded approaches may inform the development of pharmacologic strategies with enhanced predictive validity, bridging the gap between preclinical screening models and clinical efficacy, and improving the likelihood of translational success.
Materials and Methods
Cell culture
Human GBM-derived cell lines U87, U118, U138, and U251, along with their respective culture media, were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were cultured in medium supplemented with 10% fetal bovine serum (FBS), without the use of antibiotics. Cultures were maintained in a humidified incubator at 37 °C with 5% CO2, and kept subconfluent to preserve cellular integrity. Cells were passaged bi-weekly at a 1:2 ratio and used for experiments within a maximum of eight passages to ensure consistency and minimize phenotypic drift.
Formation of 3D hanging drop spheroids
Three-dimensional (3D) spheroid formation was achieved using the hanging drop technique and low-attachment culture methods, as previously described [26]. Briefly, 100 mm non-adherent petri dishes were used, with lids removed and 10 mL of sterile phosphate-buffered saline (PBS) added to the base of each dish to create a hydration chamber. Using a multichannel pipette, 40 μL droplets of cell suspension (at densities of 10,000 cells per drop) were carefully dispensed onto the inner surface of the inverted lid, ensuring adequate spacing to prevent droplet contact. Up to 50 droplets were placed per lid. The lid was then inverted over the PBS-filled chamber and incubated at 37 °C in a humidified atmosphere (5% CO₂, 95% humidity) for 5 days. Droplets were monitored daily under a microscope to assess spheroid formation. Following aggregation, spheroids were either analyzed directly or transferred to 96-well round-bottom plates (FALCON #351177) pre-coated with poly(2-hydroxyethyl methacrylate) [poly-HEMA] (P3932, Sigma-Aldrich, Oakville, ON, Canada) to prevent cell adhesion. Each well contained 250 μL of fresh complete medium to support continued growth and viability.
Microscopy and analysis of spheroids
Cell morphology was examined using a Nikon Eclipse TE2000-U inverted microscope (Nikon A1, Melville, NY, USA). Phase contrast micrographs were captured with a Q Imaging QICAM-IR Fast 1394 Digital CCD camera, operated via NIH μManager software (National Institutes of Health, Bethesda, MD). For imaging in 96-well plates, an Agilent BioTek Cytation 5 cell imaging multimode reader equipped with 4x and 10x objectives was used to acquire bright-field images of entire wells, using Gen5 software. Quantitative analysis of spheroid size was performed using QuPath version 0.5.1, in combination with the Segment Anything Model (SAM) extension [27]. This tool leverages deep learning algorithms to segment objects within images by identifying contours and morphological features, enabling precise and reproducible measurements.
Oil Red O lipid staining
Neutral lipid staining was performed using Oil Red O on both monolayer cell cultures and 3D spheroids. Prior to fixation, cells were transferred to culture plates and rinsed twice with phosphate-buffered saline (PBS) to remove residual medium and minimize staining artifacts. Cells were then fixed with 4% paraformaldehyde (PFA) for 10-15 minutes at room temperature. Following fixation, samples were rinsed with PBS and subsequently with distilled water. Excess PBS was gently blotted to prevent dilution of the alcoholic staining solution and ensure optimal lipid visualization. A freshly prepared Oil Red O working solution, composed of three parts 0.5% stock solution in isopropanol and two parts distilled water (3:2), was filtered and applied to the samples, followed by a 15-minutes incubation at room temperature. After staining, excess dye was removed by gentle washing with distilled water. Throughout the procedure, samples were kept hydrated to preserve cellular morphology and ensure specific lipid staining without precipitation artifacts. Image acquisition was conducted using the Agilent BioTek Cytation 5 cell imaging system (BioTek), utilizing both bright field contrast and fluorescence modes, with Texas Red filter cube.
Transient transfection and RNA interference
U87 cells were transiently transfected with small interfering RNA (siRNA) sequences using Lipofectamine-2000 transfection reagent (Thermo Fisher Scientific). Gene silencing was performed using 20 nM siRNA against HIF-1α (Hs_HIF1A_5 siRNA, SI02664053), PPARG (Hs_PPARG_2 siRNA, SI00071680), or a non-targeting scrambled control (AllStar Negative Control siRNA, 1027281), all synthesized by QIAGEN (Valencia, CA, USA) and annealed to form duplexes. The efficiency of gene silencing was evaluated by RT-qPCR, as described below.
Total RNA extraction, cDNA synthesis, and real-time quantitative PCR
Total RNA was extracted from 2D monolayer cultures and 3D spheroids using TRIzol reagent, following the manufacturer's protocol (Life Technologies, Gaithersburg, MD, USA). Total RNA concentration and purity were assessed using a NanoPhotometer P330 (Implen). A total of 2 μg of RNA was reverse-transcribed into cDNA using the high-capacity cDNA reverse transcription kit (Applied Biosystems; Foster City, CA, USA). Quantitative PCR (RT-qPCR) was performed using SsoFast EvaGreen Supermix (Bio-Rad; 1725204), and gene specific primer sets purchased from QIAGEN. The following QuantiTect primer sets were used: PPARG (Hs_PPARG_1_SG QT00029841), DDIT3 (Hs_DDIT3_1_SG QT00082278), FOXO1 (Hs_FOXO1_1_SG QT00044247), VEGF (Hs_VEGFA_1_SG QT01010184), GLUT1 (Hs_SLC2A1_1_SG QT00068957), HIF-1α (Hs_HIF1A_1_SG QT00083664), GAPDH (Hs_GAPDH_2_SG QT01192646) and PPIA (Hs_PPIA_4_SG QT01866137). Gene expression analysis was conducted on a Bio-Rad CFX Connect Real-Time PCR Detection System using Bio-Rad CFX Manager Software version 3.0. Relative expression levels were normalized to two housekeeping genes, GAPDH and PPIA, and calculated using the standard 2-ΔΔCq method.
Human cancer stem cell and human adipogenesis profiler PCR arrays
Premade RT2 Profiler PCR arrays for Human Cancer Stem Cells (PAHS-176Z) and Human Adipogenesis (PAHS-049ZD) were purchased from QIAGEN and used according to the manufacturer's instructions. Briefly, genomic DNA was removed prior to reverse-transcription of 0.5 μg of total RNA using the RT2 First Strand Kit (QIAGEN, 330404). Each array plate was used to analyze one cDNA sample, prepared with the RT2 SYBR Green qPCR Mastermix (QIAGEN, 330502). Gene expression analysis of 84 target genes and internal controls was performed using the GeneGlobe Data Analysis Center (QIAGEN; https://geneglobe.qiagen.com/us/analyze), applying the standard 2-ΔΔCq method for relative quantification. In figures, fold regulation values were reported: for upregulated genes, fold regulation equals fold change, for downregulated genes, fold regulation was calculated as 1/(fold change).
