3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(7):1940-1953. doi:10.7150/jca.93376 This issue Cite
Research Paper
Mendelian Randomization Validates the Immune Landscape Mediated by Aggrephagy in Esophageal Squamous Cell Carcinoma Patients from the Perspectives of Multi-omics
1. Traditional Chinese Medicine (Zhong Jing) school, Henan University of Chinese Medicine, Zhengzhou 450046, China.
2. Henan Key Laboratory of TCM Syndrome and Prescription Signaling, Henan International Joint Laboratory of TCM Syndrome and Prescription Signaling, Academy of Chinese Medical Sciences, Henan University of Chinese Medicine, Zhengzhou 450046, China.
3. Department of Orthopedic, The Third Hospital of Peking University, Beijing 100029, China.
Abstract
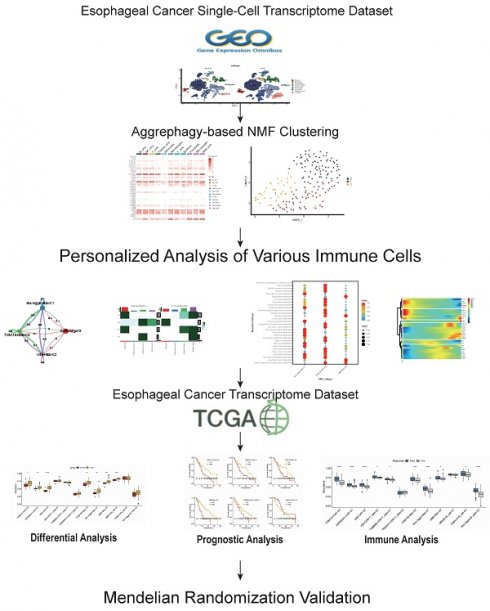
Objective: To delineate the immune landscape of ESCC patients mediated by aggrephagy through bioinformatics and identify prognostic cell cluster genes with causal attributes to esophageal cancer through Mendelian randomization.
Methods: Quality control, dimension reduction, and annotation were performed on the ESCC single-cell dataset. NMF clustering of various cell subgroups was carried out based on the expression of AGG-related genes, and AGG-related genes in each cluster were identified. Pseudo-temporal analysis was used to observe changes in the expression of AGG-related genes in each cluster. Cell communication analysis was employed to observe interactions between cell subgroups. Changes in classification, metabolism, or KEGG pathways in related subgroups were observed based on different cell characteristics. The AGG cluster attributes of TCGA and GEO samples were assessed based on GSVA, and the prognosis of each cluster was observed. The immune treatment situation and the relationship between mutation level and prognosis of AGG cluster-related samples were observed through the TIDE database and microsatellite instability. Finally, the eQTL of genes in each prognostic AGG cluster was used as an instrumental variable, with esophageal cancer as the outcome factor. Through Mendelian randomization analysis, AGG cluster-related genes with a causal relationship to esophageal cancer were established.
Results: Dimension reduction clustering of single-cell transcriptome data identified 19 different cell subgroups. After re-annotation of the 19 cell subgroups, it was found that the CAF cells, B cells, T cells, NK cells, etc., of ESCA patients were all elevated compared to the control group. CAF cells had a high degree of communication with most cells. There were significant differences in macrophage metabolism and B-cell-mediated signal transduction pathways in different AGG clusters. The TUBA1B+Mac-C0 cluster, along with other clusters, exhibits predictive prognostic and immunotherapeutic potential at the transcriptional level. Mendelian randomization analysis revealed a causal relationship between genes such as CTSZ, CTSC, DAD, COLEC12, ATOX1, within the AGG cluster, and the onset of esophageal cancer.
Conclusion: Aggrephagy mediates and influences the alterations and interactions of various immune cells in patients with ESCC. We elucidate the roles of AGG-related clusters, such as TUBA1B+Mac-C0, VIM+CD8+T_cells-C0, UBB+Mac-C2, in mediating prognosis and immune therapy in ESCC patients. Genes causally associated with the occurrence of esophageal cancer are identified within the AGG cluster, including CTSZ, CTSC, DAD, COLEC12, ATOX1, etc., offering new evidence for clinical immune therapy. These findings underscore the significance of these gene clusters in influencing both prognosis and immune responses in the context of esophageal cancer, shedding light on potential therapeutic targets and prognostic markers.
Keywords: Mendelian Randomization, Single-cell, Multi-omics, Esophageal Cancer, Causal Relationships

 Global reach, higher impact
Global reach, higher impact