Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(15):4346-4356. doi:10.7150/jca.121650 This issue Cite
Review
Emerging Roles of PTTG1/Securin in Breast Cancer
1. Breast Disease Diagnosis and Treatment Center/Department of Thyroid Surgery, Central Hospital Affiliated to Shandong First Medical University, Jinan, 250013, China.
2. Research Center of Translational Medicine, Central Hospital Affiliated to Shandong First Medical University, Jinan, 250013, China.
3. Department of Internal Medicine-Oncology, Shandong Cancer Hospital and Institute, Shandong First Medical University and Shandong Academy of Medical Sciences, Jinan, 250117, China.
*These authors contributed equally.
Received 2025-7-14; Accepted 2025-10-15; Published 2025-10-27
Abstract

Securin is a key regulator of chromosome segregation during mitosis. Dysregulation of securin triggers chromosomal instability (CIN) and aneuploidy, which are hallmarks of many solid tumors, including breast cancer (BC). Recent studies have revealed securin's multifaceted roles in the progression of BC. Overexpression of securin not only enhances the malignant behaviors of BC cells but also correlates with poor clinical outcomes in patients, suggesting its potential as both a therapeutic target and prognostic biomarker. Although interest in securin is growing, comprehensive reviews on its role in BC are sparse. In this review, we summarize the biological function of securin. We then focus on the expression patterns of securin in BC and related experimental models, and their association with CIN. Subsequently, we discuss the significance of securin as a prognostic marker for BC. Lastly, we explore how securin influences the malignant behaviors of BC cells. This review emphasizes the critical connection between CIN and BC pathobiology mediated by securin and offers insights for future research into securin-related mechanisms and therapeutic strategies.
Keywords: PTTG1, securin, breast cancer, chromosomal instability, tumor progression
Introduction
The pituitary tumor-transforming gene (PTTG) was first isolated from GH4 rat pituitary tumor cells [1]. Subsequent structural homology analyses identified PTTG1 protein as vertebrate securin, a master mitotic regulator that ensures genomic stability by controlling sister chromatid separation through the inhibition of separase activity [2]. Characterized by cell cycle-dependent expression oscillations and dynamic subcellular localization, securin is a key regulator of chromosome segregation with potential roles in cell cycle control, transcriptional regulation, and DNA damage repair [3]. Since these cellular events are frequently dysregulated in cancers, much effort has been invested in examining the role of securin in human tumors. In breast cancer (BC), recent data involve securin expression and mutations in clinical outcome, making it a valuable prognostic marker and therapeutic target. Research into securin's interaction networks, post-translational modifications, and subcellular trafficking continues to delineate its complex roles in carcinogenesis.
BC in females is the second leading cause of global cancer incidence in 2022, with an estimated 2.3 million new cases, accounting for 11.6% of all cancer diagnoses [4]. Among women, it is the most commonly diagnosed cancer and the leading cause of cancer deaths worldwide [4]. Chromosomal instability (CIN) is a characteristic of BC and is directly related to a range of clinical presentations, including disease stage, metastasis, poor prognosis, and drug resistance [5, 6]. In this review, we summarize current evidence for the biological functions of securin as a prognostic marker in BC and investigate the association between securin and CIN in BC.
The structure and function of securin
The human gene PTTG1 is located on chromosome 5 and consists of five exons and four introns. It encodes the 202-amino acid protein securin, which is 90.6% and 89.6% identical in sequence to PTTG2 and PTTG3, respectively. Securin has an amino (N)-terminal regulatory domain and a carboxyl (C)-terminal functional domain [7]. The N-terminus includes a conserved destruction box (D-box) and a KEN-box. Securin degradation is primarily regulated by the anaphase-promoting complex/cyclosomeCDC20 (APC/CCDC20) through its D-box, with the KEN-box contributing less [8]. Securin contains two proline-rich motifs within its C-terminus, which together with a specific domain in the N-terminus constitute a Src homology 3 (SH3)-binding site [9] (Figure 1). Securin can bind to various SH-3-proteins with this site, and that is of crucial significance to securin's biological function [10].
