Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(2):275-280. doi:10.7150/jca.79171 This issue Cite
Review
The Functional Role of LncRNA UCA1 in Pancreatic Cancer: a mini-review
1. Basic Medical Experimental Teaching Center, Zhejiang University, Hangzhou 310030, Zhejiang, China.
2. Key Laboratory of Gastroenterology of Zhejiang Province, Zhejiang Provincial People's Hospital, Affiliated People's Hospital, Hangzhou Medical College, Hangzhou 310014, Zhejiang, China.
3. Cancer Center, Department of Gastrointestinal and Pancreatic Surgery, Zhejiang Provincial People's Hospital, Affiliated People's Hospital, Hangzhou Medical College, Hangzhou 310014, Zhejiang, China.
4. Center for Plastic and Reconstructive Surgery, Department of Hand and Reconstruction Surgery, Zhejiang Provincial People's Hospital, Affiliated People's Hospital, Hangzhou Medical College, Hangzhou 310014, Zhejiang, China.
#These authors contributed equally to this work.
Received 2022-9-22; Accepted 2022-12-25; Published 2023-1-9
Abstract
Pancreatic cancer (PaC) is a common malignant tumor of the digestive tract, with a 5-year survival rate of less than 5% and high mortality rate in the world. LncRNAs have been showed to possess multiple biological functions in growth, differentiation, and proliferation, which play an important role in different biological processes and diseases, especially in the development of tumors. LncRNA UCA1, which is firstly identified in human bladder cancer, has been showed to be a tumor promoter in pancreatic cancer. Recent researches have showed that UCA1 might promote pancreatic carcinogenesis and progression, and correlate with drug resistance. In this review, we address the biological function and regulatory mechanism of UCA1 in pancreatic cancer, which might give a new approach for clinical diagnosis and treatment.
Keywords: Pancreatic cancer, LncRNA UCA1, Carcinogenesis, Tumor progression, Drug resistance
Introduction
Pancreatic cancer (PaC), particularly pancreatic ductal adenocarcinoma (PDAC) which account for the vast majority of pancreatic cancer, is a gastrointestinal malignancy with insidious onset, rapid progression, poor treatment effect and poor prognosis [1]. Its morbidity and mortality have been rising over the world in recent years. According to data from the American Cancer Society, the number of new cases of PaC in the United States in 2022 is expected to be 62,210, with 49,830 deaths [2]. The latest data released by National Cancer Center of China showed that the incidence of PaC has risen to ninth place, while the mortality rate has increased to sixth [3]. In 2020, the overall 5-year survival rate approached 10% for the first time, up from 5.26% in 2000 [4-6]. The survival of PaC patients has not improved significantly in the past 40 years, and it is expected to become the second leading cause of cancer related death by 2030 [7]. The etiology and pathogenesis of PaC are still not fully understood. There is a lack of efficient early detection approach and effective therapeutic options. In order to find new molecular biomarkers and therapeutic targets to improve the early diagnosis and the prognosis, the molecular mechanism of PaC pathogenesis has always been an urgent problem to be explored and studied in depth.
The long noncoding RNAs (lncRNAs) are non-protein-coding transcripts, longer than 200 nucleotides in length [8]. Many studies have found lncRNAs participated in many physiological processes by modulating gene expression at the epigenetic, transcriptional and posttranscriptional levels [8-10]. Increasing evidence indicated that several lncRNAs were linked to human disease, especially cancer, and its abnormal expression was closely related to tumor proliferation, differentiation, apoptosis and metastasis [11-16].
