Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(8):2663-2672. doi:10.7150/jca.111026 This issue Cite
Research Paper
Pretreatment Multi-sequence Contrast-Enhanced MRI to Predict Response to Immunotherapy in Unresectable Hepatocellular Carcinoma Using Transformer: A Multicenter Study
1. Department of Oncology, Fuzhou General Teaching Hospital of Fujian University of Traditional Chinese Medicine, 350001, Fuzhou, Fujian, PR China.
2. Department of Oncology, Mengchao Hepatobiliary Hospital of Fujian Medical University, 350028, Fuzhou, Fujian, PR China.
3. Department of Hepatology, Mengchao Hepatobiliary Hospital of Fujian Medical University, 350028, Fuzhou, Fujian, PR China.
4. Department of Oncology, the 900th Hospital of Joint Logistic Support Force, PLA, Fuzong Clinical College of Fujian Medical University, 350001, Fuzhou, Fujian, PR China.
5. Department of Oncology, Oriental Hospital Affiliated to Xiamen University, 350028, Fuzhou, Fujian, PR China.
#These authors contributed equally.
Received 2025-1-24; Accepted 2025-5-10; Published 2025-6-12
Abstract
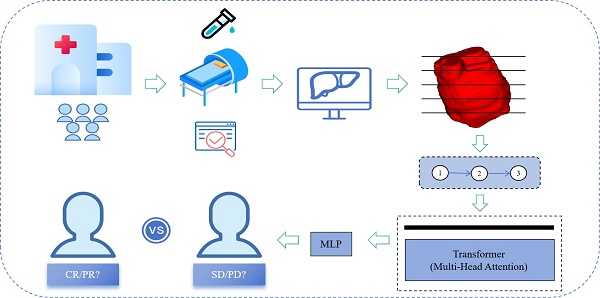
Background: Targeted combined immunotherapy (TCI) has shown certain antitumor effects in patients with unresectable hepatocellular carcinoma(uHCC), but only a subset of patients benefit. This study aims to develop a Transformer-based radiomics model to predict the objective response to combined therapy in patients with uHCC.
Methods: This multicenter, retrospective study involved 264 HCC patients who underwent contrast-enhanced MRI prior to immunotherapy. The patients were divided into a training cohort(n=180) and a validation cohort(n=84). Using a multi-instance learning approach, tumor lesions in multi-sequence MRI were segmented into cross-sectional images, and features were extracted using the ResNet50 model. The Transformer model was then trained to predict the objective response rate (ORR). The prediction process was visualized using Grad-CAM and SHAP algorithms. Model performance was assessed using ROC and DCA curves, while survival analysis was conducted using Kaplan-Meier curves.
Results: Among 264 patients, one achieved complete response (0.4%), 64 experienced partial response (24.2%). The ORR was 26.1% in the training group and 21.4% in the validation group. The model demonstrated high predictive accuracy, achieving a perfect area under the curve (AUC) of 1.000. Further validation using screenshot-based model inputs revealed an AUC of 0.929 (95% CI: 0.904, 0.947), confirming the model's clinical applicability. Kaplan-Meier analysis indicated that objective responders experienced better overall survival (OS) in both the training set (HR: 0.50, 95% CI: 0.27, 0.90) and the validation set (HR: 0.28, 95% CI: 0.08, 0.91).
Conclusion: The deep learning framework combining ResNet50 and Transformer has proven its clinical applicability in predicting and assessing the efficacy of targeted combination immunotherapy in unresectable hepatocellular carcinoma, providing crucial guidance for clinical treatment decisions.
Keywords: hepatocellular carcinoma, immunotherapy, MRI, predictive model, deep learning
Introduction
Hepatocellular carcinoma (HCC) presents a global health challenge, with projected increases in new cases and mortality rates exceeding 55% by 2040 [1-3]. In the realm of unresectable HCC (uHCC), recent years have seen significant advancements in treatment approaches, marked by the use of tyrosine kinase inhibitors (TKIs) [4] and immune checkpoint inhibitors (ICIs) [5]. Notably, Nivolumab [6] and Pembrolizumab [7] as second-line treatment options have demonstrated substantial survival benefits, with overall response rates of 20% and 17%, respectively. In 2020, following the results of the IMbrave150 trial [8], the Food and Drug Administration (FDA) approved a combination of Atezolizumab and Bevacizumab as a new standard for first-line treatment [9], improving objective response rates to 27%. However, 70% of patients still fail to respond to these therapies (targeted combined immunotherapy, TCIs), resulting in significant losses in medical resources and financial cost. Consequently, there is an urgent need for new biomarkers to predict the effectiveness of TCIs, thus enabling personalized treatment plans for patients.
Current research indicates that artificial intelligence technologies, exemplified by radiomics [10], hold significant potential in the diagnosis [11], classification [12], and prognostic marker identification [13,14] of solid tumors. Wang et al. [15] developed an MRI-based radiomics model to predict the 5-year survival rate of HCC patients. Zhang et al. [16] combined deep learning features with clinical characteristics to develop a nomogram predicting individual prognosis for HCC patients undergoing TACE and Sorafenib treatments. Recent studies by Bo et al. [17] have also explored the feasibility of radiomics models in predicting responses to Lenvatinib treatment in patients with unresectable liver cancer. Despite these advancements, current research is often limited to single MRI sequence maximum regions of interest (ROI) features and does not fully utilize multidimensional MRI data [18] to capture comprehensive lesion information.