Schematic illustration of mammalian securin protein. Mammalian securin contains a KEN box, a D-box, and a transactivating domain. DNA-binding and SH3-binding sites are indicated in red. Positions of the elements are shown by residue numbers.

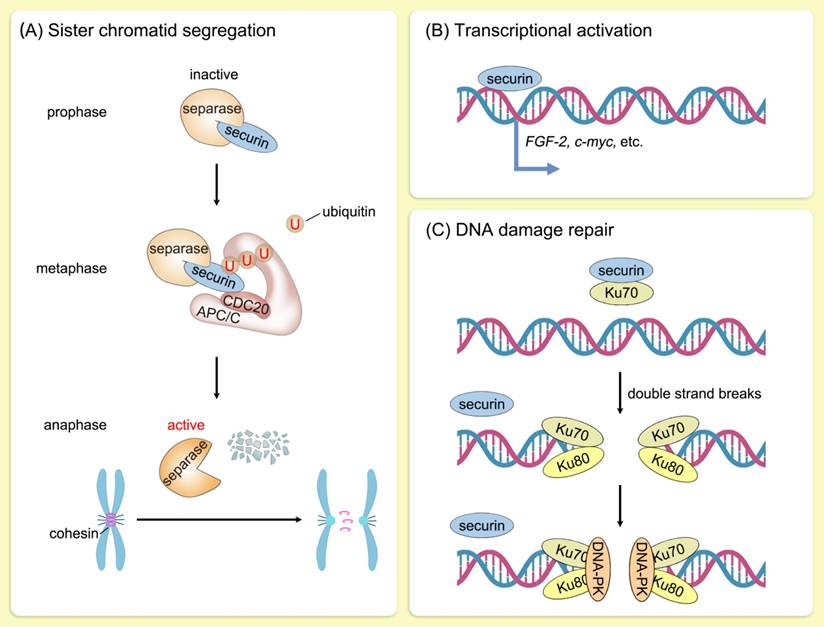
The most critical function of securin is to ensure faithful segregation of sister chromatids in mitosis. In human cells, replicated sister chromatids are held together by the cohesin complex from the S phase to early mitosis [11]. Removal of cohesin from arms of the chromosome is mediated by Sororin and wings apart-like protein homolog in prophase [12, 13], whereas removal of cohesin from centromeres is triggered by separase [14]. Securin regulates separase activity by a dual mechanism. Securin serves as a pseudosubstrate by blocking access to separase's C-terminal catalytic domain [15-17]. In addition, securin promotes nuclear translocation of separase for its acquisition of proteolytic activity [18]. During metaphase-anaphase transition, APC/CCDC20 ubiquitinates securin and releases separase to cleave centromeric cohesin [19, 20]. Sister chromatids are thus accurately segregated into two daughter cells, which is essential for maintaining genomic stability. The ubiquitination level of securin is directly regulated by its own phosphorylation and acetylation status and indirectly modulated by the APC/C activity [21-26]. Furthermore, RSUME extends the half-life of securin, and their co-expression leads to an increase in chromosomal abnormalities [27].
Furthermore, securin possesses intrinsic DNA-binding ability and functions as a transcription factor [9, 28]. The transcription of several genes, including c-myc, PKCβ-1, MEK1, MEK3, and HSP70, was activated following securin overexpression [29]. Furthermore, direct transcriptional control of the mitogenic and angiogenic factor FGF-2 by securin has been documented, dependent on the C-terminal proline-rich motifs of securin [30, 31]. Securin also directly regulates the expression and secretion of prolactin [32]. Tong et al. analyzed the influence of securin on the transcription of 20,000 genes, discovering that the majority of genes regulated by securin were involved in cell cycle, cell metabolism, or signal transduction, thus highlighting securin's role in the regulation of various cellular processes [33]. Securin is phosphorylated by MAPK, which dynamically regulates its transcriptional activation activity [34].
Securin was also identified as being involved in DNA damage repair. In mammalian cells, securin participates in DNA damage repair by interacting with Ku70, the regulatory subunit of DNA-dependent protein kinase (DNA-PK) [35, 36]. Upon DNA-damaging events, the securin-Ku70 complex is disrupted, thereby delaying mitosis onset [35] (Figure 2). In addition, decreased securin or PBF expression led to dysregulated expression of p53 target genes involved in DNA repair and apoptosis [37]. These findings further confirm the important role of securin in genomic stability.
