3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(14):2866-2875. doi:10.7150/jca.19477 This issue Cite
Research Paper
Heterochromatin Reduction Correlates with the Increase of the KDM4B and KDM6A Demethylases and the Expression of Pericentromeric DNA during the Acquisition of a Transformed Phenotype
Departamento de Genética del Desarrollo y Fisiología Molecular. Instituto de Biotecnología, Universidad Nacional Autónoma de México. 62250 Cuernavaca Morelos, México
Received 2017-2-3; Accepted 2017-6-4; Published 2017-8-24
Abstract
Cancer cells have alterations in chromatin organization, mostly a reduction in heterochromatin. How this process occurs during transformation and if it participates in the maintenance of a cancerous phenotype is not well understood. Here, using a transformation-inducible cell line, we analyzed the changes that occur in heterochromatin during transformation to a cancerous phenotype. After transformation, there is a reduction in heterochromatin bodies and a nuclear reorganization of HP1α. These occurrences correlate with reductions in H3K9me3 and H3K27me3 levels and with some of the enzymes that introduce these modifications. At the same time, there are increases in the KDM4B and KDM6A/UTX demethylases and an enhancement in the transcription of pericentromeric DNA that correlate with the reduction of H3K9me3 and the recruitment of KDM4B to these elements. The depletion of KDM4B and KDM6A/UTX has a more deleterious effect in transformed cells than in their progenitors, suggesting an important role for these enzymes in the survival of cancerous cells. These results provide new insights into heterochromatin dynamics during transformation to a cancerous phenotype as well as some of the participating mechanisms.
Keywords: Heterochromatin, Histone demethylases, Satellite DNA, Transformation.
Introduction
Constitutive heterochromatin, the most condensed form of chromatin, is present in human cells, particularly in the pericentromeric and subtelomeric regions as well as in repetitive elements along the genome. This kind of heterochromatin is enriched in nucleosomes that contain Histone 3 trimethylated at lysine 9 (H3K9me3), which is recognized by Heterochromatic Protein 1 proteins (HP1α, β and γ) [1]. Tri-methylation of H3K9 is conducted by Suppressors of Variegation 3-9 Homologs 1 and 2 (SUV3-9H-1 and SUV3-9H-2) enzymes [2]. Conversely, facultative heterochromatin is present in genes that are not expressed in particular moments of development or in specific cell types [3]. Tri-methylation of Lysine 27 of H3 (H3K27me3) is a histone mark in genes that is characteristic of this type of heterochromatin. This modification is introduced by the action of the Polycomb Repressive Complex 2 (PRC2), whose methyl transferase activity is present in its Ezh1 or Ezh2 enzymes [4, 5]. These modifications are reversible since histone demethylases that remove these specific marks have been identified. H3K9me3 can be demethylated by the KDM4A, KDM4B and KDM4C enzymes, and these enzymes also demethylate H3K36 and H1K26 [6]. H3K27me3 can be demethylated by the action of the UTX/KDM6A and KDM6B demethylases [7]. These enzymes are α-ketoglutarate-dependent dioxygenases that belong to the Jumonji family of demethylases [6, 8].
It is well documented that the chromatin in cancer cells has different features than in normal differentiated cells [9]. In general, it is less condensed and has broad changes in DNA methylation, H3K9me2, H3K9me3, H3K27me3, and histone acetylation [9]. This is linked to changes in the levels of methylation-modifying enzymes. For example, there is evidence suggesting that UTX/KDM6A may act as a tumor suppressor in T-cell acute lymphoplastic leukemia, but the demethylation activity of UTX/KDM6A is required for tumor maintenance through the activation of the NOTCH pathway and Rb dependent tumors [10-12]. It is also known that the loss of heterochromatin in cancer cells, in particular pericentromeric and telomeric chromatin, causes an increase in genome instability that favors the generation of aneuploidy, thus increasing the diversity of tumor cells [13,14]. Furthermore, there are several reports that show that the KDM4A, KDM4B and KDM4C demethylases are overexpressed in different kinds of cancer and are required for maintaining the cancerous phenotype [15].
Although it is evident that heterochromatin undergoes dramatic changes during the generation of a cancer cell, how these alterations occur during the transformation to this phenotype is not well understood. A model is needed to study chromatin changes wherein the transformation to a cancerous phenotype occurs through incubation with an inducer in a cell line that it is immortalized but not cancerous; the MCF10-Er-Src line is such a model [16]. In this model, a transformed phenotype is generated by the transient expression of Src, which induces the epigenetic switch from an immortalized human breast cell line to a stable and highly malignant transformed cell line [16]. In fact this cell line has been acceptable used as a model of oncogenesis in a variety of studies [16-19]. Using this system, we analyzed the broad heterochromatin changes that occur during cell transformation to a cancerous phenotype. After transformation, there is a reduction in heterochromatin centers as well as changes in HP1α distribution. These changes correlate with a reduction in the global levels of H3K9me3 and H3K27me3, an increase in the KDM4B and KDM6A/UTX histone demethylases and a decrease in the total levels of SUVAR39H1 as well as the components of the PRC2 complex. Furthermore, pericentromeric sequences increase transcription, correlating with a reduction in H3K9me3 and the recruitment of the KDM4B histone demethylase in these sequences. The depletion of these demethylases compromises the transformed cells. These results indicate that after the induction of a transformed phenotype, there is a reduction in heterochromatin that correlates with changes in the expression of histone modifying enzymes, allowing an increase in the transcription of heterochromatic sequences.