In silico analysis of transcript levels in clinical GBM and low-grade glioma tissues
Gene Expression Profiling Interactive Analysis (GEPIA) was used to examine RNA sequencing expression data from glioblastoma (GBM, n = 163) and low-grade glioma (LGG; n = 251) tumor samples, compared to normal brain tissue (n = 207), using data sets from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) databases [28]. GEPIA offers a suite of customizable analytical tools, including tumor/normal differential expression analysis, cancer type and pathological stage profiling, patient survival analysis, similar gene detection, correlation analysis, and dimensionality reduction (http://gepia.cancer-pku.cn/detail.php, accessed on June 28th, 2025). Differential gene expression analysis was performed using one-way ANOVA, with disease state (GBM, LGG, or normal) as grouping variable for box plot visualization.
Prognostic value of HIF-1α, CEBPD, BMP7, FOXO1, LPL, DDIT3, and PPARG in GBM patients
The prognostic significance of mRNA expression levels of HIF-1α, CEBPD, BMP7, FOXO1, LPL, DDIT3, and PPARG in GBM patients was evaluated using the GEPIA web server [28]. For each gene, expression data were retrieved from a comprehensive web-based database containing high-quality RNA sequencing datasets from GBM tissues. Overall survival analysis was performed using the log-rank test to assess the correlation between gene expression levels and patient outcomes.
Statistical data analysis
Data are presented as mean ± standard deviation (SD) from three independent experiments, unless otherwise specified. Statistical analyses were performed using non-parametric tests: the Mann-Whitney U-test for comparisons between two groups, and the Kruskal-Wallis test followed by Dunn's post hoc test for comparisons involving three or more groups. A p-value < 0.05 (*) was considered statistically significant and is indicated in the figures.
Results
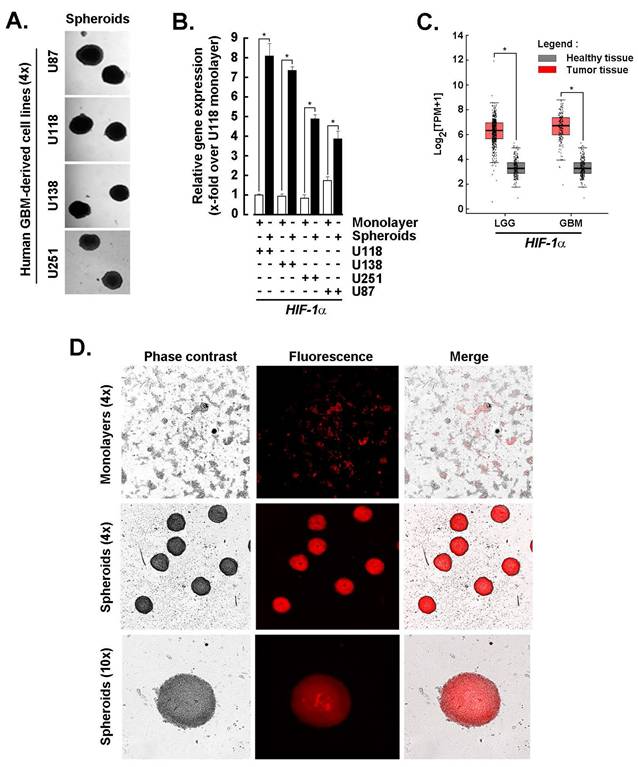
Spheroids from GBM-derived cell lines share a high HIF-1α transcriptional signature with clinical tumor tissues and maintain active adipogenesis. Monolayer cultures derived from human GBM cell lines were used to generate 3D spheroids as described in the Methods section (Fig. 1A). Total RNA was extracted, and transcript levels of HIF-1α, a key regulator of tumor survival under hostile microenvironmental conditions, were assessed by RT-qPCR. Upon spheroid formation, HIF-1α expression was significantly upregulated in U87, U118, U138, and U251 cells (Fig. 1B, black bars), compared to their respective 2D monolayer counterparts (Fig. 1B, white bars). The coordinated cellular response is likely driven by hypoxia within the spheroid core, where HIF-1α orchestrates metabolic adaptation and vascular remodeling. A similar pattern of HIF-1α upregulation was observed in clinical samples of low-grade glioma (LGG) and GBM tumors compared to normal brain tissue (Fig. 1C), further validating the relevance of the spheroid model. Additionally, Oil Red O staining of U87-derived spheroids revealed prominent lipid accumulation, indicative of active adipogenesis and a shift toward a lipogenic metabolic phenotype (Fig. 1D). These findings support the presence of a hypoxia-induced molecular signature in GBM-derived 3D spheroids and prompt further investigation into how adipogenic adaptation contributes to the chemoresistant phenotype of CSCs.
Spheroids from GBM-derived cell lines share a high HIF-1α transcriptional signature with tumor tissues and exert active adipogenesis. A) Monolayer cultures from human GBM-derived cell lines were used to generate 3D spheroids as described in the Methods section, and representative phase contrast pictures taken (Magnification = 4x). B) Total RNA was extracted from 2D monolayers (white bars) and 3D spheroids (black bars) of the indicated GBM-derived cells, and HIF-1α expression assessed by RT-qPCR as described in the Methods section (*P <0.05). C) In silico analysis of transcript levels was performed for HIF-1α using RNA extracted from clinical samples from GBM and low-grade glioma (LGG) (red boxes) and compared to healthy tissue (grey boxes) (*P <0.05). D) Active adipogenesis was assessed in 2D monolayers and 3D spheroids from U87 cells upon staining with Oil Red O as described in the Methods section.
PPARγ as a hub for the adaptive adipogenic transcriptional signature in 3D GBM-derived spheroids. A) Total RNA was extracted from 2D monolayers or 3D spheroids of the indicated GBM-derived cells. Differential adipogenesis gene arrays was performed and representative fold-change histogram profiles of gene expression in spheroids cultures versus 2D monolayers shown. B) Heat map representation of the mean expression from the conserved top 14 genes upregulated upon spheroids formation from the four tested cell lines. C) Protein-to-protein interaction network of the 14 conserved and most upregulated gene expression triggered upon 3D spheroids formation as determined with STRING.