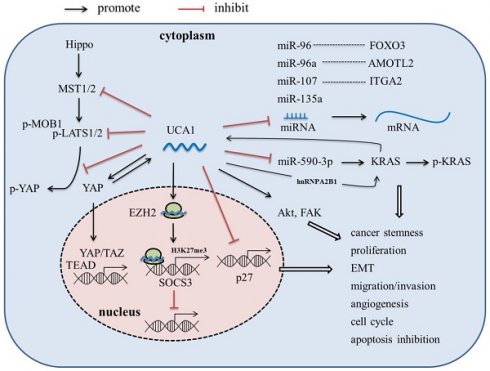
LncRNA-UCA1, firstly cloned and identified from bladder cancer cell line BLZ-211, is located on the short arm of chromosome 19, consisting of 3 exons and 2 introns with multiple stop codons without any conserved long open reading frames (ORFs) [17]. There are three transcriptional isoforms of UCA1, lncRNA UCA1 (1.4kb), lncRNA UCA1a (or denoted lncRNA CUDR, 2.2kb), and the 2.7 kb isoform (its biological function is unknown) [18, 19]. In recent years, lncRNA UCA1 has been reported to be the most abundant isoform in various malignant tumors, such as bladder cancer, breast cancer, hepatocellular carcinoma and pancreatic cancer, and play an important role in tumor invasion and metastasis, angiogenesis, immune escape and chemotherapeutic drug resistance [17, 20-30]. In this article, we review the abnormal expression, molecular mechanism (Figure 1), and clinical significance of UCA1 in pancreatic cancer, which might provide theoretical basis for the potential future clinical applications.
Expression and regulation of lncRNA UCA1 in pancreatic cancer
Several studies have showed that the expression of UCA1 is up-regulated in pancreatic tumor tissues and PaC cell lines [31-39]. In addition, UCA1 is also enriched in exosomes derived from PaC patients' serum or hypoxic PaC cell lines [24]. However, the regulatory mechanism behind the UCA1 up-regulation in PaC has not been fully elucidated yet.
Recently, Zhang et al. found that KRAS oncogene, a well-known major driver gene for PDAC, could promote UCA1 expression [31]. Besides, Yes-associated protein (YAP), the key downstream target of KRAS signaling and major downstream effector of Hippo signaling pathway, was also found able to up-regulate the expression of UCA1 in PaC [36]. Nevertheless, the specific mechanism still remains to be further studied.
The role of lncRNA-UCA1 in pancreatic carcinogenesis and progression
UCA1 contributes to pancreatic carcinogenesis
The occurrence and development of PaC is accompanied by a large number of gene mutations, and the high-frequency mutation genes KRAS, TP53, CDKN2A and SMAD4 are considered as the four major driver-genes of PDAC [40]. Among them, KRAS mutations are present in over 90% of pancreatic intraepithelial neoplasm (PanIN, a precursor lesion of PDAC) and PDAC tissues [40, 41]. KRAS is a member of the Ras GTPase family, and mutation in KRAS leads to a constitutively active, GTP-bound state, the active GTP-bound form of KRAS is necessary for the initiation, progression and metastasis of PC [42-46]. Therefore, KRAS mutation is considered to be the initiating event of pancreatic carcinogenesis.
Molecular mechanism of lncRNA UCA1 in PaC.
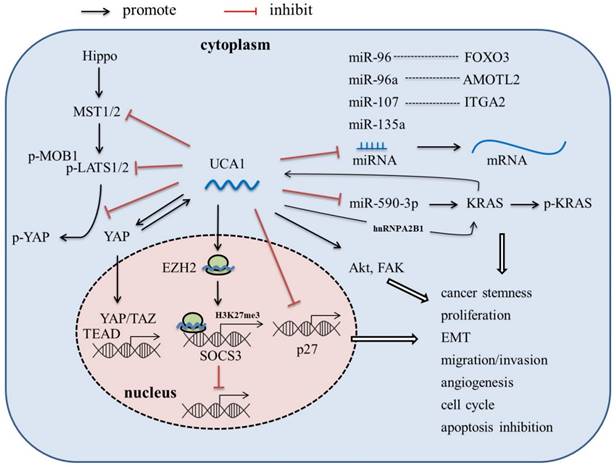
Liu et al. found that lncRNA-UCA1 could act as a competing endogenous RNA (ceRNA) to promote KRAS expression by via sponging miR-590-3p, and enhance phospho-KRAS activity by increasing the binding of hnRNPA2B1 to KRAS, while KRAS in turn increases UCA1 expression, thus enhances stemness and proliferation of PDAC cells [31]. Kras has long been considered undruggable due to the lack of pharmacologically targetable pockets, until recently a few inhibitors specific targeting to KRAS-G12C have been discovered. The role of UCA1 in regulating KRAS expression and activity suggested it could be a novel target for PDAC treatment through indirectly targeting KRAS.