The Transformer model, one of the most popular deep learning architectures, effectively integrates multidimensional medical image data through its self-attention mechanism, offering breakthroughs in tumor analysis. In various medical imaging tasks such as MRI tumor segmentation [19] and pathological image cancer classification [20], Transformers have demonstrated exceptional capabilities. However, the potential of utilizing Transformer models to predict the response of uHCC patients to TCI treatments using multidimensional MRI data remains underexplored.
This study aims to propose a multidimensional MRI-based radiomics model to predict the response of uHCC patients to TCI treatment, thereby supporting clinicians in selecting the most appropriate treatment options.
Methods
Study Design and Patients
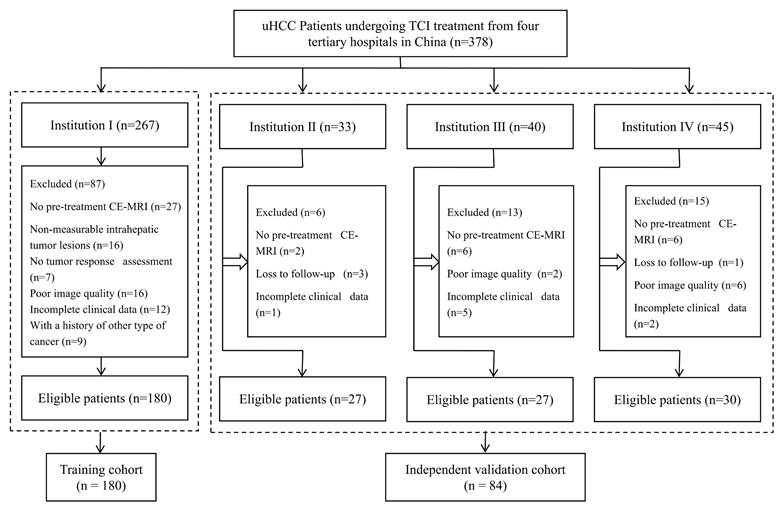
This retrospective multicenter study included 378 patients who were clinically or pathologically diagnosed with HCC between September 2017 and January 2023. These patients were recruited from four tertiary hospitals in China: Mengchao Hepatobiliary Hospital (Institution I), Xiangya Hospital (Institution II), Fujian Provincial Hospital (Institution III), and the 900th Hospital of PLA (Institution IV). All patients underwent Gd-EOB-DTPA-enhanced MRI examinations within two weeks prior to receiving combination therapy. The study was approved by the Ethics Committee of Mengchao Hepatobiliary Hospital (approval number: 2023_143_01), and the requirement for informed consent was waived.
Clinical Data Collection and Follow-up
Comprehensive clinical data were obtained from electronic medical records, and contrast-enhanced magnetic resonance images (CE-MRI) in DICOM format were retrieved from the PACS system. In line with real-world clinical settings, patients were eligible to receive various anti-PD-1 antibody therapies. Tumor response was assessed bi-monthly (±2 weeks) following targeted combination immunotherapy (TCI) using CT or MRI. The responses were classified based on the RECIST v1.1 [21] criteria into complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD). CR and PR conditions were required to be sustained for a minimum duration of two weeks. The follow-up period for this study concluded on November 15, 2023.
CE-MRI Acquisition and Image Pre-processing
All patients underwent T2-weighted imaging (T2WI)-enhanced MRI scans using a Siemens Verio 3.0T superconducting MRI scanner equipped with an 8-channel body coil. The imaging protocol included multiple phases: pre-contrast (PRE), arterial phase (AP), portal venous phase (PVP), and delayed post-contrast phase (DP). Prior to feature extraction, the images were processed with grayscale discretization and resampling to standardize data across different institutions and scanners. The resampling parameters were set at 1 mm×1 mm×1 mm, and Hounsfield units were discretized into 25 bins to address potential variations in imaging acquisition.
Regions of Interest Segmentation
Two experienced oncologists manually delineated the tumor boundaries layer-by-layer using ITK-SNAP software (version 3.8.0; http://www.itksnap.org/pmwiki/pmwiki.php) on axial T2-weighted images (T2WI) and sagittal 3D contrast-enhanced T1-weighted images (3D-CET1WI). The largest lesion in each layer was identified and saved as the ROI. In cases where there were disagreements regarding the ROI, a senior oncology expert provided the final decision. For patients with multiple tumors, the largest visible lesion on the MR images was selected as the ROI, given that multiple tumors generally indicate a higher tumor burden.