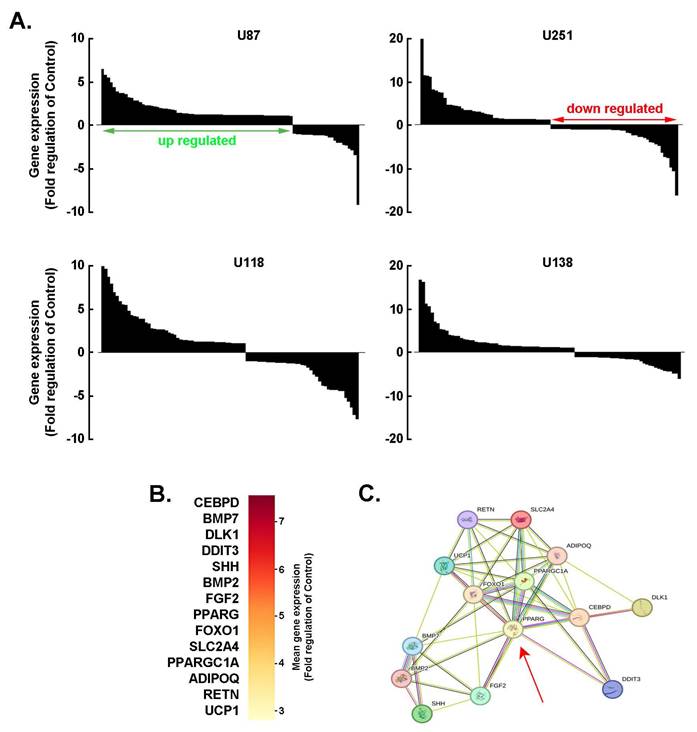
PPARγ as a hub for the adaptive adipogenic molecular signature in 3D GBM-derived spheroids. Investigating adipogenic adaptive metabolism in cancer cells is essential for understanding how tumors exploit lipid-related pathways to support growth, evade therapy, and reshape their microenvironment. By acquiring adipogenic traits, cancer cells gain metabolic flexibility, enabling them to switch between glucose and lipid utilization, thereby increasing their resilience to metabolic-targeted therapies. To explore this phenomenon, a screen of 84 adipogenesis-related genes was conducted across 3D spheroids generated from four human GBM-derived cell lines. The spheroids exhibited consistent patterns of gene upregulation and downregulation (Fig. 2A). Among the top 14 commonly upregulated genes were: CEBPD, an early adipogenic transcription factor that activates C/EBPα and PPARγ, BMP7, which promotes mesenchymal stem cells commitment to the adipogenic lineage, BMP2, which promotes adipogenic differentiation via SMAD signaling and PPARG activation, PPARG, a master regulator of adipogenesis that drives the expression of genes involved in adipocyte differentiation and lipid storage, and PPARGC1A, a coactivator of PPARγ (Fig. 2B). These genes were found to be functionally interconnected (Fig. 2C), with PPARγ emerging as a central hub linking multiple components of the adipogenic network (Fig. 2C, red arrow). Understanding this metabolic crosstalk opens new avenues for therapeutic intervention, such as targeting lipid metabolism or disrupting adipocyte-tumor signaling pathways, which may help overcome resistance and improve treatment outcomes in GBM.
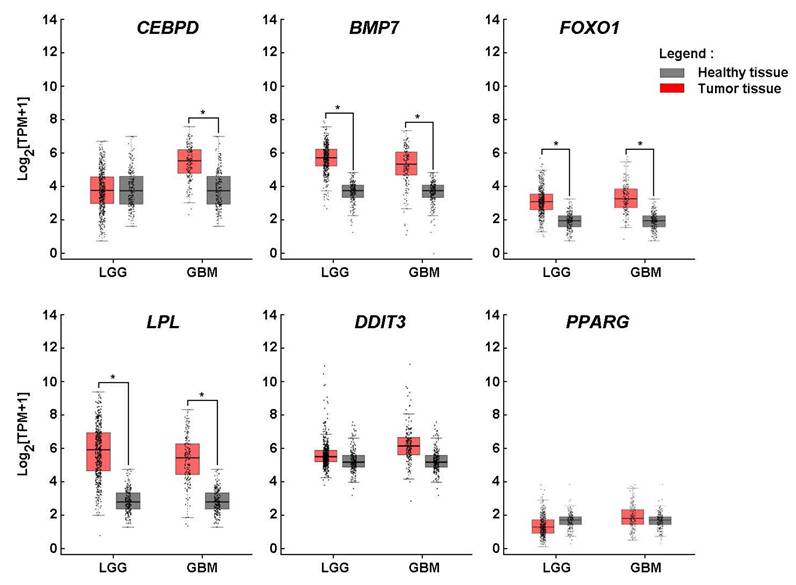
Lipid metabolic adaptation in GBM mediated by CEBPD, BMP7, FOXO1, and LPL overexpression. The upregulation of HIF-1α and adipogenesis-related genes in 3D spheroids supports the hypothesis that GBM cells may activate adipogenic pathways to promote growth, survival, and therapy resistance. When gene expression analysis was extended to clinical tissue samples from low-grade glioma (LGG) and GBM, elevated levels of CEBPD, BMP7, FOXO1, and LPL were observed in both tumor types, except for CEBPD, which was not significantly upregulated in LGG, suggesting a metabolic reprogramming that favors adipogenic and lipogenic signaling. Interestingly, while these genes were induced upon spheroid formation, no significant differences in DDIT3 and PPARG expression were detected in bulk tumor tissues compared to healthy controls (Fig. 3). This discrepancy may be attributed to tumor heterogeneity, particularly the variable expression of PPARG across GBM subtypes and cellular compartments. For instance, PPARG may be enriched in specific tumor cell populations, such as GBM stem-like cells, which are underrepresented in bulk tissue analyses. This differential interpretive approach underscores the importance of considering the distinct biological and genomic contexts in which these genes operate, and highlights the limitations of bulk tissue comparisons when investigating cell-type-specific regulatory mechanisms.
Lipid metabolic adaptation in glioblastoma mediated by CEBPD, BMP7, FOXO1, and LPL overexpression. In silico analysis of transcript levels was performed for CEBPD, BMP7, FOXO1, LPL, DDIT3, and PPARG using RNA extracted from clinical samples from GBM and low-grade glioma (LGG) (red boxes) and compared to healthy tissue (grey boxes) (*P <0.05).