Besides, UCA1 is also found to be up-regulated in plasma of malignant intraductal papillary mucinous neoplasm (IPMN) patients, when compared to benign cases [47]. Malignant IPMN has also been deemed as a precursor of PDAC, thus, it is biologically plausible that UCA1 could contribute to early pancreatic carcinogenesis.
UCA1 promotes the proliferation, invasion, migration and metastasis abilities of PaC cells
Several studies have revealed that UCA1 plays an important role in regulating PaC cells' proliferation, invasion and migration. According to Liang's report, overexpression of UCA1 could promote cell cycle progression via accelerating the transition of G0/G1 to S-G2/M, and suppress apoptosis, thus resulting in promoting of cell proliferation in PDAC [32]. On the contrary, knockdown of UCA1 expression could induce cell cycle arrest in G0/G1 phase and apoptosis [33, 34]. In addition, the study in vivo also revealed that UCA1 knockdown could significantly inhibit tumor growth in nude mice [31, 32, 37]. Furthermore, UCA1 also exerts a promotive effect on cell migration and invasion [32, 33, 35-39].
In mechanism, UCA1 mainly functions as an endogenous miRNA sponge to competitively bind to miRNAs, thereby abrogating the inhibition effect of miRNA on target genes. Zhou et al. found that high expression of lncRNA-UCA1 could down-regulate the miR-96 expression and up-regulate the FOXO3 expression, thus promoting cell proliferation, invasion, and migration in PaC cells [33]. Gong et al. showed that UCA1-miR-107-ITGA2 axis could enhance the migration and invasion ability of PaC cells via focal adhesion pathway [35]. In Liu's study, UCA1 was reported to up-regulate the expression of KRAS oncogene via sponging miR-590-3p [31]. Besides, UCA1 was shown to regulate the cell viability by sponging miR-135a [37].
In addition to the function as a miRNA sponge, UCA1 is also able to directly interact with protein, modulate the activity of the corresponding protein, and even alter cytoplasmic localization of the protein. Zhang et al. reported that UCA1 overexpression not only increased YAP expression, but also inhibited phosphorylation of YAP and promoted YAP nuclear translocation, via interaction with key proteins of Hippo pathway, including MOB1, Lats1 and YAP, resulting in improved TEAD activity [36]. YAP in turn increased UCA1 expression; however, the mechanism is still unknown [36].
LncRNA-UCA1 promotes the tumor angiogenesis in PaC
Tumor angiogenesis is an important link in tumor growth, invasion and metastasis, and plays an important role in the vast majority of solid tumors [48, 49]. Numerous studies have shown that PaC is a hypovascular and hypoxic solid tumor, and PaC tissue frequently exhibits aberrant proliferation of human vascular endothelial cells (HUVECs) [50, 51]. Moreover, it is reported that the microvessel density (MVD) in tumor tissue is positively correlated with the progression of PaC [52-55].
Exosomes are a type of small extracellular vesicle containing numerous biologically active molecules, including proteins, DNA, coding and non-coding RNA, lipids, and metabolites. Growing evidence has proved that exosomes play an important role in various aspects of cancer progress, including tumor angiogenesis [56-59]. Recently, Guo et al. found that the UCA1 was highly expressed in exosomes derived from hypoxic PC cells, and can be transferred to HUVECs via exosomes to promote migration and tube formation of HUVECs; besides, the expression of UCA1 in PaC tissue was positively correlated with MVD [24]. Further mechanistic investigation revealed that UCA1 could act as a sponge of miR-96-5p to alleviate the inhibitory effect of miR-96-5p on the expression of its target gene Angiomotin-like 2 (AMOTL2) [24]. AMOTL2 is a member of angiomotin family proteins, and is required for proliferation, migration and tube formation of HUVECs during angiogenesis [60]. Moreover, hypoxia could induce the expression of 60 kDa isoform of AMOTL2 which is able to promote tumor growth and invasion [61]. These results indicated that UCA1 may play an important role in promoting angiogenesis in PaC under hypoxic condition.