Securin and CIN in BC
Expression of securin in BC and related experimental models
Numerous studies have examined the expression of securin in BC. Compared to normal tissues, securin is highly expressed in BC tissues, and is positively correlated with the tumor pathological grade [38, 39]. Additionally, the level of securin expression correlates with the degree of malignancy in BC cell lines; it is more highly expressed in malignant BC cell lines compared to normal or less malignant BC cells [40, 41]. Among the four subtypes of BC—HER2-positive, luminal A, luminal B, and triple-negative—securin shows higher expression in estrogen receptor (ER) negative BC tissues than in ER-positive tissues [42], with significant upregulation observed in the triple-negative breast cancer (TNBC) subgroup compared to other subgroups [43, 44]. Additionally, a significant correlation between keratin 67 (Ki67) and securin was observed in invasive BC [45].
It has been reported that securin is highly expressed in mammary gland epithelial cells and is necessary for proper morphogenesis of the mammary gland in mice [46]. From the early stages (4 weeks) to the later stages (13 weeks) of mammary gland development, securin is primarily expressed in the cap and body cells of the terminal end buds, as well as the single-layer luminal epithelium in the ductal region [46]. Securin regulates a number of genes associated with proliferation and mammary gland branching morphogenesis. Pttg1-null mice displayed increased proliferation and defects in branch patterning in their mammary glands [46]. Additionally, Pttg1-null mice exhibited testicular and splenic hypoplasia, thymic hyperplasia, and thrombocytopenia [47], indicating that securin maintains global chromosomal stability and cell cycle progression during embryonic and postnatal development. To date, no mammary gland-specific Pttg1 knockout or transgenic mouse model has been developed, which could be valuable for further investigating the role of securin in the development and carcinogenesis of the mammary gland.
Biological function of securin. Schematic illustration of the biological function of securin. (A) During metaphase-anaphase transition, APC/CCDC20 ubiquitinates securin and releases separase to cleave centromeric cohesin. Sister chromatids are thus accurately segregated into two daughter cells, which is essential for maintaining genomic stability. U, ubiquitin. (B) Securin functions as a transcription factor. The transcription of a series of genes, including c-myc and FGF-2, was activated following securin upregulation. (C) Securin participates in DNA damage repair by interacting with Ku70, the regulatory subunit of DNA-dependent protein kinase (DNA-PK). Upon DNA-damaging events, the securin-Ku70 complex is disrupted, thereby delaying mitosis onset.
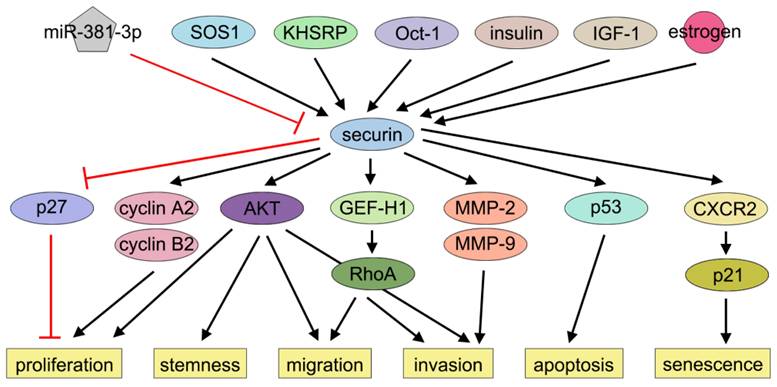
Multiple regulators of securin expression in BC have been identified. The transcription factor Oct-1 specifically bound to and transactivated the promoter of PTTG1, with a concurrent overexpression of Oct-1 and securin observed in BC [48]. Recent studies have further demonstrated that depletion of SOS1 or KHSRP downregulated securin in BC cells [49, 50]. Additionally, securin expression is modulated by certain natural hormones, such as estrogen and insulin, in MCF-7 cells [51, 52]. Some clinical chemicals also regulate securin expression. For instance, statins, which are 3-hydroxyl-3-methyl glutaryl coenzyme A reductase inhibitors, have shown unpredictable benefits in reducing BC progression and mortality [53, 54]. The stability of PTTG1 mRNA was markedly decreased by several lipophilic statins in MDA-MB-231 cells, providing insights into how statins prevent BC metastasis [41]. Furthermore, multiple noncoding RNAs have been predicted to modulate PTTG1 expression, though their biological functions require further experimental verification [43, 55].