2.5D Segmentation Methods and Feature Extraction
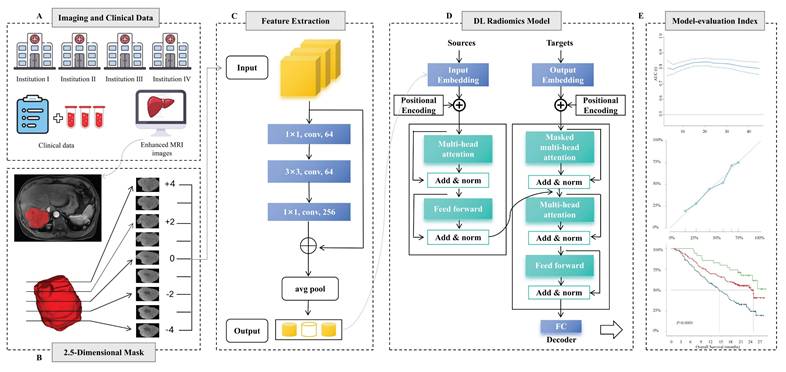
The framework for developing and validating the predictive model is illustrated in Fig. 2. To balance parameter efficiency and preserve spatial heterogeneity in 3D convolutional neural networks, an improved 2.5D feature extraction strategy was implemented. Specifically, for 3D MRI data, the tumor's largest cross-sectional layer is first located in the sagittal plane. A ROI is then extracted within a ±4 voxel layer range along the longitudinal axis, and the 2.5D image data volume is constructed by stacking along the channel dimension to generate multi-channel input data. Feature extraction is performed using a pre-trained ResNet50 network, where the convolutional layers capture local texture features, and the 1024×N-dimensional feature vector output from the global average pooling (GAP) layer serves as the high-level representation. Z-score normalization is applied using statistics from the entire dataset.
Model Construction, Validation, and Visualization
Multi-phase MRI sequences (PRE, AP, PVP, DP) were processed as independent instances, forming a 1024×N dataset (where N represents the number of instances). Feature training and efficacy prediction were conducted using an 8-head attention Transformer model[22]. The model was employed for parallel computation to capture inter-phase feature correlations and dynamically generate an attention weight matrix to adaptively select key imaging biomarkers. Initially, the data passed through a linear layer to reduce the dimensionality to 512×N, then passes through two layers of self-attention neural networks, followed by a multilayer perceptron (MLP) layer that outputs patient-level prediction labels and linear prediction values followed by two layers of self-attention neural networks. The training process employs the AdamW optimizer (learning rate = 1×10-5, weight decay = 1×10-5), with a maximum training period of 50 epochs. Early stopping is applied, halting training if the loss function does not change for five consecutive epochs. To enhance clinical interpretability, Grad-CAM is integrated to generate class activation heatmaps for localizing spatially sensitive regions. Additionally, SHAP value back-projection is used to map the contribution of higher-order features back to the original image space.
Statistical Analysis
Categorical variables were analyzed using the Chi-square test or Fisher's exact test, while continuous variables were compared using the Mann-Whitney U test and the Kruskal-Wallis test. A significance level of P <0.05 was applied. Model performance was evaluated through area under the curve (AUC) , with net benefits assessed using Decision Curve Analysis (DCA). Overall survival (OS) curves were plotted using the Kaplan-Meier method. All statistical analyses were conducted using R software (version 4.3.1) and Python (version 3.9).
Results
Patient Characteristics
A total of 264 eligible patients were included in the study, with 180 from Institution I comprising the training cohort, and 84 from other hospitals forming the independent validation cohort (Fig. 1). Of these, 206 (78%) received first-line therapy, 55 (20.8%) underwent second-line therapy, and 3 (1.2%) were treated with third-line therapy. In the training cohort, patients exhibited more adverse prognostic indicators (PS Score, Child-Pugh class, BCLC stage), a higher number of intrahepatic tumors, and more frequent portal vein invasion, all statistically significant (P < 0.05). No significant differences were observed between the training and validation cohorts in other baseline demographic and disease characteristics (Table 1). The ORR for the two cohorts were 26.1% (47/180) and 21.4% (18/84), respectively (P = 0.108).
In the univariate analysis of the entire cohort (Table S1), we examined the relationships between adverse tumor response and several clinical factors: Child-Pugh class B (OR 0.57; 95% CI: 0.26, 1.23), HBV infection (OR 0.72; 95% CI: 0.28, 1.85), portal hypertension (OR 0.77; 95% CI: 0.44, 1.34), and the presence of three or more tumors (OR 0.62; 95% CI: 0.33, 1.17). However, none of these relationships reached statistical significance (P > 0.05).
Flowchart of inclusion and exclusion criteria for eligible patients. uHCC=unresectable hepatocellular carcinoma; TCI=Targeted Combined Immunotherapy; CE-MRI=Contrast-enhanced MR images.