UCA1 increases drug resistance in PaC
Drug resistance is a major cause of cancer treatment failure. A large number of lncRNAs have also been shown to induce drug resistance in cancer cells [62-64]. Overexpression of UCA1 has been reported to be associated with resistance to chemotherapeutic drugs, including 5-fluorouracil, cisplatin, gemcitabine, paclitaxel, docetaxel, gefitinib, cetuximab, doxorubicin, daunorubicin, tamoxifen, temozolomide, and trastuzumab, in many kinds of tumor cells [65-80]. Recently, the role of UCA1 in the chemoresistance of PaC has also been investigated. Chi et al. showed that exosomal UCA1 derived from hypoxia-induced pancreatic stellate cells could promote gemcitabine resistance in PaC, via the SOCS3/EZH2 axis [81]. Besides, Liang et al also reported that UCA1 overexpression also could induce resistance to 5-Fu in PDAC cells [32]. It is generally believed that UCA1 promotes drug resistance by directly binding to specific miRNAs and acting as a “sponge”. However, the underlying molecular mechanisms by which UCA1 promoted drug resistance in PaC still remain to be investigated in depth.
Future clinical applications of UCA1 in PaC
UCA1 as biomarker for PaC diagnosis
Early diagnosis is the key to successful treatment of cancer. A number of studies have revealed the high expression of UCA1 in PaC tissues as well as in serum of PaC patients. Particularly, the expression of UCA1 has also been found up-regulated in plasma of patients with malignant IPMN (a PDAC precursor) compared to benign cases; UCA1, along with other seven lncRNAs, performed greater accuracy in discriminating between benign and malignant IPMNs than the standard clinical and radiologic features, with an AUC value of 0.77 [47]. Nevertheless, it is reported that UCA1 could be released into exosomes, and the level of exosomal UCA1 in serum of PC patients were significantly higher than in healthy controls, with an AUC value of 0.78 [24]. Thus, UCA1 has the potential to be an early diagnostic biomarker for PaC.
UCA1 as biomarker for PaC prognosis
The increased expression of UCA1 has been reported to be significantly associated with poor PaC prognosis. Guo et al. revealed that the elevated UCA1 level in serum exosomes is significantly associated with tumor size (p = 0.038), lymphatic invasion (p = 0.018), late TNM stage (p = 0.017) [24]. Besides, Chen et al. reported that UCA1 expression in PaC tissues is also significantly correlated with tumor size (p = 0.021), depth of invasion (p = 0.033), tumor stage (p = 0.013) and CA19-9 level (p = 0.034) [34]. Patients with high UCA1 expression in cancer tissue or serum had relatively short overall survival [24, 34]. Multivariate Cox analysis results showed that the high expression of UCA1 is an independent prognostic factor in PaC [34, 39]. In adition, several LncRNA prognostic Models, such as a three-lnRNA penal (UCA1, AC009014.3, and RP11-48O20.4), a seven m6A-related lncRNA penal (UCA1, LINC01094, CASC19, LINC02323, PRECSIT, ITGB1-DT, and NRAV), have been constructed and showed noticeable potential prognostic value [82,83].
UCA1 as potential targets for pancreatic cancer therapy
At present, multiple studies on UCA1 in various tumor types have proved the possibility of UCA1 as a target for cancer treatment. Down-regulating UCA1 not only significantly inhibits cell proliferation in vitro and tumor growth in vivo, but also increases the sensitivity of cancer cells to different drugs and improves chemotherapeutic effect in various human cancer, including pancreatic cancer, which suggesting that UCA1 may become a potential tumor therapeutic target. However, none of these novel findings are yet to be assessed in clinical trials, and further clinical trials are needed to validate these findings in the future.
Conclusions
LncRNA-UCA1 is considered to be the most important lncRNA associated with PaC prognosis. UCA1 participated in the regulation of the key of PaC progression, including cancer cell growth, invasion, migration, metastasis and angiogenesis. Hence, UCA1 could be a potential target for PaC therapy, which requires more in-depth mechanism research as theoretical support. Patients with elevated UCA1 expression had shorter overall survival, suggesting that UCA1 might be an important independent predictor of poor survival. In conclusion, UCA1 shows great potential as a diagnostic, predictive or prognostic biomarker, and a therapeutic target in PaC, and the mechanisms needs to be elucidated in greater detail, which might provide new ideas and solutions for the diagnosis and treatment of PaC.