Effects of securin on CIN in BC
CIN refers to an increased frequency of chromosomal alterations, including both numerical and structural aberrations [6]. CIN enables cancer cells to rapidly increase genomic complexity through the simultaneous acquisition of small- and large-scale losses, gains, and rearrangements of DNA, and is a hallmark of cancer [56]. CIN promotes tumor subclone diversification by amplifying intratumoral heterogeneity, while also enhancing phenotypic plasticity in response to selective pressures [57]. Paradoxically, CIN can suppress cancer cell survival due to prolonged mitotic duration and the generation of inviable karyotypes [58, 59]. Thus, understanding how cancer cell populations reach an optimal equilibrium for CIN rate is of considerable scientific interest.
Approximately 60-80% of BC deviate from a diploid karyotype, and invasive ductal carcinomas present aneuploidy significantly more often than other types of BC [60]. In terms of the receptor status, ER-negative and HER2-positive were associated with a higher degree of CIN [61-63]. Among luminal BC, luminal B had a higher rate of CIN than luminal A [62]. The causes of CIN are diverse, including mitotic errors, replication stress, impairment of homologous recombination, and telomere dysfunction [64]. CIN is associated with a wide range of clinical features of BC including disease stage, metastasis, prognosis, and therapeutic resistance [5, 6]. Although high levels of CIN have generally been linked with poor clinical outcomes in BC, some studies have found that extreme CIN correlates with a good prognosis [63, 65]. This paradox implies that the relationship between CIN and BC patient prognosis is complex. Therefore, identification of primary regulators controlling CIN in BC and classification of patients based on their degree of CIN is crucial for developing new therapeutic strategies and enabling personalized therapy. Accordingly, a number of therapeutics directed against CIN-related mechanisms are under development or have already been used clinically.
Considering the crucial role of securin in ensuring accurate chromosome separation, dysregulation of securin has been identified as a potential driver of CIN. However, to date, only a few studies have investigated the association between securin and CIN in BC. Ogbagabriel et al. found that securin overexpression correlated with the degree of nuclear pleomorphism, which is associated with CIN [66]. Karra et al. reported that high-level securin expression predicted an increased risk of aneuploidy in BC, highlighting the link between securin expression and aneuploidy [67, 68]. Talvinen et al. found that securin positivity predicted the occurrence of aneuploid DNA content [69]. Securin-induced aneuploidy was prevented by p53-dependent apoptosis in MCF-7 cells, and p53 suppresses hormone-induced tumor risk and the incidence of aneuploidy by inhibiting securin expression [70, 71]. Collectively, it is confirmed that securin promotes CIN through p53 in BC, but whether securin has other p53-independent mechanisms requires further investigation. Indirect evidence is provided by research into securin's transcription activation targets, such as FGF-2, c-myc, and SP1, all of which have been shown to associate with CIN in cancer [72-74]. Future research should concentrate on clarifying the p53-independent pathways by which securin causes CIN, perhaps through validating the cellular functions of its transcriptional targets or revealing new effectors. Additionally, exploring the therapeutic value of securin and its downstream effectors to suppress CIN and aneuploidy in BC is promising.