Materials and Methods
Cell cultures
The cell line MCF10A ER-Src was kindly donated by Dr. Kevin Struhl (Harvard Medical School). Treatment of such cells with tamoxifen (TAM) rapidly induces Src and morphological transformation is observed within 24-36 hours [16, 20]. MCF-10A ER-Src cells were cultured as indicated in [16]. Cells were treated with 1μM tamoxifen when 80% confluent for transformation. MDA-MB-231 and MCF7 breast cancer cell lines were grow in DMEM, 10% FBS and pen/step.
Western blot analysis
Nuclear extracts were prepared as described in [21]. Cells were washed twice with PBS1X and then resuspended and incubated for 15' in cold lysis buffer A plus inhibitors, followed by adding 10% Triton X100, cytoplasmic lysates were cleared by centrifugation. The pellet was resuspended and incubated for 15' in Nuclear Lysis Buffer (NLB), and then a spin down at high speed was necessary for recovering the nuclear proteins [21]. The proteins were quantitated and separated in SDS-polyacrylamide gels and transferred to nitrocellulose membranes, following overnight blocking with 10-20% non-fat milk at 4ºC. Membranes were incubated with the primary antibody at different dilutions at room temperature for 2 h and then incubated with a horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature and detected by Chemiluminescence (Thermoscientific). Antibodies used were: HP1α (ab77256-abcam), H3K9me3 (07-442-millipore), H3K9me2 (ab1220-abcam), H3K9ac (ab4441-abcam), H3K27me3 (ab6002-abcam), KDM4A (PA5-25057-thermo scientific), KDM4B (ab27531-abcam), KDM4C (PA5-23065-thermo-scientific), KDM6A/UTX (ABE409-millipore), EZH2 (MABE362-millipore), SUZ12 (sc-46264-Santa Cruz) H3 (ab1791-abcam), Actin (JLA20-C-DSHB), Tubulin (E7-C- DSHB).
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed as described in ChIP assay kit (Upstate Biotech). For each condition 2 mg of soluble chromatin fragments of 250-500 bp in length were incubated with 50 μg of Dynabeads for 2-4h at 4º C in rotation. The dynabeads were discarded and the pre-cleared chromatin was incubated with 2 μg of anti-H3K9me3 or anti-KDM4B antibodies [22] and rabbit anti-IgG antibody for 16 h at 4 °C. To recover the DNA-protein complexes, 25 μg of Dynabeads were added (previously blocked). Reverse cross-linking was performed by incubation at 65°C for 12h, followed by 4ºC incubation for 10 min. Samples were treated with RNase A for 1 h at 37 °C proteinase K was added and incubated for 2 h at 42 °C. DNA fragments were purified and resuspended in water.
Immunofluorescence
40-50% confluent cells were cultured on Nunc-Lab-TekII Chamber Slides. The immunofluorescences were performed according to the manufacturer's instructions (http://www.abcam.com/protocols/immunocytochemistry-immunofluorescence-protocol). Cells were incubated overnight with the corresponding antibodies. The chambers were washed six times with PBS1X and incubated with Alexa Fluor 488 or Alexa Fluor 568-conjugated secondary antibodies. Nuclei were stained with Hoechst (dilution 1:20000) for 10 min. Samples were examined with Confocal Olympus FV1000 multi-photonic microscope.
RT-PCR
Total RNA was extracted with TRIZOL. cDNA synthesis was performed in a reaction mix containing 1 μg of total RNA, oligo-dT, random primer and M-MLV Reverse Transcriptase (Invitrogen). The DNA was used to perform PCRs reactions using the following primers (SATa, SATIII and mcBox) reported in [14].
siRNA assays
siRNA-silencing was performed according to Dharmacon instructions. The cells were seeded in a 6 well plates (60% confluence) and incubated for 12h in starvation medium and then independently the siRNA and the DharmaFECT reagent 1 were diluted in serum free medium and incubated for 5 min at room temperature, after that time those were mixed and incubated once more for 25 min at room temperature and finally added to the cells. The siRNA was used in a 25 and 50nM final concentration and incubated at 37ºC in 5% CO2 for 48-72h, ready to be analysed by flow cytometry for apoptosis and viability assays.
Intra cellular protein staining for flow cytometry, Viability and apoptosis assays
The intracelular protein staining was performed according to the protocols previously described by [23]. The Viability and apoptosis assays were performed according to the protocols previously described in Affymetrix cat.#65-0865 and Biolegend cat.#640918 respectively. Cells were harvested and stained using Anexin pacific blue to labelled apoptotic cells. The cells were stained with viability solution to label dead cells. Briefly, the cells were wash twice and resuspended in fixable viability dye for 30 min at 4ºC protected from light, then the dye was removed with flow staining buffer, later the cells were resuspend in Annexin V binding buffer containing Annexin V and incubate for 15 min at RT (25°C) in the dark; the cells were washed with binding buffer and then fixed with PFA 2% at 37ºC for 10min, finally the cells were washed newly with FACS juice and stored at 4oC until the analysis. The cells and the data were analysed with FACSCanto II flow cytometer, and FLOWJO software respectively.