Baseline characteristics of patients in the training cohort and validation cohort
| Variables | Training cohort (N=180) | Validation cohort (N=84) | P value |
|---|---|---|---|
| Age(yr) | 55.5 ± 12.3 | 54.2 ± 12.0 | 0.411 |
| Sex | 0.728 | ||
| Female | 27 (15.0%) | 14 (16.7%) | |
| Male | 153 (85.0%) | 70 (83.3%) | |
| HBV | 0.209 | ||
| No | 13 (7.2%) | 10 (11.9%) | |
| Yes | 167 (92.8%) | 74 (88.1%) | |
| Child-Pugh class | 0.003 | ||
| A | 135 (75.0%) | 76 (90.5%) | |
| B | 45 (25.0%) | 8 (9.5%) | |
| Log AFP | 2.1 ± 1.3 | 2.4 ± 1.3 | 0.221 |
| Portal hypertension | 0.002 | ||
| No | 79 (43.9%) | 54 (64.3%) | |
| Yes | 101 (56.1%) | 30 (35.7%) | |
| PVTT | <0.001 | ||
| No | 76 (42.2%) | 57 (67.9%) | |
| Yes | 104 (57.8%) | 27 (32.1%) | |
| PS Score | <0.001 | ||
| 0 | 41 (22.8%) | 43 (51.2%) | |
| 1 | 104 (57.8%) | 40 (47.6%) | |
| 2 | 35 (19.4%) | 1 (1.2%) | |
| Largest tumor size(cm) | 8.1 ± 4.6 | 7.7 ± 4.6 | 0.513 |
| Tumor number | <0.001 | ||
| 1 | 56 (31.1%) | 33 (39.3%) | |
| 2 | 43 (23.9%) | 6 (7.1%) | |
| 3 | 32 (17.8%) | 5 (6.0%) | |
| >3 | 49 (27.2%) | 40 (47.6%) | |
| Extrahepatic metastasis | 0.779 | ||
| No | 119 (66.1%) | 57 (67.9%) | |
| Yes | 61 (33.9%) | 27 (32.1%) | |
| BCLC stage | 0.003 | ||
| A | 8 (4.4%) | 14 (16.7%) | |
| B | 32 (17.8%) | 15 (17.9%) | |
| C | 140 (77.8%) | 55 (65.5%) | |
| Response | 0.108 | ||
| CR | 0 (0.0%) | 1 (1.2%) | |
| PR | 47 (26.1%) | 17 (20.2%) | |
| SD | 44 (24.4%) | 30 (35.7%) | |
| PD | 89 (49.4%) | 36 (42.9%) | |
| Treatment line | 0.205 | ||
| 1 | 145 (80.6%) | 61 (72.6%) | |
| 2 | 34 (18.9%) | 21 (25.0%) | |
| 3 | 1 (0.6%) | 2 (2.4%) |
Differences are compared using the chi-square test (or Fisher's exact test) for categorical measures and Kruskal-Wallis test for continuous measures. AFP=α-fetoprotein; PVTT=portal vein tumor thrombus; PS=Eastern Cooperative Oncology Group performance status; BCLC=Barcelona Clinic Liver Cancer.
Deep Learning Features Predicted the Objective Tumor Response
Based on expert evaluations using RECIST v1.1 criteria, patients were stratified into two groups: those with objective responses (CR/PR group) and those without (SD/PD group). Fig. S1. A, B illustrate the distribution of deep learning scores for patients in both the training and validation sets. Notably, patients in the CR and PR groups received high scores, whereas those in the SD and PD groups were assigned low scores. The analysis revealed significant differences between these groups. Importantly, the model demonstrated high concordance between its predictions of TCI responses in uHCC patients and the actual tumor response outcomes.
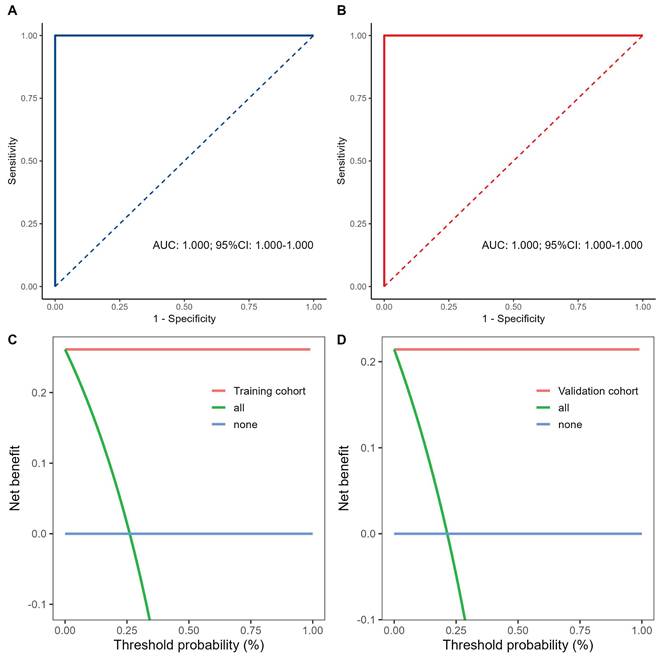
Fig. 3. A and B illustrate the ROC curves for the TCI model in the training and validation cohorts, with both achieving AUC values of 1.000. Fig. 3. C and D show the DCA results, indicating that the proposed model yields superior net benefits within a reasonable range of threshold probabilities.
We performed random occlusions of series and layers in MRI scans to evaluate model performance sensitivity (Table 2). The occlusions of the PRE and AP series resulted in AUC decreases of 3.8% and 3.1%, respectively; however, these decreases were not statistically significant compared to the optimal AUC of 1.000 in the complete dataset. Moreover, occlusions in other layers minimally affected the AUC, demonstrating the model's robustness to partial data occlusion. This study confirms that the multi-instance learning and Transformer strategy effectively maintain high stability and performance, even with incomplete medical imaging data.