Securin as a prognostic marker of BC
Expression levels
Numerous studies have confirmed the prognostic value of securin in BC. By analyzing 90 BC and 18 normal breast tissues, Ogbagabriel et al. reported that securin overexpression correlated with the mitotic index, lymph node invasion, degree of nuclear pleomorphism, and ER-α expression [66]. Talvinen et al. focused on the expression of securin in invasive BC and found that securin immunopositivity was an independent prognosticator, predicting the survival of patients based on histological type, Ki-67 proliferation status, and tumor size [75, 76]. Karra et al. conducted a study on 603 BC patients and found that securin was a strong independent prognostic marker for survival [67]. Through comprehensive genomic analysis of Middle Eastern female patients with BC, Colak et al. identified age-specific gene signatures while systematically investigating molecular alterations associated with cancer progression in young women. Although PTTG1 was not incorporated into the age-specific gene subset, it was one of the 16 genes involved in tumorigenesis, invasion, and progression [77]. Vihervuori et al. investigated 179 TNBC patients with complete clinical data, noting that high expression of securin was significantly associated with a low fraction of tumor-infiltrating lymphocytes and CD8+ T cells, suggesting that the level of securin expression might be useful to evaluate the tumor inflammatory response in TNBC [78].
Meanwhile, the prognostic value of securin has been evaluated using publicly available clinical data by various researchers. They found that BC patients with higher levels of securin exhibited significantly poorer overall survival (OS), relapse-free survival (RFS), and distant metastasis-free survival (DMFS) compared to patients with relatively lower levels of securin [38-40, 43, 44, 49, 51, 79-81]. Additionally, high expression of securin was prevalent in metastatic BC tissues, suggesting its potential as a biomarker for BC metastasis [39, 82].
Furthermore, several studies have confirmed the prognostic accuracy of securin in BC when combined with other genes/proteins. Chen et al. detected circulating tumor cells (CTCs) in the peripheral blood of female BC patients using a panel of four genes: PTTG1, BIRC5, UBCH10, and TK1. They found that tumor size, histological grade, lymph node metastasis, and TNM stage were significantly correlated with the positive detection rate of the multimarker panel [83]. Talvinen et al. observed that combined detection of cdc27 and securin predicted cancer death in BC [69]. Karra et al. investigated 445 patients with BC and found that high expression levels of Cdc20 and securin demonstrated a 6.8-fold increased mortality risk [68]. Notably, this dual protein overexpression pattern was predominantly observed in TNBC patients and was particularly associated with the subgroup exhibiting extremely short survival (on average 2.4 years) [68]. Repo et al. developed a prognostic model combining securin, separase, and Cdk1 for predicting tumor size, histological grade, axillary lymph node status, and the risk of mortality, based on a study comprised of 1,135 BC patients [84]. Additionally, the OncoMasTR Molecular Score (OMm) is identified as a combination of master transcriptional regulators: FOXM1, PTTG1, and ZNF367. OMclin1 combines OMm with nodal status, tumor size, and grade, and OMclin2 is a simpler form of OMclin1 that excludes tumor grade [85]. Buus et al. reported that OMm, OMclin1, and OMclin2 were highly prognostic for early and late distant recurrence in women with early-stage ER-positive BC receiving 5 years of endocrine therapy [85]. Collectively, securin expression serves as a strong prognosticator of BC outcomes.
Single nucleotide polymorphism (SNP)
SNPs are single-base differences in the DNA sequence that occur with a frequency of ≥1% and may serve as biological markers for disease-associated genes [86]. Functional polymorphisms that modulate gene expression may underlie interindividual variation in BC susceptibility and clinical outcomes [87]. Lo et al. examined SNPs in several mitotic checkpoint genes in 698 primary BC patients and 1,492 healthy controls, and observed that for the SNP rs2910203 in the PTTG1 gene, carriers of the C1892G genotype had a significantly higher risk of BC [88]. Additionally, a combined effect of SNPs in TTK, BUB1B, and PTTG1 on BC risk was confirmed through genotype/haplotype analysis [88]. Brendle et al. investigated whether SNPs in CIN-related genes affect BC risk and clinical outcomes in a Swedish cohort of BC cases and found that for the SNP rs1862392 in the PTTG1 gene, carriers of the TA genotype were more likely to have tumors with regional lymph node metastasis compared to carriers of the wild-type genotype [89]. These findings suggest that functional polymorphisms in PTTG1 influence BC susceptibility and progression. Future studies should confirm the efficiency of these SNPs as biomarkers and investigate how they mechanistically regulate securin function.