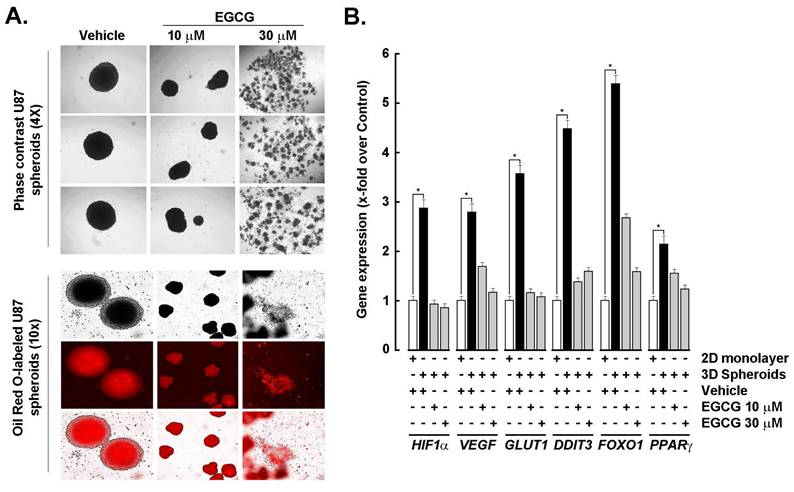
EGCG transcriptional inhibition of HIF-1α and adipogenic signature in 3D GBM spheroids. (-)-Epigallocatechin-3-gallate (EGCG), the predominant catechin in green tea, inhibits adipogenesis through multiple molecular mechanisms, making it a promising candidate for both anti-obesity and anti-cancer therapies. To investigate whether EGCG could modulate the transcriptional and signaling landscape associated with 3D spheroid formation, spheroids were generated from 2D monolayer cultures in the presence or absence of EGCG. EGCG treatment resulted in a dose-dependent reduction in spheroid size: 10 μM EGCG led to a noticeable decrease, while 30 μM disrupted spheroid integrity (Fig. 4A, upper panels). Concurrently, Oil Red O staining revealed diminished lipid accumulation in EGCG-treated spheroids, indicating suppression of active adipogenesis (Fig. 4A, lower panels). At the molecular level, EGCG dose-dependently inhibited the spheroid-induced upregulation of HIF-1α, as well as angiogenic markers VEGF and GLUT1, and adipogenic markers DDIT3, FOXO1, and PPARG (Fig. 4B). These findings suggest that EGCG interferes with hypoxia-driven metabolic reprogramming and adipogenic signaling, thereby disrupting the cellular adaptations that support GBM spheroid growth and stemness.
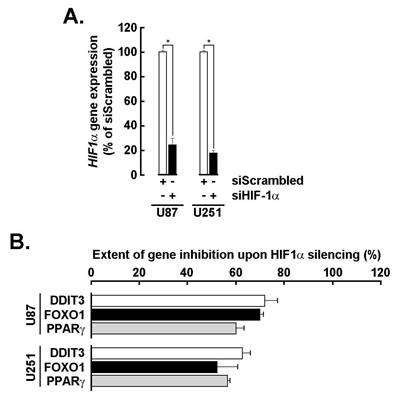
Transient siRNA-mediated HIF-1α silencing inhibits the adipogenic signature in GBM spheroids. To more specifically assess the role of HIF-1α in the acquisition of the adipogenic signature, transient gene silencing was performed in U87 and U251 cells (Fig. 5A). Knockdown of HIF-1α resulted in a marked reduction in the expression of adipogenic markers DDIT3, FOXO1, and PPARG in 3D spheroids (Fig. 5B), supporting its upstream regulatory role in driving adipogenic reprogramming. It is therefore reasonable to infer that the EGCG-mediated suppression of HIF-1α contributes to the downstream inhibition of these adipogenic effectors [29], further reinforcing the compound's potential to disrupt hypoxia-induced metabolic adaptation in GBM.
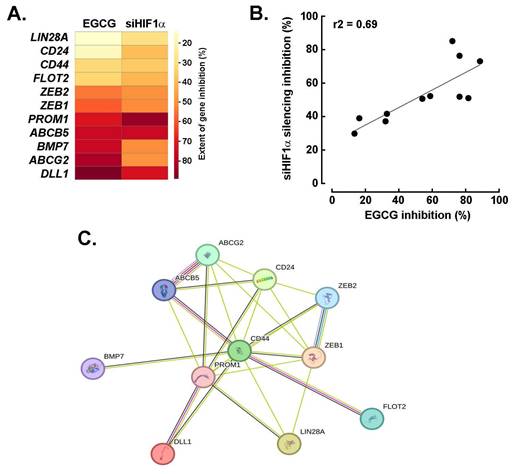
EGCG and silencing of HIF-1α share a common transcriptional inhibition activity that impacts the acquisition of a CSCs transcriptional signature in U87 spheroids. To investigate the acquisition of a cancer stem cell (CSC) phenotype in 3D spheroids, gene expression profiling was performed using a targeted array. Among the genes found to be downregulated, both EGCG treatment and HIF-1α silencing inhibited the expression of 11 common targets, with the most significantly affected being ABCB5, BMP7, ABCG2, and DLL1 (Fig. 6A). Interestingly, the extent of inhibition across these shared targets was well correlated, ranging from 20 to 80% reduction in expression (Fig. 6B). Further analysis using the STRING database confirmed coherent interrelationships among these genes, highlighting functional connectivity within the CSC-associated network (Fig. 6C).
EGCG inhibits HIF-1α and adipogenic signature in GBM spheroids. A) Monolayer cultures from the human U87 GBM-derived cell line were used to generate 3D spheroids in the absence (vehicle) or presence of the indicated EGCG concentrations as described in the Methods section and representative phase contrast pictures taken (Upper panels; Magnification = 4x). Similarly, treated cells were stained with Oil Red O and representative phase contrast and fluorescent pictures taken (Magnification = 10x). B) Total RNA was extracted from 2D monolayers (white bars) and 3D spheroids (black bars) of the indicated GBM-derived cells and HIF-1α, VEGF, GLUT1, DDIT3, FOXO1, and PPARG expression assessed by RT-qPCR as described in the Methods section (*P < 0.05).