Acknowledgements
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (grant number LY19H160033, LY20H160042 and LQ20H160060) and the Zhejiang Province Bureau of Health (2019KY031, 2021KY531, 2018ZA011, and 2021ZQ010).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Barros AG, Pulido CF, Machado M, Brito MJ, Couto N, Sousa O. et al. Treatment optimization of locally advanced and metastatic pancreatic cancer (Review). Int J Oncol. 2021 59(6)
2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7-33
3. Xia C, Dong X, Li H, Cao M, Sun D, He S. et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl). 2022;135(5):584-590
4. Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020;395(10242):2008-2020
5. Park W, Chawla A, O'Reilly EM. Pancreatic Cancer: A Review. JAMA. 2021;326(9):851-862
6. Saluja A, Maitra A. Pancreatitis and Pancreatic Cancer. Gastroenterology. 2019;156(7):1937-1940
7. Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913-21
8. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629-41
9. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172(3):393-407
10. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904-14
11. Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015;116(4):737-50
12. Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of Long Noncoding RNAs. Annu Rev Immunol. 2017;35:177-198
13. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29(4):452-463
14. Liu SJ, Dang HX, Lim DA, Feng FY, Maher CA. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer. 2021;21(7):446-460
15. Chen S, Shen X. Long noncoding RNAs: functions and mechanisms in colon cancer. Mol Cancer. 2020;19(1):167
16. Huang Z, Zhou JK, Peng Y, He W, Huang C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer. 2020;19(1):77
17. Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ. et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12(16):4851-8
18. Xue M, Chen W, Li X. Urothelial cancer associated 1: a long noncoding RNA with a crucial role in cancer. J Cancer Res Clin Oncol. 2016;142(7):1407-19
19. Wang H, Guan Z, He K, Qian J, Cao J, Teng L. LncRNA UCA1 in anti-cancer drug resistance. Oncotarget. 2017;8(38):64638-64650
20. Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582(13):1919-27
21. Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL. et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6(10):7899-917
22. Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M. et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 2014;5:e1008
23. Luan Y, Li X, Luan Y, Zhao R, Li Y, Liu L. et al. Circulating lncRNA UCA1 Promotes Malignancy of Colorectal Cancer via the miR-143/MYO6 Axis. Mol Ther Nucleic Acids. 2020;19:790-803
24. Guo Z, Wang X, Yang Y, Chen W, Zhang K, Teng B. et al. Hypoxic Tumor-Derived Exosomal Long Noncoding RNA UCA1 Promotes Angiogenesis via miR-96-5p/AMOTL2 in Pancreatic Cancer. Mol Ther Nucleic Acids. 2020;22:179-195
25. Yang A, Liu X, Liu P, Feng Y, Liu H, Gao S. et al. LncRNA UCA1 promotes development of gastric cancer via the miR-145/MYO6 axis. Cell Mol Biol Lett. 2021;26(1):33
26. Fang Z, Zhao J, Xie W, Sun Q, Wang H, Qiao B. LncRNA UCA1 promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by sunppressing miR-184 expression. Cancer Med. 2017;6(12):2897-2908
27. Zhang ZS, Wang J, Zhu BQ, Ge L. Long noncoding RNA UCA1 promotes multiple myeloma cell growth by targeting TGF-beta. Eur Rev Med Pharmacol Sci. 2020;24(24):12623