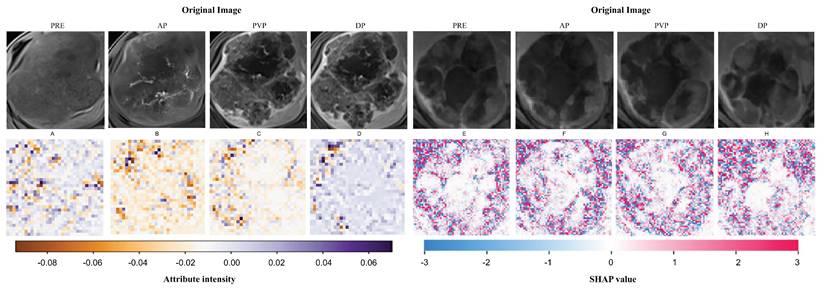
To demonstrate the interpretability of the deep learning model, heatmaps were generated for two patients selected from the validation set using the Grad-CAM and SHAP algorithms. These heatmaps (Fig. 4) highlight the image regions that contribute most significantly to the network's decision-making process. Notably, hotspot regions are concentrated around the tumor, whereas areas of necrosis or liquefaction contribute less to the efficacy predictions. This aligns with the common understanding that regions of high malignancy are closely associated with prognosis.
The change of AUC after random occlusion series or layers of MRI
| Cohort | Model | AUC | 95%CI | change |
|---|---|---|---|---|
| Series | Baseline | 1.000 | (1.000, 1.000) | 0.0% |
| -PRE | 0.962 | (0.922, 0.992) | -3.8% | |
| -AP | 0.969 | (0.939, 0.993) | -3.1% | |
| -PVP | 0.990 | (0.971, 1.000) | -1.0% | |
| -DP | 1.000 | (1.000, 1.000) | 0.0% | |
| Layers | -0 | 0.992 | (0.975, 1.000) | -0.8% |
| ±2 | 1.000 | (1.000, 1.000) | 0.0% | |
| ±4 | 0.992 | (0.975, 1.000) | -0.8% |
Radiomic Features Associated with OS and PFS
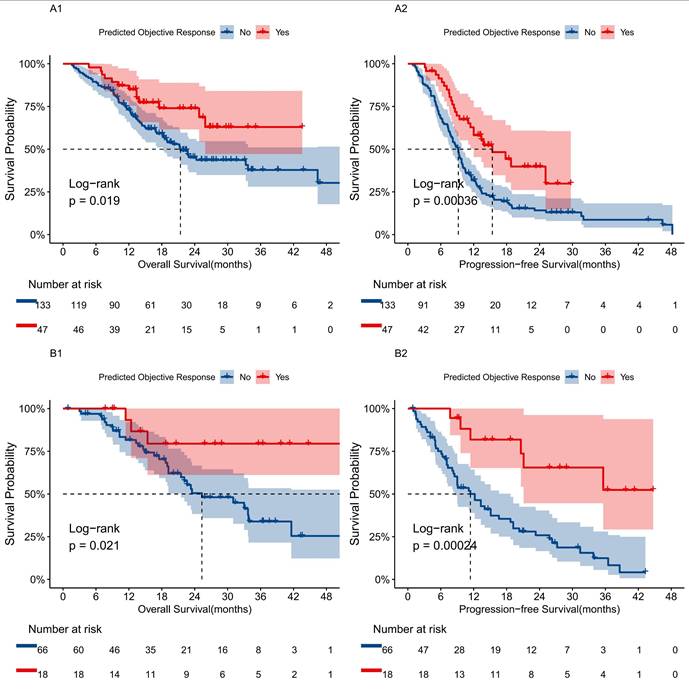
We evaluated survival outcomes based on the presence of objective responses in patients, finding that those predicted to respond favorably demonstrated significantly better prognoses than non-responders. Kaplan-Meier analysis revealed that the mortality and disease progression rates for the predicted responder group were 27.7% and 53.2%, respectively, compared to 50.4% and 85.7% for the non-responder group (Fig. 5). The responder group showed significant survival advantages in OS (HR 0.50; 95% CI: 0.27, 0.90, P=0.0019) and PFS (HR 0.46; 95% CI: 0.30, 0.72, P=0.00036). Similar results were observed in the validation set.
Workflow of developing deep learning models. A Clinical data and image acquisition. B 2.5D ROI segmentation, the largest tumor area was segmented into regions of interest (ROIs) for contrast-enhanced MRI, extracting the largest ROI layer along with its longitudinal layers from -4 to +4. C Deep learning feature extraction, utilize RESNET50 to extract features from 2.5D slices, selecting the deep learning features with the highest relevance through five-fold cross-validation. D Construct a predictive model based on the Transformer architecture. E Model performance verification, validate the efficacy of the model using Receiver Operating Characteristic (ROC) curves, Decision Curve Analysis (DCA) curves, and Kaplan-Meier curves.
Performance and comparison of prediction models in the training and validation cohorts. A, B Analysis and comparison of the receiver operating characteristic curves for the predictive model in the training and validation sets. C, D Decision curve analysis of the predictive model in the training and validation cohorts.
Visualization of feature mappings derived through ResNet50. A-D Visualization analysis of Patient A's full-sequence CE-MRI was conducted using the Grad-CAM algorithm. E-H Visualization analysis of Patient B's full-sequence CE-MRI was conducted using the SHAP algorithm. The full sequence is ordered as PRE, AP, PVP and DP.