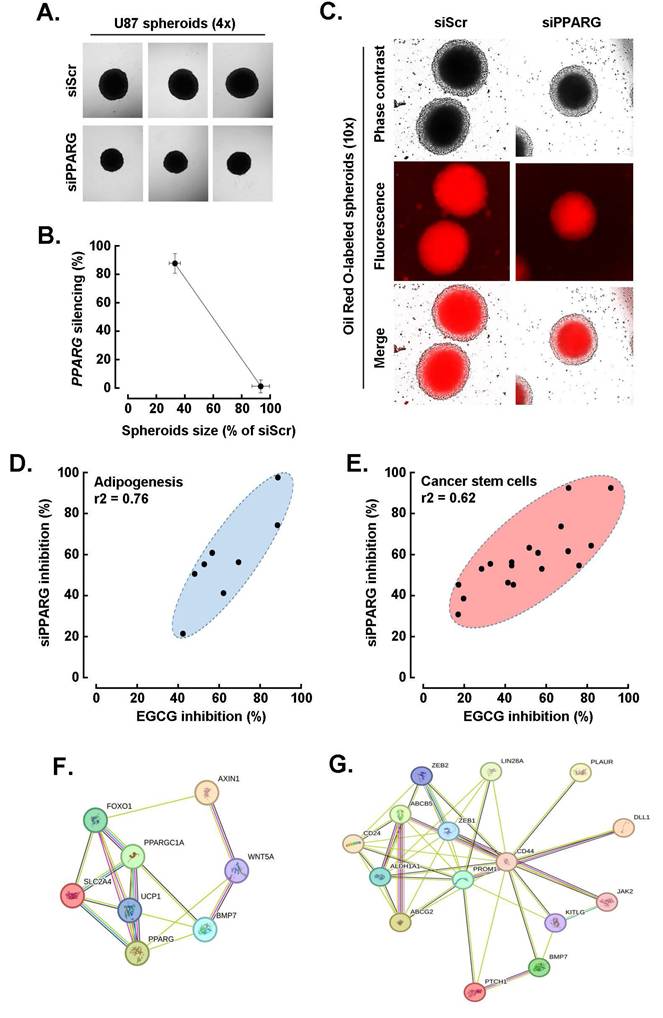
EGCG targeting of PPARG alters both the CSCs and adipogenic transcriptional signature in U87 spheroids. Building on the approach used for HIF-1α, we next investigated the interplay between EGCG and PPARγ in regulating the CSC and adipogenic transcriptional phenotype induced during U87 spheroid formation. To this end, PPARG was transiently silenced, and its impact on spheroid formation was assessed. Representative phase contrast images (Fig. 7A) revealed a significant reduction in spheroid size following PPARG knockdown, which correlated with the extent of gene repression confirmed by qPCR (Fig. 7B). Additionally, Oil Red O staining showed a peripheral reduction in lipid accumulation in PPARG-silenced cells, indicating suppressed adipogenesis (Fig. 7C). Expression analysis of the top eight adipogenesis-related genes, including PPARG, FOXO1, SLC2A4, and BMP7, demonstrated a consistent pattern of inhibition following either EGCG treatment or PPARG silencing, with a strong correlation (Fig. 7D, r2 = 0.76). A similar trend was observed for the top fifteen CSC-related genes, such as PROM1, ABCG2, ABCB5, and CD44, which also showed correlated downregulation under both conditions (Fig. 7E, r2 = 0.62). STRING network analysis further revealed functional crosstalk among the modulated adipogenesis-related genes (Fig. 7F) and CSC-associated genes (Fig. 7G), highlighting the interconnected transcriptional landscape influenced by PPARγ and EGCG during spheroid formation.
HIF-1α transient silencing inhibits PPARG, DDIT3, and FOXO1 transcript levels. A) Transient gene silencing of HIF-1α was performed in U87 and U251 cells as described in the Methods section and extent of gene silencing assessed by RT-qPCR (*P <0.05). B) Total RNA was extracted from U87 and U251 3D spheroids where HIF-1α was silenced, and impact on DDIT3, FOXO1, and PPARG expression assessed by RT-qPCR as described in the Methods section.
EGCG and silencing of HIF-1α share a common transcriptional inhibition activity that impacts the acquisition of a CSCs transcriptional signature in U87 spheroids. A) Representative heat map of the extent to which EGCG and HIF-1a silencing alters those eleven common genes induced expression and that characterize the acquisition of a CSCs transcriptional phenotype in spheroids. B) Correlation of the extent of inhibition of genes from A) between EGCG-treated cells and cells where HIF-1α was silenced. C) Protein-to-protein interaction network of the 11 most upregulated genes for which expression was altered by both EGCG and upon HIF-1α silencing in spheroids as determined with STRING.
EGCG targeting of PPARG alters both the cancer stem cell and adipogenic transcriptional signature in U87 spheroids. A) Transient PPARG silencing was performed in monolayer cultures from human U87 GBM-derived cell lines and 3D spheroids generated as described in the Methods section, and representative phase contrast pictures taken and compared to control (siScrambled-transfected cells, siScr; Magnification = 4x). B) Correlation between 3D spheroids size and extent of PPARG gene silencing. C) Similarly, treated cells were stained with Oil Red O and representative phase contrast and fluorescent pictures taken (Magnification = 10x). D) Modulation of the top eight adipogenesis-related genes for which induction upon spheroids formation was inhibited by both EGCG treatment and PPARG silencing. E) Modulation of the top fifteen CSC-related genes for which induction upon spheroids formation was inhibited by both EGCG treatment and PPARG silencing. F) Protein-to-protein interaction network of the eight most upregulated adipogenic genes, and of G) the fifteen most upregulated CSC genes, for which expression was altered by both EGCG and upon PPARG silencing in spheroids as determined with STRING.
Discussion
Glioblastoma (GBM) is characterized by pronounced metabolic plasticity, enabling tumor cells to dynamically adapt to hostile microenvironmental conditions, such as hypoxia, nutrient deprivation, and oxidative stress. These metabolic adaptations drive extensive transcriptional reprogramming, reinforcing pro-survival and therapy-resistant phenotypes. To investigate the molecular interplay between metabolic shifts and stemness-associated transcriptional networks, we employed a 3D GBM spheroid culture system that recapitulates key features of in vivo tumor physiology. This model was used to examine the induction of adipogenic transcriptional signatures and their convergence with stem cell-associated gene expression programs, both of which contribute to chemoresistance and tumor persistence. Furthermore, we evaluated the modulatory effects of EGCG, a bioactive phytochemical derived from dietary green tea, on the coordinated transcriptional regulation mediated by HIF-1α and PPARγ. EGCG attenuated the expression of adipogenic and stemness-related markers, underscoring its potential as a chemopreventive agent capable of disrupting transcriptional and metabolic vulnerabilities in GBM. These findings support the development of integrative therapeutic strategies that simultaneously target metabolic flexibility and transcriptional resilience, with the goal of improving treatment outcomes and reducing tumor recurrence.
Enhanced adipogenic signaling within tumor cells promotes lipid accumulation, creating intracellular reservoirs that can sequester lipophilic chemotherapeutic agents, and reduce their cytotoxic efficacy [14]. This lipid-enriched microenvironment also contributes to the stabilization of HIFs, which reinforce cancer stem-like phenotypes and further enhance therapeutic resistance [30]. Notably, the ATP-binding cassette transporter G2 (ABCG2), a key efflux protein implicated in multidrug resistance, is significantly upregulated in 3D spheroids and cancer stem-like cells. Its expression is closely associated with adipogenic molecular pathways [31]. Hypoxia-induced lipid accumulation observed in malignancies such as breast, ovarian, and GBM, correlates with sustained stemness and diminished treatment responsiveness. In parallel, adipokines such as leptin and interleukin-6, upregulated under hypoxic conditions, promote tumor progression and facilitate immune evasion [32]. In solid tumors, hypoxic stress can induce adipogenic-like metabolic traits that critically influence tumor plasticity and drug resistance. Consistent with the inhibitory effects of EGCG on PPARG transcript levels, pharmacologic inhibition of PPARγ or lipid metabolic pathways has been shown to restore chemosensitivity in preclinical models [33]. Collectively, these findings highlight the pivotal role of an adipogenic transcriptional program in remodeling the TME to favor chemoresistance. Moreover, the diet-derived phytochemical EGCG demonstrates potential in targeting adipogenic-like metabolic reprogramming, offering a promising chemopreventive strategy against transcriptionally driven therapy resistance in GBM.