28. Gao H, Yang JY, Tong LX, Jin H, Liu CZ. Long noncoding RNA UCA1 promotes proliferation and metastasis of thyroid cancer cells by sponging miR-497-3p. Eur Rev Med Pharmacol Sci. 2020;24(14):7555
29. Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY, Liu Q. et al. The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol Cancer. 2019;18(1):115
30. Ramli S, Sim MS, Guad RM, Gopinath SCB, Subramaniyan V, Fuloria S. et al. Long Noncoding RNA UCA1 in Gastrointestinal Cancers: Molecular Regulatory Roles and Patterns, Mechanisms, and Interactions. J Oncol. 2021;2021:5519720
31. Liu Y, Feng W, Gu S, Wang H, Zhang Y, Chen W. et al. The UCA1/KRAS axis promotes human pancreatic ductal adenocarcinoma stem cell properties and tumor growth. Am J Cancer Res. 2019;9(3):496-510
32. Liang X, Qi M, Wu R, Liu A, Chen D, Tang L. et al. Long non-coding RNA CUDR promotes malignant phenotypes in pancreatic ductal adenocarcinoma via activating AKT and ERK signaling pathways. Int J Oncol. 2018;53(6):2671-2682
33. Zhou Y, Chen Y, Ding W, Hua Z, Wang L, Zhu Y. et al. LncRNA UCA1 impacts cell proliferation, invasion, and migration of pancreatic cancer through regulating miR-96/FOXO3. IUBMB Life. 2018;70(4):276-290
34. Chen P, Wan D, Zheng D, Zheng Q, Wu F, Zhi Q. Long non-coding RNA UCA1 promotes the tumorigenesis in pancreatic cancer. Biomed Pharmacother. 2016;83:1220-1226
35. Gong J, Lu X, Xu J, Xiong W, Zhang H, Yu X. Coexpression of UCA1 and ITGA2 in pancreatic cancer cells target the expression of miR-107 through focal adhesion pathway. J Cell Physiol. 2019;234(8):12884-12896
36. Zhang M, Zhao Y, Zhang Y, Wang D, Gu S, Feng W. et al. LncRNA UCA1 promotes migration and invasion in pancreatic cancer cells via the Hippo pathway. Biochim Biophys Acta Mol Basis Dis. 2018;1864(5 Pt A):1770-1782
37. Zhang X, Gao F, Zhou L, Wang H, Shi G, Tan X. UCA1 Regulates the Growth and Metastasis of Pancreatic Cancer by Sponging miR-135a. Oncol Res. 2017;25(9):1529-1541
38. da Paixao VF, Sosa OJ, da Silva Pellegrina DV, Dazzani B, Correa TB, Riserio Bertoldi E. et al. Annotation and functional characterization of long noncoding RNAs deregulated in pancreatic adenocarcinoma. Cell Oncol (Dordr). 2022;45(3):479-504
39. Fu XL, Liu DJ, Yan TT, Yang JY, Yang MW, Li J. et al. Analysis of long non-coding RNA expression profiles in pancreatic ductal adenocarcinoma. Sci Rep. 2016;6:33535
40. Cao L, Huang C, Cui Zhou D, Hu Y, Lih TM, Savage SR. et al. Proteogenomic characterization of pancreatic ductal adenocarcinoma. Cell. 2021;184(19):5031-5052 e26
41. Kanda M, Matthaei H, Wu J, Hong SM, Yu J, Borges M. et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142(4):730-733 e9
42. Waters AM, Der CJ. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb Perspect Med. 2018 8(9)
43. Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M. et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496(7443):101-5
44. Rozeveld CN, Johnson KM, Zhang L, Razidlo GL. KRAS Controls Pancreatic Cancer Cell Lipid Metabolism and Invasive Potential through the Lipase HSL. Cancer Res. 2020;80(22):4932-4945
45. Mann KM, Ying H, Juan J, Jenkins NA, Copeland NG. KRAS-related proteins in pancreatic cancer. Pharmacol Ther. 2016;168:29-42
46. Humpton TJ, Alagesan B, DeNicola GM, Lu D, Yordanov GN, Leonhardt CS. et al. Oncogenic KRAS Induces NIX-Mediated Mitophagy to Promote Pancreatic Cancer. Cancer Discov. 2019;9(9):1268-1287