Survival prognosis analysis using a transformer model for uHCC patients receiving TCI treatment. A1, B1 Kaplan-Meier curves for OS between predicted tumor response and non-response groups in the training and validation cohorts based on the transformer model. A2, B2 Kaplan-Meier curves for PFS between predicted tumor response and non-response groups in the training and validation cohorts based on the transformer model.
Assessment of the Model's Clinical Applicability
To assess the clinical applicability of the model, JPEG images of tumor target lesions captured via screenshots were used as input. Fig. S2. A and B compare the model's performance using screenshot images versus original image inputs, with AUC values of 0.929 (95% CI: 0.904, 0.947) and 1.000, respectively. The confusion matrix in Supplementary Fig. S2. C indicates that the model based on screenshot images is reasonably accurate in predicting tumor response, though a few misclassifications were observed.
Discussion
In this study, we developed a Transformer model based on CE-MRI to predict long-term survival and treatment responses in uHCC patients undergoing TCI. Validated with a multicenter dataset, our model demonstrated a strong correlation between deep learning features and both OS and PFS following treatment. We visualized the prediction process using Grad-CAM and SHAP algorithms, and assessed the impact of using raw MR images versus screenshot images as inputs on model performance. To the best of our knowledge, this may be the first successful application of a multi-instance learning and Transformer strategy in the field of HCC MRI radiomics.
Although TCI protocols are effective in some uHCC patients, less than 30% experience benefits [6-8]. Previous studies have identified various biomarkers related to immunotherapy prognosis, including tumor characteristics (PD-1/PD-L1 expression [23], TMB [24]), tumor microenvironment (CD3/CD8 tumor-infiltrating lymphocytes [25], dMMR/MSI [26]), peripheral blood markers (NLR/PLR [27], ctDNA [28] ALFP score [29]), and gut microbiota [30]. Despite this, the high heterogeneity of HCC and complex immune response mechanisms [31-33] challenge the practicality and efficacy of these biomarkers.
Radiomics-based approaches have shown promising potential, including models proposed by Xu et al. [34], Wei et al. [35], and Shen et al. [36]. Notably, the first two studies reported AUC values of 0.820 and 0.882, respectively, for predicting treatment response. A recent multi-parametric MRI fusion model proposed by Kang et al. [37] achieved an AUC of 0.869 for similar tasks. Additionally, a recent study [38] using a machine learning classifier to predict the effects of LPI treatment, achieved an AUC of 0.893, further validating the applicability of radiomics in predicting immunotherapy outcomes. In comparison with these studies, the significant increase in AUC observed in this study can be attributed to the use of more advanced strategies. The combination of MIL and Transformer models enables dynamic establishment of image feature correlations across anatomical regions, thereby more effectively integrating complementary information from multi-sequence MRI. In a multicenter study [39] of over 13,000 individuals assessing microsatellite instability in colorectal cancer HE-WSI, researchers extracted essential features from each patch and synthesized them into WSI labels using a five-fold cross-validation Transformer model. The findings indicated that both the negative and positive predictive values of the model achieved 0.99, reaching clinical-grade accuracy. This study confirms the efficacy of this advanced methodological integration in pathological research.
Our dataset included patients undergoing various lines of combination immunotherapy, enhancing the clinical generalizability of findings. Although traditional clinical-pathological features (such as AFP levels, extrahepatic metastasis, and maximum tumor diameter) showed no significant correlation with treatment response (P >0.05), the radiomics-based predictive model maintained excellent stability in validation across three independent medical centers, confirming its robustness against equipment variations. In this study, there was no statistically significant difference in predictive performance between screenshot JPG data and original DICOM images (AUC: 0.89 vs. 0.91, P =0.12), consistent with the findings of Sedlaczek et al. [40], indicating that screenshot-based analysis can reduce data coordination costs and streamline clinical translation pathways. Through visual analysis, we explored potential connections between deep learning features, tumor heterogeneity, and the immune microenvironment. However, these associations need further validation through additional genomic and histopathological studies.
This study has several limitations. First, as a multicenter study, differences in MRI scanner performance and imaging techniques, despite efforts to standardize images, may have influenced feature extraction. Second, subjectivity involved in manually delineating ROIs and the potential loss of crucial information related to the tumor microenvironment represent additional limitations. In future studies, we plan to expand the cohort to five centers and apply an automatic segmentation algorithm based on 3D U-Net [41] to systematically assess quantitative feature variations in the tumor/liver parenchyma. Third, the retrospective nature of the study and the relatively limited sample size also introduce potential selection bias. Future work should focus on larger, prospective studies to optimize and refine the model.
This study preliminarily validates the efficacy of combining multi-instance learning with the Transformer method in radiomics, demonstrating that the model, which utilizes screenshot data, achieves high accuracy in predicting treatment outcomes. Currently, the use of radiomics technology for predicting HCC immunotherapy outcomes is confined to small-scale exploratory studies. Future research will necessitate larger sample sizes and more extensive data from multiple centers to further confirm the reliability of these findings. Specifically, robust validation of the stability of MRI screenshot data could significantly facilitate clinical applications.
Supplementary Material
Supplementary figures and table.
Acknowledgements
We are grateful for the support of professional knowledge from Fujian HCC-biomarker Study Group. We also thank the patients and their families.