Results
Induction of cell line transformation impairs heterochromatin structure and partial relocalization HP1α
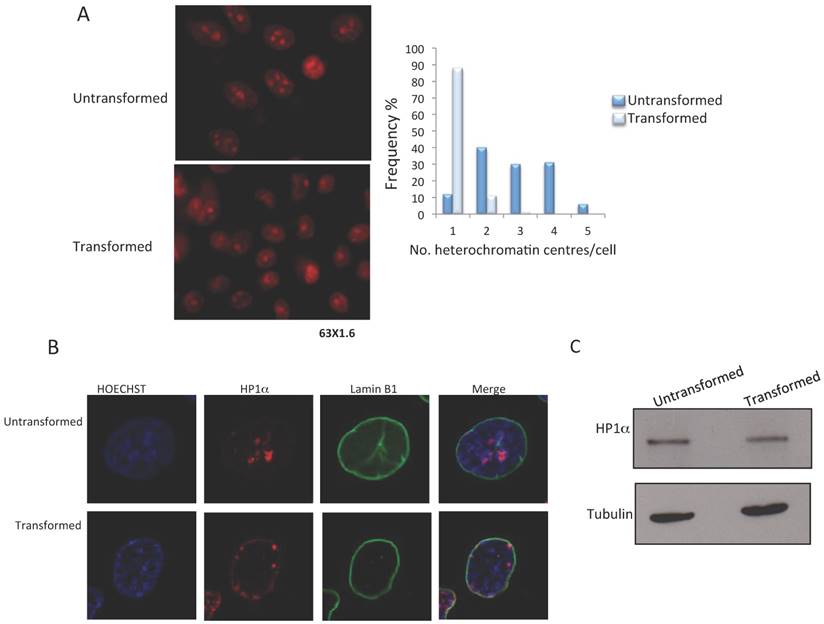
The MCF10-Er-Src line can be induced to transform to a malignant phenotype through the activation of Src by incubation with tamoxifen [16]. Using this model, we determined the changes that occur in heterochromatin after transformation. The first approximation was to analyze the effect on heterochromatic centers. Ethidium bromide staining of transformed and non-transformed cells shows that there is a reduction in the number of heterochromatic centers in cells induced with tamoxifen after 36-48 hrs (Fig. 1A and B). We observed between 2-5 heterochromatic centers in the non-transformed cells and a reduction to 1-2 centers in the transformed cells (Fig. 1B). This result suggests that there is a rapid rearrangement and reduction in heterochromatin when the cancerous phenotype is induced in MCF10-Er-Src cells.
Since HP1α is located in constitutive heterochromatin, we decided to determine if there are changes in the levels or distribution of HP1α changes in the levels or distribution of in transformed cells. HP1α can be identified in heterochromatic centers in non-transformed cells; however, there is a change in the nuclear location of HP1α in the tamoxifen treated cells (Fig. 1C). Some of the HP1α is located in the nuclear periphery, and some HP1α remains in the heterochromatic bodies (Fig. 1B). These changes in HP1α distribution occur without affecting HP1α levels (Fig. 1C). Thus, the generation of a transformed phenotype in MCF10-Er-Src cells correlates with board changes in heterochromatin organization, suggesting a reduction in constitutive heterochromatin.
Global levels of H3K9me3 and H3K27me3 are reduced in transformed MCF10-Er-Src cells
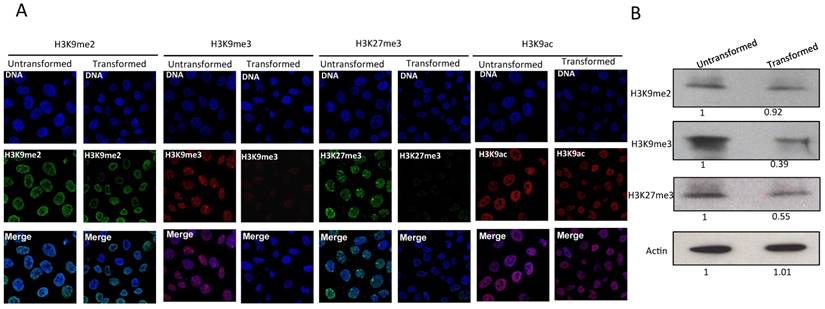
Since the levels of heterochromatic centers are reduced and the nuclear distribution of HP1α changes after transformation, we next analyzed the histone marks H3K9me3, H3K9me2 and H3K27me3. H3K9me3 and H3K9me2 are recognized by HP1α, and H3K27me3 is a mark present in genes that are not expressed. Fig. 2A shows immunostains of non-transformed and transformed cells. H3K9me3 and H3K27me3 can be detected forming foci in the nuclei of normal cells [24-26]. In the case of H3K27me3, a predominant focus near the nuclear membrane can be visualized that corresponds to the inactive X chromosome. Intriguingly, a reduction in the levels of H3K9me3 and H3K27me3 is observed in transformed cells (Fig. 2A). The H3K27me3 signal of the inactive X chromosome is also reduced. The levels of H3K9me2 are not reduced in transformed cells (Fig. 2A). Conversely, significant changes in the levels of H3K9Ac are not detected (Fig. 2A). The reduction in the levels of H3K9me3 and H3K27me3 was confirmed by Western blot analysis using nuclear protein from non-transformed and transformed cells (Fig. 2B). These results suggest that the decrease in heterochromatin is linked to a reduction of both facultative and constitutive histone marks.