47. Permuth JB, Chen DT, Yoder SJ, Li J, Smith AT, Choi JW. et al. Linc-ing Circulating Long Non-coding RNAs to the Diagnosis and Malignant Prediction of Intraductal Papillary Mucinous Neoplasms of the Pancreas. Sci Rep. 2017;7(1):10484
48. Moriya J, Minamino T. Angiogenesis, Cancer, and Vascular Aging. Front Cardiovasc Med. 2017;4:65
49. Hirte HW. Novel developments in angiogenesis cancer therapy. Curr Oncol. 2009;16(3):50-4
50. Tao J, Yang G, Zhou W, Qiu J, Chen G, Luo W. et al. Targeting hypoxic tumor microenvironment in pancreatic cancer. J Hematol Oncol. 2021;14(1):14
51. Ren B, Cui M, Yang G, Wang H, Feng M, You L. et al. Tumor microenvironment participates in metastasis of pancreatic cancer. Mol Cancer. 2018;17(1):108
52. Bolm L, Zghurskyi P, Lapshyn H, Petrova E, Zemskov S, Vashist YK. et al. Alignment of stroma fibers, microvessel density and immune cell populations determine overall survival in pancreatic cancer-An analysis of stromal morphology. PLoS One. 2020;15(7):e0234568
53. Xiang T, Xia X, Yan W. Expression of Matrix Metalloproteinases-2/-9 is Associated With Microvessel Density in Pancreatic Cancer. Am J Ther. 2017;24(4):e431-e434
54. Ntellas P, Dadouli K, Perivoliotis K, Sogka E, Pentheroudakis G, Ioannou M. et al. Microvessel Density and Impact of Angiogenesis on Survival of Resected Pancreatic Cancer Patients: A Systematic Review and Meta-analysis. Pancreas. 2019;48(2):233-241
55. Wei Y, Song S, Duan N, Wang F, Wang Y, Yang Y. et al. MT1-MMP-Activated Liposomes to Improve Tumor Blood Perfusion and Drug Delivery for Enhanced Pancreatic Cancer Therapy. Adv Sci (Weinh). 2020;7(17):1902746
56. Richards KE, Zeleniak AE, Fishel ML, Wu J, Littlepage LE, Hill R. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene. 2017;36(13):1770-1778
57. Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA. et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546(7659):498-503
58. Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK. et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17(6):816-26
59. Zhou W, Zhou Y, Chen X, Ning T, Chen H, Guo Q. et al. Pancreatic cancer-targeting exosomes for enhancing immunotherapy and reprogramming tumor microenvironment. Biomaterials. 2021;268:120546
60. Wang Y, Li Z, Xu P, Huang L, Tong J, Huang H. et al. Angiomotin-like2 gene (amotl2) is required for migration and proliferation of endothelial cells during angiogenesis. J Biol Chem. 2011;286(47):41095-104
61. Mojallal M, Zheng Y, Hultin S, Audebert S, van Harn T, Johnsson P. et al. AmotL2 disrupts apical-basal cell polarity and promotes tumour invasion. Nat Commun. 2014;5:4557
62. He J, Zhu S, Liang X, Zhang Q, Luo X, Liu C. et al. LncRNA as a multifunctional regulator in cancer multi-drug resistance. Mol Biol Rep. 2021;48(8):1-15
63. Yuan L, Xu ZY, Ruan SM, Mo S, Qin JJ, Cheng XD. Long non-coding RNAs towards precision medicine in gastric cancer: early diagnosis, treatment, and drug resistance. Mol Cancer. 2020;19(1):96
64. Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv L. et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer. 2020;19(1):62
65. Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu Y. et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep. 2016;6:23892
66. Xian Z, Hu B, Wang T, Zeng J, Cai J, Zou Q. et al. lncRNA UCA1 Contributes to 5-Fluorouracil Resistance of Colorectal Cancer Cells Through miR-23b-3p/ZNF281 Axis. Onco Targets Ther. 2020;13:7571-7583
67. Fu J, Pan J, Yang X, Zhang Y, Shao F, Chen J. et al. Mechanistic study of lncRNA UCA1 promoting growth and cisplatin resistance in lung adenocarcinoma. Cancer Cell Int. 2021;21(1):505