Funding
This work was supported by the Central Guidance on Local Science and Technology Development Fund of Fujian Province (2023L3028), the Natural Science Foundation of Fujian Province (2021J01281), the Research Initiation Foundation of Mengchao Hepatobiliary Hospital of Fujian Medical University (QDZJ-2022-001 and 2025-QDZJ-001), the Health and Wellness Youth Scientific Research Project of Fujian Province (2023QNA081), the Science and Technology Innovation Joint Foundation of Fujian Province (2024Y9421), the Sister City Scientific and Technological Innovation Cooperation Special Fund of Fuzhou (2024-Y-023).
Statement of Ethics
The study protocol was conducted in accordance with the principles of the World Medical Association Declaration of Helsinki and was approved by the Mengchao Hepatobiliary Hospital Research Ethics Committee (Approval Numbers: 2023_143_01). The requirement for informed consent was waived (retrospective computational analysis of images) by the Mengchao Hepatobiliary Hospital Research Ethics Committee. The requirement for informed consent was waived.
Author Contributions
Study concept and design: CJL, LLB; data analysis and interpretation: CJL, LLB, GXY; resources: CJL, CJ, YYM, LLF, GSM; funding acquisition: CX; writing-original draft: CJL, LLB; writing-review & editing: all authors; final approval of manuscript: all authors.
Data Availability Statement
Raw data of MRI used in this study are unable to be shared as the requirement of Hospital Research Ethics Committees. Derived data (e.g., extracted radiomic features) are available upon reasonable request from the corresponding authors.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Laversanne M, Sung H. et al. Global cancer statistics 2022: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229-263
2. Rumgay H, Arnold M, Ferlay J. et al. Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol. 2022;77(6):1598-1606
3. Verslype C, Rosmorduc O, Rougier P, ESMO Guidelines Working Group. Hepatocellular carcinoma: ESMO-ESDO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2012;23(Suppl 7):vii41-vii48
4. Villanueva A, Llovet JM. Targeted therapies for hepatocellular carcinoma. Gastroenterology. 2011;140(5):1410-1426
5. Wang Y, Jiang M, Zhu J. et al. The safety and efficacy of lenvatinib combined with immune checkpoint inhibitors therapy for advanced hepatocellular carcinoma. Biomed Pharmacother. 2020;132:110797
6. Yau T, Kang Y, Kim T. et al. Efficacy and safety of nivolumab plus ipilimumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib. JAMA Oncol. 2020;6(11):e204564
7. Zhu AX, Finn RS, Edeline J. et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 2018;19(7):940-952
8. Galle PR, Finn RS, Qin S. et al. Patient-reported outcomes with atezolizumab plus bevacizumab versus sorafenib in patients with unresectable hepatocellular carcinoma (IMbrave150): an open-label, randomised, phase 3 trial. Lancet Oncol. 2021;22(7):991-1001
9. Casak SJ, Donoghue M, Fashoyin-Aje L. et al. FDA approval summary: atezolizumab plus bevacizumab for the treatment of patients with advanced unresectable or metastatic hepatocellular carcinoma. Clin Cancer Res. 2021;27(7):1836-1841
10. Lambin P, Rios-Velazquez E, Leijenaar R. et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441-446
11. Liu X, Khalvati F, Namdar K. et al. Can machine learning radiomics provide pre-operative differentiation of combined hepatocellular cholangiocarcinoma from hepatocellular carcinoma and cholangiocarcinoma to inform optimal treatment planning? Eur Radiol. 2021;31(1):244-255
12. de Jesus FM, Yin Y, Mantzorou-Kyriaki E. et al. Machine learning in the differentiation of follicular lymphoma from diffuse large b-cell lymphoma with radiomic [18F]FDG PET/CT features. Eur J Nucl Med Mol Imaging. 2022;49(5):1535-1543
13. Dissaux G, Visvikis D, Da-Ano R. et al. Pretreatment 18F-FDG PET/CT radiomics predict local recurrence in patients treated with stereotactic body radiotherapy for early-stage non-small cell lung cancer: a multicentric study. J Nucl Med. 2020;61(6):814-820
14. Yusufaly TI, Zou J, Nelson TJ. et al. Improved prognosis of treatment failure in cervical cancer with nontumor PET/CT radiomics. J Nucl Med. 2022;63(7):1087-1093
15. Wang X, Long L, Cui Y. et al. MRI-based radiomics model for preoperative prediction of 5-year survival in patients with hepatocellular carcinoma. Br J Cancer. 2020;122(7):978-985
16. Zhang L, Xia W, Yan Z. et al. Deep learning predicts overall survival of patients with unresectable hepatocellular carcinoma treated by transarterial chemoembolization plus sorafenib. Front Oncol. 2020;10:593292
17. Bo Z, Chen B, Zhao Z. et al. Prediction of response to lenvatinib monotherapy for unresectable hepatocellular carcinoma by machine learning radiomics: a multicenter cohort study. Clin Cancer Res. 2023;29(9):1730-1740
18. Zech CJ, Ba-Ssalamah A, Berg T. et al. Consensus report from the 8th international forum for liver magnetic resonance imaging. Eur Radiol. 2020;30(1):370-382