Reduction of heterochromatin bodies and delocalization of HP1α in transformed MCF10A-Er-Src cells. A, Ethidium Bromide staining of non-transformed and transformed cells. Heterochromatin bodies (2-4) are evident in the nuclei of non-transformed cells. A reduction in the number of heterochromatin bodies is observed in the nuclei of transformed cells. The plot indicates the frequency of the number of heterochromatic bodies in non-transformed and transformed cells. B, HP1α and nuclear laminin B1 immunostaining of non-transformed and transformed cells. Note the change in the HP1α distribution in the transformed cells. C, HP1α western blot experiments from non-transformed and transformed cells. A tubulin antibody was used as loading control.
The levels H3K9me3 and H3K27me3 marks are reduced in the MCF10A-Er-Src cells after transformation. A, Immunostaining detection of H3K9me2, H3K9me3, H3K27me3 and H3K9ac in non-transformed and transformed cells. Note the reduction in the signal of H3K9me3 and H3K27me3 marks in the transformed cells. The reduction of the signal is homogeneous in both cases, although the presence of H3K9me3 and H3K27 bodies is maintained in the transformed cells. B, Western blot experiments using antibodies that recognize H3K9me3, H3K9me2, H3K27me3 from non-transformed and transformed cells. An Actin antibody was used as loading control. The numbers at the bottom in the western blots indicate the relative levels of the protein signal versus the acting levels determined by densitometry adjusted to 1 in the nontransformed cells.
Transformation causes an increase in KDM4B and UTX/KDM6A and a reduction in SUV39H1 and PRC2 levels
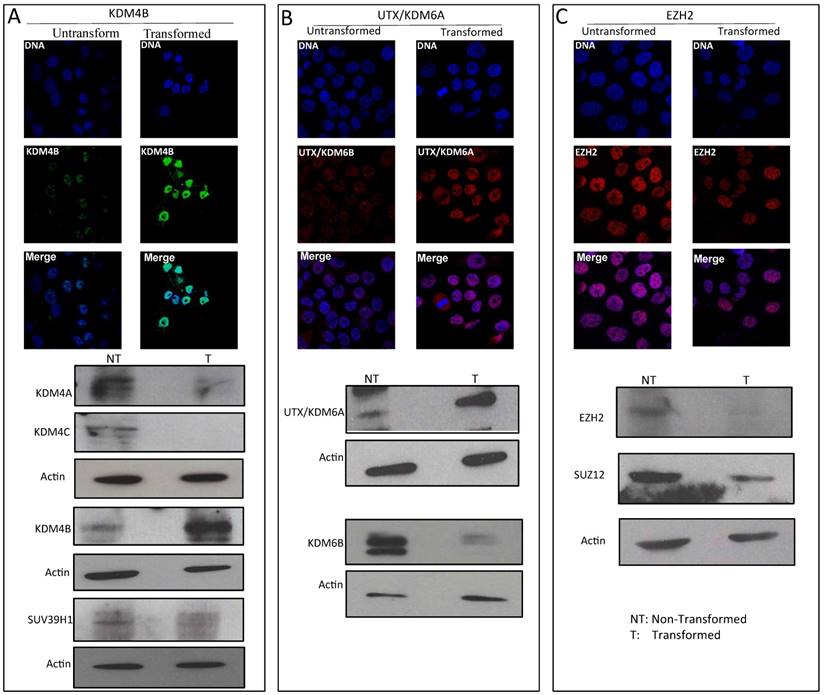
We have observed that a reduction in global heterochromatin in transformed cells correlates with a decrease in H3K9me3 and H3K27me3 levels. Therefore, we determined the effect of transformation on the histone methyl-transferases and demethylases that modulate these modifications. For H3K9me3, we determined the total levels of the stress-inducible KDM4B demethylase as well as the levels of the SUV39H1 methyl-transferase. In the transformed cells, there is an important increase in the levels of KDM4B compared with the non-transformed cells (Fig. 3A). In addition, Western blot analysis confirmed the KDM4B immunostaining data but showed a significant reduction in the levels of other H3K9me3 demethylases (Fig. 3A). Conversely, the levels of SUV39H1 decrease in the transformed cells (Fig. 3A). Thus, the reduction in the H3K9me3 level correlates with an increase of KDM4B and with the decrease of one of the methyl-transferases that incorporates methyl groups into H3K9.
To investigate H3K27me3 demethylases and methyl-transferases, we analyzed the levels of UTX/KDM6A and KDM6B as well as the levels of ENZh2 and SUZ12, which are part of PRC2. Fig. 3B shows that the levels of UTX/KDM6A are increased, but that the levels of KDM6B are reduced in the transformed cells. Interestingly, the levels of the PRC2 components decrease in the cancerous cell nuclei (Fig. 3C). Collectively, these results indicate that in transformed cells, there are changes in the levels of enzymes that affect the methylation state of the K9 and K27 residues of H3. In particular, the data suggest that there is a reduction in the levels of SUVAR39H1 and an increase of KDM4B, which may explain the reduction in the total levels of H3K9me3 in the transformed cells. Conversely, a reduction in the levels of PRC2 and an increase in UTX/KDM6A may be linked to the decrease in the levels of H3K27me3. Other factors that participate in these modifications also undergo changes during transformation; however, these factors do not correlate with the global changes observed in H3K9me3 and H3K27me3.