Beyond metabolic reprogramming, CSC plasticity plays a critical role in shaping therapeutic outcomes by enabling resistance mechanisms, driving tumor recurrence, and promoting metastatic dissemination, each representing major barriers to sustained remission. CSCs exhibit intrinsic resistance to chemotherapy and radiotherapy, largely due to their quiescent state, enhanced DNA repair capacity, and elevated expression of drug efflux transporters [34]. Additionally, CSCs also exert immunomodulatory effects within the TME, dampening host immune surveillance and compromising the efficacy of immunotherapeutic strategies. Conventional treatment modalities, which primarily target proliferative tumor cells, often fail to eliminate this resilient CSC population. As a result, therapeutic relapse remains a significant challenge. Our findings support the development of future therapeutic strategies aimed at disrupting stemness-associated signaling pathways or leveraging chemopreventive agents, particularly those derived from dietary sources, for their capacity to prevent the acquisition of CSC characteristics and enhance treatment responsiveness [35].
Targeting CSC plasticity represents a promising therapeutic strategy to overcome treatment resistance and improve clinical outcomes. Approaches aimed at disrupting epithelial-mesenchymal transition (EMT) and reversing stemness-associated traits, particularly through inhibition of key transcriptional regulators such as SNAIL, ZEB, and TWIST, have gained traction for their potential to reduce tumor aggressiveness and therapy resistance [36]. Diet-derived phytochemicals, including curcumin, resveratrol, sulforaphane, and EGCG, have demonstrated the ability to inhibit EMT and downregulate these transcription factors, thereby attenuating CSC-related phenotypes and enhancing sensitivity to conventional therapy [37-39]. Beyond direct effects on cancer cells, modulation of the CSC niche, including immune infiltrates, cancer-associated fibroblasts, and extracellular matrix components, can further influence stemness dynamics. For example, resveratrol and sulforaphane have shown efficacy in suppressing HIF-1α activity and reprogramming immune responses, thereby compromising CSC survival and reducing tumor persistence [40]. Differentiation therapy is another emerging approach, wherein agents such as retinoic acid and genistein promote the transition of CSCs into non-stem-like phenotypes that are more susceptible to standard treatments [6, 41]. Collectively, these strategies underscore the multifaceted potential of combining phytochemical-based interventions with conventional therapies to effectively target CSC-driven resistance and prevent tumor relapse.
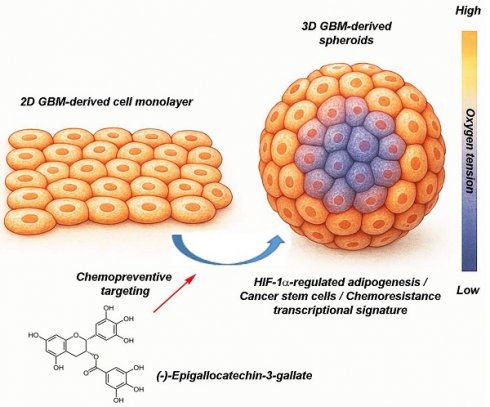
EGCG targets 3D spheroids by disrupting hypoxia-driven processes, cancer stem cells, and adipogenesis-mediated resistance-linked transcription. This figure presents two in vitro culture models of GBM, a classical 2D cell monolayer and a more physiologically relevant 3D spheroid culture. The 3D spheroid model better mimics the TME, particularly through the presence of an oxygen gradient, with normoxic conditions at the periphery and hypoxia toward the core. This spatial heterogeneity enables more accurate assessment of tumor behavior and treatment response. The diagram also illustrates the chemopreventive property of (-)-Epigallocatechin-3-gallate (EGCG), a green tea-derived polyphenol, which targets key pathways linked to metabolic rewiring and tumor chemoresistance. Specifically, EGCG is depicted as inhibiting HIF-1α-regulated adipogenesis, and modulating transcriptional profiles associated with cancer stem cells and therapy resistance. These insights underscore the necessity of using complex 3D models in preclinical research and highlight EGCG as a promising candidate in overcoming GBM treatment challenges.
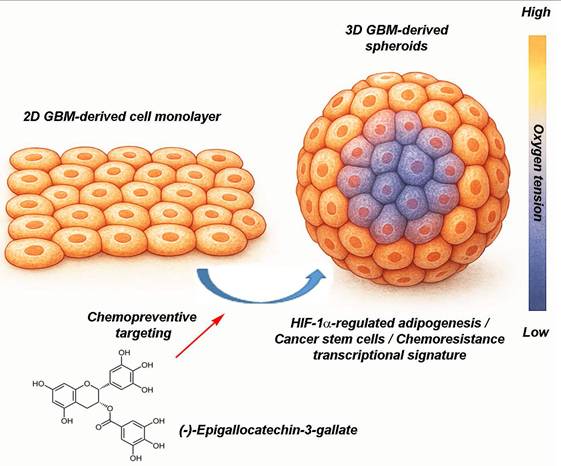
The integration of diet-derived phytochemicals into pharmacological studies using 3D GBM spheroid models represents a significant advancement in preclinical therapeutic research. Unlike conventional 2D monolayer cultures, 3D spheroid systems more accurately recapitulate the TME, including spatial gradients of oxygen, nutrients, and drug diffusion. These features enhance the physiological relevance and predictive validity of experimental outcomes. Emerging preclinical evidence suggests that phytochemicals such as EGCG interfere with adipogenic transcriptional pathways mediated by PPARγ and C/EBPα, which contribute to metabolic reprogramming and the stabilization of stem-like phenotypes in GBM. This dual impact, on both CSC-associated signaling and adipogenic transcriptional programs, offers a novel therapeutic strategy, particularly within 3D spheroid systems that mimic key aspects of in vivo tumor biology. Such models not only improve translational relevance but may accelerate the identification of effective chemopreventive or combination treatment regimens targeting metabolic and stemness-linked vulnerabilities in GBM.
GBM also remains one of the most treatment-resistant forms of brain cancer, largely due to the overexpression of ATP-binding cassette transporters such as ABCG2. These transporters actively efflux chemotherapeutic agents from cancer cells, thereby reducing intracellular drug concentrations and diminishing therapeutic efficacy. In this context, EGCG has shown promising potential to overcome drug resistance. Clinically, EGCG is recognized for its ability to sensitize GBM cells to chemotherapy, particularly temozolomide, by downregulating drug efflux mechanisms, enhancing intracellular drug retention and increasing cytotoxicity in GSCs. At the molecular level, EGCG modulates ABCG2 expression and function through multiple pathways. It suppresses transcription factors such as NFκB and HIF-1α, both of which are known to upregulate ABCG2. Additionally, EGCG exerts epigenetic effects, including alterations in DNA methylation and histone acetylation, which may contribute to the silencing of ABCG2 gene expression. EGCG has also been shown to reduce stemness markers and impair neurosphere formation, indirectly suppressing ABCG2 levels in CSC populations. Moreover, EGCG induces oxidative stress and apoptosis in GBM cells, disrupting survival pathways that support ABCG2 expression. Together, these molecular insights, summarized in Fig. 8, underscore EGCG's potential as an adjunctive therapy aimed at improving treatment outcomes in GBM by targeting multidrug resistance mechanisms.