Subcellular localization
Accumulating evidence has demonstrated that the subcellular localization of securin also has a prognostic impact on BC. However, inconsistencies remain among the conclusions of relevant studies. Gurvits et al. reported that cytoplasmic securin expression in BC cells was associated with aggressive subtypes and high mortality rates [90]. Securin exhibited low (or absent) nuclear expression in benign breast epithelia and luminal carcinomas, whereas HER2-amplified and triple-negative carcinomas showed marked cytoplasmic overexpression [90]. Similarly, Repo et al. found that high expression and cytoplasmic localization of securin were directly associated with aggressive tumor features and poorer patient survival in BC [91]. In patients with TNBC, cytoplasmic securin localization correlated with a significantly elevated mortality risk and a reduced estimated 5-year survival rate compared to patients with predominantly nuclear securin expression [91]. In contrast, another study conducted by Repo et al. in 2020 reported that securin was predominantly localized in the nucleus, though both nuclear and/or cytoplasmic immunoreactivity were occasionally observed in invasive BC [84]. Future studies analyzing additional BC samples across different subtypes are needed to clarify the correlation between securin subcellular localization patterns and clinical outcomes. In addition, it has been reported that SPTBN1 mediates the cytoplasmic constraint of securin and impaired its oncogenic activity in human seminoma [92]. Considering that SPTBN1 also regulates cell growth and EMT in BC [93], it is imperative to conduct a combined analysis of the expression and subcellular localization of both securin and SPTBN1 in BC.
DNA methylation
DNA methylation, one of the most widely studied epigenetic modifications, refers to the transfer of a methyl group from S-adenosylmethionine to cytosine residues within CpG dinucleotides by DNA methyltransferases. DNA hypermethylation and hypomethylation are both implicated in the development and prognosis of BC [94]. Qi et al. identified aberrantly expressed hub genes potentially regulated by DNA methylation in BC and found that PTTG1 was one of the 12 hub genes whose promoter was hypomethylated in BC and was accountable for significantly poor clinical outcomes [95]. These findings suggest that DNA methylation is potentially a major cause of abnormal expression of PTTG1 in BC.
Effects of securin on BC progression
Cell proliferation, stemness, migration, and invasion
Over the past decades, several studies have shown that securin is associated with BC cell proliferation. Xie and Wang demonstrated that securin overexpression promoted MCF-7 cell proliferation by inducing nuclear exclusion of the cyclin-dependent kinase inhibitor 1B (p27), thereby alleviating p27-mediated G1-phase arrest [96]. Khazaei et al. reported that securin knockdown by siRNA inhibited MDA-MB-231 cell proliferation [42]. Meng et al. found that securin enhanced BC cell viability and proliferation by regulating cyclin (CCNA2 and CCNB2) expression [51].
Securin also plays a role in regulating the stemness, migration, and invasion of BC cells. Liao et al. reported that knockdown of securin reduced motility, invasion, and metastasis of BC cells by suppressing Rho guanine nucleotide exchange factor-H1 (GEF-H1) expression and RhoA activation. In contrast, overexpression of securin had the opposite effects [97]. Yoon et al. reported that securin promoted BC cell migration and invasion through epithelial-to-mesenchymal transition (EMT). This study further revealed that downregulation of securin reduced the self-renewal capacity and tumorigenic potential of BT549 cells, while securin overexpression had the opposite effects. Mechanistically, these effects were partially attributed to AKT activation, a key signaling pathway governing EMT and stemness in cancer cells [40]. Yin et al. reported that knockdown of securin markedly inhibited MDA-MB-231 cell invasion and the activities of MMP-2 and MMP-9, while ectopic expression of securin promoted BC cell invasion [41]. In the investigation of bisphenol A (BPA), a mass-produced industrial compound implicated in BC progression, Deng et al. observed that BPA exposure significantly promoted BC cell proliferation and migration but not invasion. Subsequent bioinformatic analysis identified securin as one of the key downstream targets of BPA. BPA elevated securin protein level by suppressing miR-381-3p, and securin knockdown attenuated BPA-induced MCF-7 cell proliferation and migration [55]. Xing et al. demonstrated that securin acts as a downstream effector of SOS1, modulating cancer stemness and M2 macrophage polarization [49]. Li et al. performed single-cell RNA sequencing on 16 paired samples of primary BC and metastatic lymph nodes and observed that dual-positive RAC2/securin BC stem cells, which exhibit the highest stem-like traits, were markedly enriched in metastatic lymph nodes. These cells may drive tumor metastasis by activating metastasis-promoting transcription factors and signaling cascades [98]. Future research could focus on the influence of securin on BC cell cycle regulation and the effects of its post-translational modifications on cell proliferation, stemness, migration, and invasion.