KDM4B is recruited to heterochromatin, correlating with an increase in the transcription of heterochromatic sequences in transformed cells
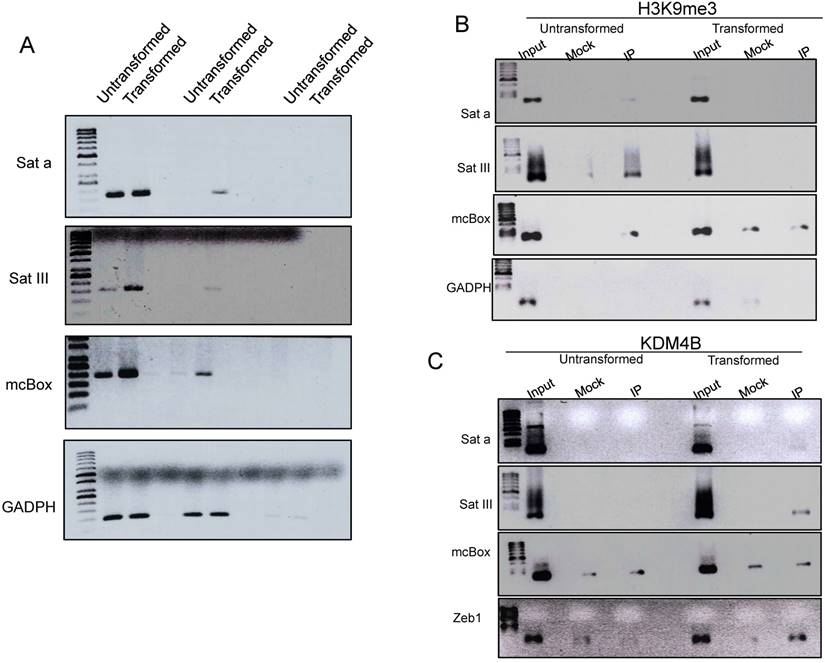
A characteristic of pericentromeric heterochromatin is that it is enriched in H3K9me3. These DNA satellite sequences are only transcribed at low levels in normal cells, but an increase in their transcription has been observed in cancer cells (14). Since we observed a decrease of this mark in the transformed cells, we decided to investigate whether there is also an enhancement in the transcription of Sat a, SatIII and mcBox satellite sequences [27, 28]. We observed an increase in the transcript levels of all these elements in the transformed cells (Fig. 4A). These results suggest that the reduction in the level of H3K9me3 in heterochromatic centers increases the transcription rate of constitutive heterochromatin in transformed cells.
Variation in the levels of enzymes that modify histone H3 in transformed MCF10A-Er-Src cell line. A, Immunostaining to detect KDM4B demethylase and western blots detecting KDM4A, KDM4B and KDM4C histone demethylases as well as the methyl transferase SUV39H1 levels in non-transformed and transformed cells. B, Immunostaining to detect UTX/KDM6A and western blot experiments detecting UTX/KDM6A and KDM6B in non-transformed and transformed cells. C, Immunostaining to detect the methyl transferase EZH2 and western blots to detect EZH2 and SUZ12 in transformed and no-transformed cells. Acting was used as loading control in all the western blot experiments. NT=Non-transformed cells; T= Transformed cells.
To further analyze this possibility, we performed ChIP experiments to test if there is a reduction in H3K9me3 levels in these satellite sequences as well as an increase in the occupancy of KDM4B that may be removing this repressive mark. Fig. 4B shows that in non-transformed cells, the H3K9me3 mark is present in the three satellite sequences and, as expected, it is not present in the GADPH gene. Interestingly, in transformed cells, this H3 modification is not detected, at least no more than the mock antibody. Intriguingly, when we analyzed the non-transformed cells, the presence of KDM4B in the satellite sequences is not detected; however, KDM4B is recruited to these repetitive elements after transformation induction (Fig. 4B) Collectively, these experiments suggest that there is a recruitment of KDM4B to the satellite sequences that demethylate H3K9, favoring the transcription of these repetitive elements in transformed cells.
Knock down of UTX/KDM6A and KDM4B preferentially kills transformed cells
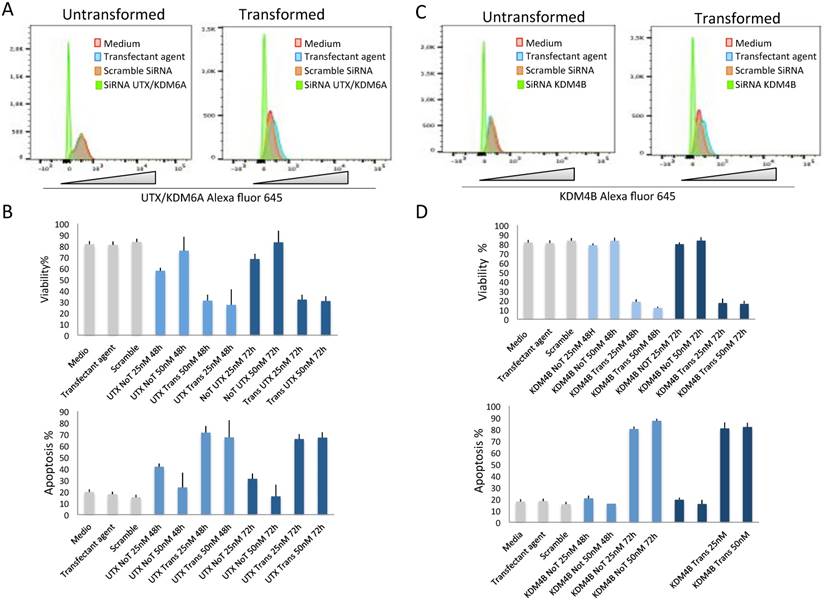
The fact that the levels of UTX/KDM6A and KDM4B increase after transformation and that in the case of KDM4B this increase correlates with its occupancy in repetitive DNA sequences in which its transcription levels are enhanced opens the question of whether these demethylases are required for the maintenance of transformed cells. To resolve this question, we transfected transformed and non-transformed cells with dsRNA against the UTX/KDM6A and KDM4B transcripts. The levels of UTX/KDM6A and KDM4B as well as the viability of the cells were analyzed after 48 and 72 hrs. At these two time points, there is a clear reduction in the two demethylases in both non-transformed and transformed cells transfected with the dsRNAs (Fig. 5A and 5C). Interestingly, the reduction of both enzymes significantly reduced the viability of the transformed cells compared with the non-transformed cells (Fig. 5B and 5D). This reduction in viability correlates with an increase in apoptosis in the transformed cells (Fig. 5B and 5D) and suggests that the transformed cells require high levels of UTX/KDM6A and KDM4B to survive.