Conclusions
This study underscores the value of physiologically relevant 3D GBM spheroid models as platforms for investigating the molecular mechanisms underlying tumor plasticity and therapeutic resistance. By integrating the use of EGCG, a diet-derived phytochemical, we demonstrate its potential as a chemopreventive agent capable of disrupting tumor-promoting transcriptional programs. Specifically, EGCG attenuated the induction of a hybrid adipogenic/CSC transcriptional signature, which is associated with tumor cell adaptation to hypoxic and nutrient-deprived microenvironments. These findings suggest that compounds such as EGCG may serve not only as adjuncts to conventional therapies, but also as modulators of tumor cell resilience by targeting key metabolic and stemness-associated pathways. Importantly, this chemopreventive strategy aligns with the broader concept of leveraging nutritional compounds to inform precision medicine approaches and enhance therapeutic efficacy in GBM. Future translational efforts may benefit from combining EGCG with agents specifically designed to eradicate CSCs, potentially overcoming treatment resistance and reducing tumor recurrence.
Abbreviations
ATCC: American type culture collection; C/EBPα: CCAAT/enhancer-binding protein alpha; CSC: Cancer stem cells; EGCG: (-)-Epigallocatechin-3-gallate; EMT: Epithelial-mesenchymal transition; FBS: Fetal bovine serum; GEPIA: Gene expression profiling interactive analysis; GBM: Glioblastoma; GSC: Glioblastoma stem cells; GTEx: Genotype-tissue expression; HIF-1α: Hypoxia inducible factor-1 alpha; LGG: Low-grade glioma; PBS: Phosphate-buffered saline; PPARG: Peroxisome proliferator-activated receptor gamma; siRNA: Small interfering RNA; TME: Tumor microenvironment.
Acknowledgements
BA holds an Institutional Research Chair in Cancer Prevention and Treatment at UQAM.
Funding
This work was funded by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC, RGPIN-2024-04541) to BA.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author contributions
Conceptualization, Abdallah Fallah, Borhane Annabi; Data curation, Abdallah Fallah, Alain Zgheib, Maellis Payet-Desruisseaux, Alain Zgheib; Formal analysis, Abdallah Fallah, Alain Zgheib, Bogdan Alexandru Danalache, Nicoletta Eliopoulos, Borhane Annabi; Funding acquisition, Borhane Annabi; Investigation, Abdallah Fallah, Maellis Payet-Desruisseaux, Alain Zgheib, Bogdan Alexandru Danalache; Methodology, Alain Zgheib, Bogdan Alexandru Danalache; Supervision, Borhane Annabi; Writing - original draft, Abdallah Fallah, Nicoletta Eliopoulos, Borhane Annabi; Writing - review & editing, Abdallah Fallah, Alain Zgheib, Maellis Payet-Desruisseaux, Nicoletta Eliopoulos, Borhane Annabi. All authors read and approved the final manuscript.
Author information (ORCID)
Borhane Annabi: https://orcid.org/0000-0002-5082-7183.
Nicoletta Eliopoulos: https://orcid.org/0000-0002-5172-8742.
Bogdan Alexandru Danalache: https://orcid.org/0000-0003-1254-3689.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chu X, Tian W, Ning J, Xiao G, Zhou Y, Wang Z. et al. Cancer stem cells: advances in knowledge and implications for cancer therapy. Signal Transduct Target Ther. 2024;9(1):170
2. Dakal TC, Bhushan R, Xu C, Gadi BR, Cameotra SS, Yadav V. et al. Intricate relationship between cancer stemness, metastasis, and drug resistance. MedComm (2020). 2024;5(10):e710
3. El-Tanani M, Rabbani SA, Satyam SM, Rangraze IR, Wali AF, El-Tanani Y. et al. Deciphering the role of cancer stem cells: Drivers of tumor evolution, therapeutic resistance, and precision medicine strategies. Cancers (Basel). 2025;17(3):382
4. Pan Y, Yuan C, Zeng C, Sun C, Xia L, Wang G. et al. Cancer stem cells and niches: challenges in immunotherapy resistance. Mol Cancer. 2025;24(1):52
5. Duan H, Liu Y, Gao Z, Huang W. Recent advances in drug delivery systems for targeting cancer stem cells. Acta Pharm Sin B. 2021;11(1):55-70
6. Mai Y, Su J, Yang C, Xia C, Fu L. The strategies to cure cancer patients by eradicating cancer stem-like cells. Mol Cancer. 2023;22(1):171
7. Galdieri L, Jash A, Malkova O, Mao DD, DeSouza P, Chu YE. et al. Defining phenotypic and functional heterogeneity of glioblastoma stem cells by mass cytometry. JCI Insight. 2021;6(4):e128456
8. Visioli A, Trivieri N, Mencarelli G, Giani F, Copetti M, Palumbo O. et al. Different states of stemness of glioblastoma stem cells sustain glioblastoma subtypes indicating novel clinical biomarkers and high-efficacy customized therapies. J Exp Clin Cancer Res. 2023;42(1):244
9. Steponaitis G, Tamasauskas A. Mesenchymal and proneural subtypes of glioblastoma disclose branching based on GSC associated signature. Int J Mol Sci. 2021;22(9):4964
10. Renoult O, Laurent-Blond M, Awada H, Oliver L, Joalland N, Croyal M. et al. Metabolic profiling of glioblastoma stem cells reveals pyruvate carboxylase as a critical survival factor and potential therapeutic target. Neuro Oncol. 2024;26(9):1572-1586
11. Vijayanathan Y, Ho IAW. The impact of metabolic rewiring in glioblastoma: The immune landscape and therapeutic strategies. Int J Mol Sci. 2025;26(2):669
12. Zhou W, Wahl DR. Metabolic abnormalities in glioblastoma and metabolic strategies to overcome treatment resistance. Cancers (Basel). 2019;11(9):1231
13. Lee H, Kim D, Youn B. Targeting oncogenic rewiring of lipid metabolism for glioblastoma treatment. Int J Mol Sci. 2022;23(22):13818
14. Lu L, Zhang Y, Yang Y, Jin M, Ma A, Wang X. et al. Lipid metabolism: the potential therapeutic targets in glioblastoma. Cell Death Discov. 2025;11(1):107