Apoptosis
Contradictory evidence persists regarding securin's regulation of apoptotic pathways in BC cells. Yu et al. demonstrated that securin overexpression induced p53-dependent apoptosis in MCF-7 cells [71]. Similarly, Hamid and Kakar reported that securin overexpression elevated Bax expression and triggered apoptosis through p53 in MCF-7 cells [99]. However, Xie and Wang found that knockdown of securin did not alter apoptosis in MCF-7 cells [96].
Several possibilities could account for these contradictions. For instance, functional redundancy within the PTTG gene family may enable compensatory mechanisms, where paralogous genes maintain apoptotic homeostasis when securin is silenced. In addition, the efficiency of the shRNA used for securin knockdown must be systematically evaluated, as insufficient knockdown may fail to reach the threshold required to impact apoptosis. Alternatively, supraphysiological overexpression of securin might induce non-specific cellular stress responses, leading to cell death. Collectively, further investigations across diverse BC cell lines under varying experimental conditions are warranted to clarify the influence of securin on cell apoptosis.
Senescence
Cellular senescence, a state of prolonged cell cycle arrest, is considered an important tumor-suppressive mechanism [100, 101]. Chen et al. and Yu et al. demonstrated that securin played a crucial role in determining radiosensitivity and cell fate. Radiation induced apoptosis in wild-type cells but induced senescence in securin-null cells. Moreover, the knockdown of securin switched irradiation-induced apoptosis to senescence in MDA-MB-231 cells [102, 103]. Ruan et al. reported that securin overexpression reinforced senescence through CXCR2 signaling, while escape from CXCR2/p21-dependent senescence surveillance was essential for securin's oncogenic effects [104] (Figure 3). Thus, securin acts as a key regulator of radiation-induced cell fate decisions in BC. More studies need to elucidate how securin controls the apoptosis-senescence transition following irradiation and how this pathway can be therapeutically modulated.
Chemosensitivity and resistance
Studies have confirmed that securin is associated with sensitivity or resistance to multiple anticancer drugs. Ghayad et al. explored new biomarkers of endocrine resistance in ERα-positive BC and found that PTTG1 was significantly overexpressed in samples from patients who relapsed after tamoxifen treatment compared to those who did not. Furthermore, PTTG1 was identified as an independent prognostic marker correlated with significantly shorter RFS [105]. Yu et al. reported that gefitinib, a tyrosine kinase inhibitor targeting EGFR, downregulates securin at the protein level by reducing its stability without affecting gene transcription. Securin overexpression increased both proliferation rates and resistance to gefitinib-induced death in several cancer cell lines, indicating that high securin levels may confer cellular resistance to gefitinib [106]. Yin et al. demonstrated that ectopic expression of securin partially reversed simvastatin-mediated inhibition of cell invasion in MDA-MB-231 cells [41]. Bashari et al. investigated candidate genes associated with drug resistance and susceptibility in TNBC using data from PharmacoDB and found that high expression levels of PTTG1 were significantly correlated with sensitivity to IKK 16 and bromopyruvic acid [44]. Targeted strategies for inhibiting securin transcription or decreasing protein stability in conjunction with EGFR inhibitors or tamoxifen may address both the underlying mechanisms and translation opportunities against securin-mediated chemoresistance.
Effects of securin on BC progression. Securin exerts multiple effects, such as proliferation, migration, invasion, and apoptosis via many signaling pathways in BC.