An increase in transcription of percentromeric sequences in the transformed MCF10A-ER-Src cells correlates with a reduction in H3K9me3 and the presence of KDM4B in these repetitive elements. A, Semi quantitative RT-PCR experiments to determine the transcript levels of Sat a, SatIII and mcBox repetitive DNA elements in non-transformed and transformed cells. GADPH transcript was used as internal control. The number of amplification cycles is indicated at the bottom of the figure. B, ChIP-PCR experiments to determine the presence of H3K9me3 at the Sat a, Sat III and mcBox DNA elements in non-transformed and transformed cells. The input, mock and immunoprecipitation lanes are indicated. GADPH was used as negative control. C, ChIP-PCR experiments to determine the occupancy of KDM4B demethylase at the Sat a, Sat III and mcBox DNA elements in not-transformed and transformed cells. The promoter of the Zeb1 gene was used as a positive control, since it is known that the activation of this gene in cancer cells recruits KDM4B [37]. GADPH was used a negative control. The mock signal is used to determine nonspecificity in each ChIP experiment.
Discussion
It is known that the global configuration of chromatin is altered in cancer cells, which is observed as a preferential reduction in the levels of heterochromatin. For instance, recent analysis has found the loss of heterochromatin in large extensions of the genome in cancer cells [29, 30]. In addition, mutations in several factors involved in heterochromatin maintenance have been related to the cancerous phenotype [28, 29]. Interestingly, there are examples in which mutations that favor the formation or the decondensation of heterochromatin favor some types of tumors and suppress the cancerous phenotype in others [31-35, 36]. However, many of the studies analyzing the changes in heterochromatin in cancer cells are not based on comparative studies between the cancerous cell and its progenitor. In this work, we used the MCF10-Er-Src cell line to compare the changes that occur in heterochromatin during the generation of a transformed phenotype. In fact we corroborate the advantage to perform comparative experiments between a malignant cell and its direct precursor when we compare the levels of KDM4B as well as the transcript levels of the Sat a, SatIII and mcBox satellite sequences in other breast cancer lines. In particular we analyzed the MCF7 cell line that is positive for the Estrogen Receptor (ER) and the triple negative MDA-MB-231 together with the non-cancerous cell line MFC10A. Intriguingly, when we analyzed the levels of KDM4B, we found that also in these lines the KDM4B protein levels are higher than in the MCF10A non-cancerous line (Fig. S1). However, when we determined the Sat a, SatIII and mcBox transcript levels, we found that only Sat a and SatIII in the MDA-MB-231 and in the transformed MCF10A-ER-SRc cell lines are increased, if compared with the MCF10A line (Fig. 1S). From these experiments it is clear that comparative experiments between cell lines that have different origin are hard to be conclusive, since in the absence of the precursor cells for each cell line, the levels of KDM4B and of the satellite sequences before the transformation of these cells are unknown. However, our comparative results using the MCF10A-Er-Src line after and before transformation indicates that there are severe alterations in heterochromatin in the transformed cells, such as the reduction of heterochromatin centers and a partial delocation of HP1α.
The depletion of KDM4B and UTX/KDM6A preferentially kills transformed cells. A, Flow citometry quantification of cells stained with an antibody against UTX/KDM6A after the introduction of SiRNA (50 nM) against the UTX/KDM6A transcript. Note the high reduction of cells expressing the demethylase basal levels if compared with the controls. Scramble SiRNA and the transfectant agent were used as controls. B, Quantification of cell viability and apoptosis by flow citometry of transformed (indicated as UTX Trans) and non-transformed (indicated as UTX No T) cells at 25 and 50 nM of the UTX/KDM6A siRNA. Scramble SiRNA and the transfectant agent were used as controls. C, Flow citometry quantification of cells stained with an antibody against UTX/KDM6A after the introduction of SiRNA (50 nM) against the KDM6B transcript. Note the high reduction of cells expressing the demethylase if compared with the controls. Scramble SiRNA and the transfectant agent were used as controls. D, Quantification of cell viability and apoptosis by flow citometry of transformed (indicated as KDM4B Trans) and non-transformed cells (indicated as KDM4b no T) at 25 and 50 nM of the KDM4B siRNA. Scramble SiRNA and the transfectant agent were used as controls.