68. Zhou H, Shen Q, Fu J, Jiang F, Wang L, Wang Y. Analysis of lncRNA UCA1-related downstream pathways and molecules of cisplatin resistance in lung adenocarcinoma. J Clin Lab Anal. 2020;34(8):e23312
69. Liu X, Huang Z, Qian W, Zhang Q, Sun J. Silence of lncRNA UCA1 rescues drug resistance of cisplatin to non-small-cell lung cancer cells. J Cell Biochem. 2019;120(6):9243-9249
70. Pan J, Li X, Wu W, Xue M, Hou H, Zhai W. et al. Long non-coding RNA UCA1 promotes cisplatin/gemcitabine resistance through CREB modulating miR-196a-5p in bladder cancer cells. Cancer Lett. 2016;382(1):64-76
71. Wang J, Ye C, Liu J, Hu Y. UCA1 confers paclitaxel resistance to ovarian cancer through miR-129/ABCB1 axis. Biochem Biophys Res Commun. 2018;501(4):1034-1040
72. Xu T, Yan S, Wang M, Jiang L, Ma P, Lu B. et al. LncRNA UCA1 Induces Acquired Resistance to Gefitinib by Epigenetically Silencing CDKN1A Expression in Non-small-Cell Lung Cancer. Front Oncol. 2020;10:656
73. Chen X, Wang Z, Tong F, Dong X, Wu G, Zhang R. lncRNA UCA1 Promotes Gefitinib Resistance as a ceRNA to Target FOSL2 by Sponging miR-143 in Non-small Cell Lung Cancer. Mol Ther Nucleic Acids. 2020;19:643-653
74. Yuan HH, Zhang XC, Wei XL, Zhang WJ, Du XX, Huang P. et al. LncRNA UCA1 mediates Cetuximab resistance in Colorectal Cancer via the MiR-495 and HGF/c-MET Pathways. J Cancer. 2022;13(1):253-267
75. Wo L, Zhang B, You X, Hu Y, Gu Z, Zhang M. et al. Up-regulation of LncRNA UCA1 by TGF-beta promotes doxorubicin resistance in breast cancer cells. Immunopharmacol Immunotoxicol. 2022;44(4):492-499
76. Yao Q, Zhang L, Wang Y, Liu J, Yang L, Wang Y. LncRNA UCA1 elevates the resistance of human leukemia cells to daunorubicin by the PI3K/AKT pathway via sponging miR-613. Biosci Rep. 2021 41(6)
77. Li Z, Yu D, Li H, Lv Y, Li S. Long noncoding RNA UCA1 confers tamoxifen resistance in breast cancer endocrinotherapy through regulation of the EZH2/p21 axis and the PI3K/AKT signaling pathway. Int J Oncol. 2019;54(3):1033-1042
78. Xu CG, Yang MF, Ren YQ, Wu CH, Wang LQ. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur Rev Med Pharmacol Sci. 2016;20(20):4362-4368
79. Cheng M, Wang Q, Chen L, Zhao D, Tang J, Xu J. et al. LncRNA UCA1/miR-182-5p/MGMT axis modulates glioma cell sensitivity to temozolomide through MGMT-related DNA damage pathways. Hum Pathol. 2022;123:59-73
80. Zhu HY, Bai WD, Ye XM, Yang AG, Jia LT. Long non-coding RNA UCA1 desensitizes breast cancer cells to trastuzumab by impeding miR-18a repression of Yes-associated protein 1. Biochem Biophys Res Commun. 2018;496(4):1308-1313
81. Chi Y, Xin H, Liu Z. Exosomal lncRNA UCA1 Derived From Pancreatic Stellate Cells Promotes Gemcitabine Resistance in Pancreatic Cancer via the SOCS3/EZH2 Axis. Front Oncol. 2021;11:671082
82. Li M, Li H, Chen Q, Wu W, Chen X, Ran L. et al. A Novel and Robust Long Noncoding RNA Panel to Predict the Prognosis of Pancreatic Cancer. DNA Cell Biol. 2020;39(7):1282-1289
83. Huang B, Liu J, Lu J, Gao W, Zhou L, Tian F. et al. Aerial View of the Association Between m6A-Related LncRNAs and Clinicopathological Characteristics of Pancreatic Cancer. Front Oncol. 2021;11:812785
Author contact
![]() Corresponding authors: Hui-Ju Wang, E-mail: wanghuijuedu.cn; Liang Wang, E-mail: wangliangedu.cn.
Corresponding authors: Hui-Ju Wang, E-mail: wanghuijuedu.cn; Liang Wang, E-mail: wangliangedu.cn.

 Global reach, higher impact
Global reach, higher impact