15. Liu J, Cao S, Imbach KJ, Gritsenko MA, Lih TM, Kyle JE. et al. Multi-scale signaling and tumor evolution in high-grade gliomas. Cancer Cell. 2024;42(7):1217-1238.e19
16. Shen S, Vagner S, Robert C. Persistent cancer cells: The deadly survivors. Cell. 2020;183(4):860-874
17. Li Y, Pan Y, Zhao X, Wu S, Li F, Wang Y. et al. Peroxisome proliferator-activated receptors: A key link between lipid metabolism and cancer progression. Clin Nutr. 2024;43(2):332-345
18. Kim HY, Jang HJ, Muthamil S, Shin UC, Lyu JH, Kim SW, Go Y, Park SH, Lee HG, Park JH. Novel insights into regulators and functional modulators of adipogenesis. Biomed Pharmacother. 2024Aug;177:117073
19. Chen Z, Han F, Du Y, Shi H, Zhou W. Hypoxic microenvironment in cancer: molecular mechanisms and therapeutic interventions. Signal Transduct Target Ther. 2023;8(1):70
20. He Q, Gao Z, Yin J, Zhang J, Yun Z, Ye J. Regulation of HIF-1{alpha} activity in adipose tissue by obesity-associated factors: adipogenesis, insulin, and hypoxia. Am J Physiol Endocrinol Metab. 2011;300(5):E877-85
21. Doktorova H, Hrabeta J, Khalil MA, Eckschlager T. Hypoxia-induced chemoresistance in cancer cells: The role of not only HIF-1. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2015;159(2):166-77
22. Mangani S, Kremmydas S, Karamanos NK. Mimicking the complexity of solid tumors: How spheroids could advance cancer preclinical transformative approaches. Cancers (Basel). 2025;17(7):1161
23. Pinto B, Henriques AC, Silva PMA, Bousbaa H. Three-dimensional spheroids as in vitro preclinical models for cancer research. Pharmaceutics. 2020;12(12):1186
24. Wahab SMR, Islam F, Gopalan V, Lam AK. The Identifications and Clinical Implications of Cancer Stem Cells in Colorectal Cancer. Clin Colorectal Cancer. 2017Jun;16(2):93-102
25. Garnique AdMB, Parducci NS, de Miranda LBL, de Almeida BO, Sanches L, Machado-Neto JA. Two-dimensional and spheroid-based three-dimensional cell culture systems: Implications for drug discovery in cancer. Drugs and Drug Candidates. 2024;3(2):391-409
26. Ohguro H, Watanabe M, Sato T, Nishikiori N, Umetsu A, Higashide M, Yano T, Suzuki H, Miyazaki A, Takada K, Uhara H, Furuhashi M, Hikage F. Application of Single Cell Type-Derived Spheroids Generated by Using a Hanging Drop Culture Technique in Various In Vitro Disease Models: A Narrow Review. Cells. 2024Sep14;13(18):1549
27. Sugawara K. Training deep learning models for cell image segmentation with sparse annotations. bioRxiv preprint (2023) doi: https://doi.org/10.1101/2023.06.13.54478.
28. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556-W560
29. Zhang Q, Tang X, Lu Q, Zhang Z, Rao J, Le AD. Green tea extract and (-)-epigallocatechin-3-gallate inhibit hypoxia- and serum-induced HIF-1alpha protein accumulation and VEGF expression in human cervical carcinoma and hepatoma cells. Mol Cancer Ther. 2006;5(5):1227-38
30. Jun JC, Rathore A, Younas H, Gilkes D, Polotsky VY. Hypoxia-inducible factors and cancer. Curr Sleep Med Rep. 2017;3(1):1-10
31. Kukal S, Guin D, Rawat C, Bora S, Mishra MK, Sharma P. et al. Multidrug efflux transporter ABCG2: expression and regulation. Cell Mol Life Sci. 2021;78(21-22):6887-6939
32. Kim JW, Kim JH, Lee YJ. The role of adipokines in tumor progression and its association with obesity. Biomedicines. 2024;12(1):97
33. Monsalve FA, Pyarasani RD, Delgado-Lopez F, Moore-Carrasco R. Peroxisome proliferator-activated receptor targets for the treatment of metabolic diseases. Mediators Inflamm. 2013;2013:549627
34. Phi LTH, Sari IN, Yang YG, Lee SH, Jun N, Kim KS. et al. Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. 2018;2018:5416923
35. Chan MM, Chen R, Fong D. Targeting cancer stem cells with dietary phytochemical - Repositioned drug combinations. Cancer Lett. 2018;433:53-64
36. Bhat GR, Sethi I, Sadida HQ, Rah B, Mir R, Algehainy N. et al. Cancer cell plasticity: from cellular, molecular, and genetic mechanisms to tumor heterogeneity and drug resistance. Cancer Metastasis Rev. 2024;43(1):197-228
37. Sravani ANKV, Thomas J. Targeting epithelial-mesenchymal transition signaling pathways with Dietary Phytocompounds and repurposed drug combinations for overcoming drug resistance in various cancers. Heliyon. 2025;11(3):e41964
38. Gresseau L, Roy ME, Duhamel S, Annabi B. A signaling crosstalk links SNAIL to the 37/67 kDa laminin-1 receptor ribosomal protein SA and regulates the acquisition of a cancer stem cell molecular signature in U87 glioblastoma neurospheres. Cancers (Basel). 2022;14(23):5944
39. Djediai S, Gonzalez Suarez N, El Cheikh-Hussein L, Rodriguez Torres S, Gresseau L, Dhayne S. et al. MT1-MMP cooperates with TGF-β receptor-mediated signaling to trigger SNAIL and induce epithelial-to-mesenchymal-like transition in U87 glioblastoma cells. Int J Mol Sci. 2021;22(23):13006
40. Naujokat C, McKee DL. The "big five" phytochemicals targeting cancer stem cells: Curcumin, EGCG, sulforaphane, resveratrol and genistein. Curr Med Chem. 2021;28(22):4321-4342
41. Arima Y, Nobusue H, Saya H. Targeting of cancer stem cells by differentiation therapy. Cancer Sci. 2020;111(8):2689-2695
Author contact
![]() Corresponding authors: Laboratoire d'Oncologie Moléculaire, Université du Québec à Montréal, C.P. 8888, Succ. Centre-ville, Montréal, Québec, CANADA, H3C 3P8, Phone: (514) 987-3000 ext. 7610, E-mail: annabi.borhaneca.
Corresponding authors: Laboratoire d'Oncologie Moléculaire, Université du Québec à Montréal, C.P. 8888, Succ. Centre-ville, Montréal, Québec, CANADA, H3C 3P8, Phone: (514) 987-3000 ext. 7610, E-mail: annabi.borhaneca.

 Global reach, higher impact
Global reach, higher impact