It is known that HP1α binds chromatin, recognizing H3K9me2 or H3K9me3 [1, 9]. We observed that the levels of H3K9me3 were reduced after transformation, suggesting that the delocation of HP1α from the heterochromatin bodies may be due to the reduction in H3K9me3 levels. In addition, the reduction in H3K9me3 in the transformed cells also correlates with an increase of the H3K9me3 demethylase KDM4B. The levels of the KDM4B enzyme rise significantly 36-48 hrs after the induction of transformation in the MCF10a-Er-Src cells, showing a rapid response during transformation to a cancerous phenotype. Contrary to KDM4B, the levels of KDM4A and KDM4C, which also demethylate H3K9me3 and H39me36, are reduced in the cancerous cells (Fig. 3A). These results show that these three demethylases are differentially regulated during acquisition of the transformed phenotype. This suggests that different factors may associate with these enzymes for performance of their functions and that the reduction in H3K9me3 levels may be controlled in part by KDM4B. However, we cannot disregard the possibility that the reduction in H3K9me3 levels may also be caused by an effect on the H3K9 methyl transferase SUV39H1, since the levels of this enzyme were also affected. Based on these results, we suggest that the combination of an increase of KDM4B and a reduction of SUV39H1 cause an important reduction in the global levels of tri-methylated H3K9 in transformed cells.
Repetitive DNA elements are organized as heterochromatin. Although these elements are transcribed, the loss of heterochromatin increases their transcription, generating genome instability [30, 35, 36]. In addition, it has been reported that mutations in the BRCA1 gene linked to breast cancer induce heterochromatin decondensation in repetitive DNA, increasing its transcription and promoting the generation of cancer [14]. It has also been reported that there is an expansion of SatII sequences mediated by an increase in transcription of this repetitive element in colon cancer cells [37]. Intriguingly, we found that the expression of the Sat a, SatIII and mcBox satellite sequences is increased in the transformed cells and that this increase correlates with a decrease in the H3K9me3 mark in these sequences. Furthermore, the occupancy of KDM4B is increased in these elements after transformation, suggesting that KDM4B is recruited to these DNA sequences, demethylating H3K9me3 and favoring the transcription of these sequences. Based on these observations, it could be important in future experiments to determine if the increase in the presence of transcripts of these repetitive elements contribute to generation of the cancerous phenotype. Additionally, it will be important to determine what recruits KDM4B to these sequences in the transformed cells as well as the occupancy of SUV39H1 in these regions. These observations agree with recent reports that show that KDM4B may participate in the generation of cancer [38, 39].
We also observed a global reduction in the tri-methylation of H3K27. The decrease in this histone mark correlates with an enrichment of the UTX/KDM6A demethylase, although there is also a reduction of KDM6B. Intriguingly, the levels of EzH2 and Suz12, components of PRC2, also decrease in the transformed cells. As mentioned before, there is evidence that some tumors are linked to an increase in PRC2 activities but that others are associated with a reduction of PRC2 [40]. These results suggest that the global reduction in H3K27me3 occurs with an increase in the UTX/KDM6A demethylase and a decrease in the levels of PRC2 in the MCF10A-Er-Src transformed cells, which may be related to the expression of genes that are necessary for the induction and maintenance of the transformed phenotype.
The high levels of KDM4B and UTX/KDM6A in the transformed cells seem to be necessary for viability of the transformed MCF10A-Er-Src cells since the reduction of both enzymes is more deleterious in these cells than in the progenitor cells. These results suggest that the decondensation of heterochromatic regions is required for the survival of cancerous cells. However, at this point, we do not know if the relaxation of heterochromatin is maintained in mamospheres derived from these cells or in these cells when they are introduced into animals that allow the development of tumors [16]. Although it is not possible to generalize that the relaxation of the heterochromatin is required for the acquisition of the cancerous phenotype in other cancer models, our results strongly suggest that dramatic changes in heterochromatin configuration occur soon after or during transformation and that these changes are important for the survival of cancer cells. In addition, these studies allow further exploration of the mechanisms that cause alterations to heterochromatin in the early stages of transformation, for instance to determine how KDM4B is recruited to heterochromatin. Furthermore, we have identified important candidates that participate in the heterochromatin dynamics during transformation that are suitable for global genomic analysis before and after the acquisition of a cancerous phenotype.
Supplementary Material
Figure S1.
