3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(15):4585-4594. doi:10.7150/jca.59933 This issue Cite
Research Paper
Hallmark microRNA signature in liquid biopsy identifies hepatocellular carcinoma and differentiates it from liver metastasis
1. OncoSeek Limited, Hong Kong Science and Technology Parks, Hong Kong Special Administrative Region, People's Republic of China.
2. Department of Clinical Oncology, LKS Faculty of Medicine, The Hong Kong Special Administrative Region, People's Republic of China.
* Contributed equally to this work.
Abstract
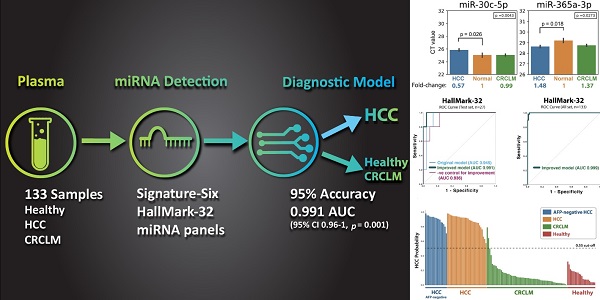
Purpose: This study aims to develop a liquid biopsy assay to identify HCC and differentially diagnose hepatocellular carcinoma (HCC) from colorectal carcinoma (CRC) liver metastasis. Methods: Thirty-two microRNAs (“HallMark-32” panel) were designed to target the ten cancer hallmarks in HCC. Quantitative PCR and supervised machine learning models were applied to develop an HCC-specific diagnostic model. One hundred thirty-three plasma samples from intermediate-stage HCC patients, colorectal cancer (CRC) patients with liver metastasis, and healthy individuals were examined. Results: Six differentially expressed microRNAs (“Signature-Six” panel) were identified after comparing HCC and healthy individuals. The microRNA miR-221-3p, miR-223-3p, miR-26a-5p, and miR-30c-5p were significantly down-regulated in the plasma of HCC samples, while miR-365a-3p and miR-423-3p were significantly up-regulated. Machine learning models combined with HallMark-32 and Signature-Six panels demonstrated promising performance with an AUC of 0.85-0.96 (p ≤ 0.018) and 0.84-0.93 (p ≤ 0.021), respectively. Further modeling improvement by adjusting sample quality variation in the HallMark-32 panel boosted the accuracy to 95% ± 0.01 and AUC to 0.991 (95% CI 0.96-1, p = 0.001), respectively. Even in alpha fetoprotein (AFP)-negative (< 20ng/mL) HCC samples, HallMark-32 still achieved 100% sensitivity in identifying HCC. The Cancer Genome Atlas (TCGA, n=372) analysis demonstrated a significant association between HallMark-32 and HCC patient survival. Conclusion: To the best of our knowledge, this is the first report to utilize circulating miRNAs and machine learning to differentiate HCC from CRC liver metastasis. In this setting, HallMark-32 and Signature-Six are promising non-invasive tests for HCC differential diagnosis and distinguishing HCC from healthy individuals.
Keywords: Hepatocellular carcinoma, miRNA signature, Liquid biopsy, Machine learning, HCC diagnosis, HCC screening

 Global reach, higher impact
Global reach, higher impact