19. Conze P, Andrade-Miranda G, Le Meur Y. et al. Dual-task kidney mr segmentation with transformers in autosomal-dominant polycystic kidney disease. Comput Med Imaging Graph. 2024;113:102349
20. Cai H, Feng X, Yin R. et al. MIST: multiple instance learning network based on swin transformer for whole slide image classification of colorectal adenomas. J Pathol. 2023;259(2):125-135
21. Eisenhauer EA, Therasse P, Bogaerts J. et al. New response evaluation criteria in solid tumours: revised recist guideline (version 1.1). Eur J Cancer. 2009;45(2):228-247
22. Aguilera AC, Olmos PM, Artés-Rodríguez A. et al. Regularizing transformers with deep probabilistic layers. Neural Netw. 2023;161:565-574
23. Wang W, Li R, Li H. et al. Addition of immune checkpoint inhibitor showed better efficacy for infiltrative hepatocellular carcinoma receiving hepatic arterial infusion chemotherapy and lenvatinib: a multicenter retrospective study. Immunotargets Ther. 2024;13:399-412
24. Xu Q, Xu H, Deng R. et al. Multi-omics analysis reveals prognostic value of tumor mutation burden in hepatocellular carcinoma. Cancer Cell Int. 2021;21(1):342
25. Liu C, Zhu S, Dong Y. et al. The potential predictive biomarkers for advanced hepatocellular carcinoma treated with anti-angiogenic drugs in combination with pd-1 antibody. Front Immunol. 2022;13:930096
26. Qian B, Lin H, Lan T. et al. Risk predictive model based on three immune-related gene pairs to assess prognosis and therapeutic sensitivity for hepatocellular carcinoma. World J Surg Oncol. 2022;20(1):252
27. Yu JI, Park HC, Yoo GS. et al. Clinical significance of systemic inflammation markers in newly diagnosed, previously untreated hepatocellular carcinoma. Cancers (Basel). 2020;12(5):1300
28. Wang D, Xu Y, Goldstein JB. et al. Preoperative evaluation of microvascular invasion with circulating tumour DNA in operable hepatocellular carcinoma. Liver Int. 2020;40(8):1997-2007
29. Gong B, Wang X, Guo W. et al. Prognosis of patients with hepatocellular carcinoma treated with transarterial chemoembolization(MC-hccAI 001): development and validation of the ALFP score. J Hepatocell Carcinoma. 2023;10:1341-1351
30. Ma J, Li J, Jin C. et al. Association of gut microbiome and primary liver cancer: a two-sample mendelian randomization and case-control study. Liver Int. 2023;43(1):221-233
31. Yu SJ, Ma C, Heinrich B. et al. Targeting the crosstalk between cytokine-induced killer cells and myeloid-derived suppressor cells in hepatocellular carcinoma. J Hepatol. 2019;70(3):449-457
32. Yang C, Zhang S, Cheng Z. et al. Multi-region sequencing with spatial information enables accurate heterogeneity estimation and risk stratification in liver cancer. Genome Med. 2022;14(1):142
33. Ma L, Heinrich S, Wang L. et al. Multiregional single-cell dissection of tumor and immune cells reveals stable lock-and-key features in liver cancer. Nat Commun. 2022;13(1):7533
34. Xu B, Dong S, Bai X. et al. Tumor radiomic features on pretreatment mri to predict response to lenvatinib plus an Anti-PD-1 antibody in advanced hepatocellular carcinoma: a multicenter study. Liver Cancer. 2022;12(3):262-276
35. Wei Y, Yang M, Xu L. et al. Novel computed-tomography-based transformer models for the noninvasive prediction of pd-1 in pre-operative settings. Cancers (Basel). 2023;15(3):658
36. Shen L, Jiang Y, Lu L. et al. Dynamic prognostication and treatment planning for hepatocellular carcinoma: A machine learning-enhanced survival study using multi-centric data. The Innovation Medicine. 2025;3:100125
37. Kang W, Tang P, Luo Y. et al. Multiparametric MRI-based Machine Learning Radiomics for Predicting Treatment Response to Transarterial Chemoembolization Combined with Targeted and Immunotherapy in Unresectable Hepatocellular Carcinoma: A Multicenter Study. Acad Radiol. 2024;32:S1076-6332 (24)00825-0
38. Hua Y, Sun Z, Xiao Y. et al. Pretreatment CT-based machine learning radiomics model predicts response in unresectable hepatocellular carcinoma treated with lenvatinib plus PD-1 inhibitors and interventional therapy. J Immunother Cancer. 2024;12(9):e010330
39. Wagner SJ, Reisenbüchler D, West NP. et al. Transformer-based biomarker prediction from colorectal cancer histology: a large-scale multicentric study. Cancer Cell. 2023;41(9):1650-1661
40. Sedlaczek OL, Kleesiek J, Gallagher FA. et al. Quantification and reduction of cross-vendor variation in multicenter DWI MR imaging: results of the Cancer Core Europe imaging task force. Eur Radiol. 2022;32(12):8617-8628
41. Isensee F, Jaeger PF, Kohl SAA. et al. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18(2):203-211
Author contact
![]() Corresponding author: Phone: +1-362-508-2108; Fax: 0086-591-88113085; Email address: cxiongzpcedu.cn.
Corresponding author: Phone: +1-362-508-2108; Fax: 0086-591-88113085; Email address: cxiongzpcedu.cn.

 Global reach, higher impact
Global reach, higher impact