Acknowledgements
This work was supported by grants from CONACyT 219673, CONACyT 250588 and PAPIIT/UNAM IN200315. We thank Arturo Pimentel, Andres Saralegui, Chris Wood and LNMA for the advice in the use of the confocal microscopes.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Eissenberg JC, Elgin SC. HP1a: a structural chromosomal protein regulating transcription. Trends Genet. 2014;30:103-10
2. Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications - writers that read. EMBO Rep. 2015;16:1467-81
3. Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL. et al. LINE-1 Activity in Facultative Heterochromatin Formation during X Chromosome Inactivation. Cell. 2016;141:956-69
4. Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503-18
5. Blackledge NP, Rose NR, Klose RJ. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol. 2015;16:643-9
6. Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297-311
7. Shpargel KB, Starmer J, Yee D, Pohlers M, Magnuson T. KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS Genetics. 2014:10 e1004507
8. Holowatyj A, Yang ZQ, Pile LA. Histone lysine demethylases in Drosophila melanogaster. Fly. 2015;9:36-44
9. Déjardin J. Switching between Epigenetic States at Pericentromeric Heterochromatin. Trends Genet. 2015;31:661-72
10. Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F. et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood. 2015;125:13-21
11. Benyoucef A, Palii GG, Wang C, Porter CJ, Chu, A Dai F. et al. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes Dev. 2016;30:508-21
12. Herz HM, Madden LD, Chen Z, Bolduc C, Buff E, Gupta R. et al. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol Cell. Biol. 2010;30:2485-97
13. Millanes-Romero A, Herranz N, Perrera V, Iturbide A, Loubat-Casanovas J, Gil J. et al. Regulation of heterochromatin transcription by Snail1/LOXL2 during epithelial-to-mesenchymal transition. Mol Cell. 2013;52:746-57
14. Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N. et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477:179-84
15. Berry WL, Janknecht R. KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res. 2013;73:2936-42
16. Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693-706
17. Fleming JD, Giresi PG, Lindahl-Allen M, Krall EB, Lieb JD, Struhl K. STAT3 acts through pre-existing nucleosome-depleted regions bound by FOS during an epigenetic switch linking inflammation to cancer. Epigenetics Chromatin. 2015:14 8:7
18. Janzer A, German NJ, Gonzalez-Herrera KN, Asara JM, Haigis MC, Struhl K. Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. K. Proc Natl Acad Sci U S A. 2014;111:10574-79
19. Hirsch HA, Iliopoulos D, Joshi A, Zhang Y, Jaeger SA, Bulyk M, Tsichlis PN, Shirley Liu X, Struhl K. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell. 2010;17:348-61
20. Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256-268
21. Schreiber E, Matthias P, Müller MM, Schaffner W. Rapid detection of octamer binding proteins with 'mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419
22. Gao C, Wang L, Milgrom E, Shen WC, On the mechanism of constitutive Pdr1 activator-mediated PDR5 transcription in Saccharomyces cerevisiae. evidence for enhanced recruitment of coactivators and altered nucleosome structures. J Biol Chem. 2004;279:42677-86
23. Krutzik PO, Clutter MR, Trejo A, Nolan GP. Fluorescent cell barcoding for multiplex flow cytometry. Curr. Protoc. Cytom. 2011 Chapter 6:Unit 6.31
24. Riclet R, Chendeb M, Vonesch JL, Koczan D, Thiesen HJ, Losson R, Cammas F. Disruption of the interaction between transcriptional intermediary factor 1{beta} and heterochromatin protein 1 leads to a switch from DNA hyper- to hypomethylation and H3K9 to H3K27 trimethylation on the MEST promoter correlating with gene reactivation. Mol Biol Cell. 2009;20:296-305
25. Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P. et al. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934-47
26. Oliveira C, Wright JM. Molecular cytogenetic analysis of heterochromatin in the chromosomes of tilapia, Oreochromis niloticus (Teleostei: Cichlidae). Chromosome Res. 1998;6:205-11
27. Valgardsdottir R, Chiodi I, Giordano M, Rossi A, Bazzini S, Ghigna C. et al. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res. 2008;36:423-34
28. Ferreira D, Meles S, Escudeiro A, Mendes-da-Silva A, Adega F, Chaves R. Satellite non-coding RNAs: the emerging players in cells, cellular pathways and cancer. Chromosome Res. 2015;23:479-93
29. Rafique S, Thomas JS, Sproul D, Bickmore WA. Estrogen-induced chromatin decondensation and nuclear re-organization linked to regional epigenetic regulation in breast cancer. Genome Biol. 2015;16:145
30. McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol. 2011;18:867-74
31. Nizialek AE, Sankunny M, Niazi F, Eng C. Cancer-predisposition gene KLLN maintains pericentric H3K9 trimethylation protecting genomic stability. Nucleic Acids Res. 2016;44:3586-94
32. Morgan SA, Shilatifard A. Chromatin signatures of cancer. Genes Dev. 2015;29:238-49
33. Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D. et al. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299-310
34. Wassef M, Margueron R. The Multiple Facets of PRC2 Alterations in Cancers. J Mol Biol. 2016;429:1978-1993
35. Frescas D, Guardavaccaro D, Kuchay SM, Kato H, Poleshko A, Basrur V. et al. KDM2A represses transcription of centromeric satellite repeats and maintains the heterochromatic state. Cell Cycle. 2008;7:3539-47
36. Bierhoff H, Postepska-Igielska A, Grummt I. Noisy silence: non-coding RNA and heterochromatin formation at repetitive elements. Epigenetics. 2014;9:53-61
37. Bersani F, Lee E, Kharchenko PV, Xu AW, Liu M, Xega K. et al. Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc Natl Acad Sci U S A. 2015;112:15148-53
38. Wilson C, Qiu L, Hong Y, Karnik T, Tadros G, Mau B. et al. The histone demethylase KDM4B regulates peritoneal seeding of ovarian cancer. Oncogene. 2016;36:2565-76
39. Li S, Wu L, Wang Q, Li Y, Wang X. KDM4B promotes epithelial-mesenchymal transition through up-regulation of ZEB1 in pancreatic cancer. Acta Biochim Biophys Sin. 2015;47:997-1004
40. Serresi M, Gargiulo G, Proost N, Siteur B, Cesaroni M, Koppens M, Xie H. et al. Polycomb Repressive Complex 2 Is a Barrier to KRAS-Driven Inflammation and Epithelial-Mesenchymal Transition in Non-Small-Cell Lung Cancer. Cancer Cell. 2016;29:17-31
Author contact
![]() Corresponding author: email: mariozunam.mx
Corresponding author: email: mariozunam.mx

 Global reach, higher impact
Global reach, higher impact