Adverse reactions following radiotherapy
The relationship between PTTG1 and adverse reactions after radiotherapy has produced conflicting conclusions. Suga et al. identified haplotypes of SNPs associated with the risk of early adverse skin reactions (EASRs) after radiotherapy in 399 Japanese patients with BC, and found that the GTTG haplotype in PTTG1 was associated with a significantly reduced risk of EASRs [107]. Aguiar et al. evaluated the association between SNPs and acute radiation dermatitis (RD) in patients with BC and found that four SNPs in PTTG1 were associated with RD [108]. However, Murray et al. and Mumbreker et al., analyzed distinct cohorts of BC patients, reported no significant association between SNPs in the PTTG1 gene and EASRs following radiotherapy [109, 110]. Discrepancies among these findings could be attributed to limited sample sizes and geographic heterogeneity among patient populations. Nevertheless, a general consensus has emerged that complex clinical manifestations, such as EASRs, arise from the interplay of multiple molecular pathways and pathophysiological mechanisms. Therefore, further studies are needed to clarify the connection between genetic variations in PTTG1 and adverse reactions following radiotherapy in BC patients.
Conclusions
In this review, we systematically examined the multiple roles of securin in BC. While substantial progress has been made over the past decades in delineating securin's involvement in the regulation of CIN and BC progression, several uncharted areas demand further exploration to fully realize its diagnostic, prognostic, and therapeutic potential. Future investigations should rigorously characterize disparities in the pathobiological role of securin across geographically diverse populations and molecular subtypes of BC, with particular attention to variations in genetic backgrounds, environmental exposures, and healthcare disparities that may influence its oncogenic functions, therapeutic responses, and prognostic significance. Additionally, expanded studies focusing on the pharmacogenomic landscape of securin, especially its role in modulating therapeutic sensitivity and resistance, will be valuable. SECURA-3, a novel RNA aptamer targeting securin developed in 2020, displayed high detection sensitivity in MCF-7 and HeLa cells [111]. Future studies can focus on exploiting its diagnostic potential and evaluating its therapeutic applications.
Elucidating the multidimensional protein interactome through which securin governs CIN—including separase regulation, DNA damage response coordination, and cell cycle checkpoint control—is equally critical for clarifying its roles in BC progression. To this end, integrating high-throughput interactome screening, multi-omics profiling, and single-cell sequencing approaches may provide key insights. As targeting CIN gains increasing therapeutic momentum, decoding securin's interaction network holds promise for breakthroughs in BC management. Furthermore, the generation of tissue-specific Pttg1 knockout or transgenic mouse models is necessary to better elucidate the physiological and pathological roles of securin in the mammary gland.
Acknowledgements
Funding
This work was supported by the Medical and Health Science and Technology Development Plan of Shandong Province (202402050868, 202304080737), project ZR2022LZL005 supported by Shandong Provincial Natural Science Foundation, the Postdoctoral Innovation Project of Shandong Province (SDCX-ZG-202201001), and the Foundation for High-Level Talents in Medical and Health of Jinan (202412).
Author contributions
Data authentication is not applicable. All authors have read, have contributed equally and approved the final version of the manuscript.
Competing interests
The authors have declared that no competing interest exists.
References
1. Pei L, Melmed S. Isolation and characterization of a pituitary tumor-transforming gene (PTTG). Mol Endocrinol. 1997;11:433-41
2. Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418-22
3. Vergani E, Teveroni E, Mancini F, Di Nicuolo F, Raia S, Chiloiro S. et al. Pituitary tumor-transforming gene 1 and endocrine cancers: an up-to-date review through history, current insights and future perspectives. Endocr Relat Cancer. 2025;32:e240163
4. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-63
5. Tijhuis AE, Johnson SC, McClelland SE. The emerging links between chromosomal instability (CIN), metastasis, inflammation and tumour immunity. Mol Cytogenet. 2019;12:17
6. Milán M. Chromosomal instability in development and disease: Beyond cancer evolution. Curr Opin Cell Biol. 2025;95:102537
7. Zhang X, Horwitz GA, Prezant TR, Valentini A, Nakashima M, Bronstein MD. et al. Structure, expression, and function of human pituitary tumor-transforming gene (PTTG). Mol Endocrinol. 1999;13:156-66
8. Tian W, Li B, Warrington R, Tomchick DR, Yu H, Luo X. Structural analysis of human Cdc20 supports multisite degron recognition by APC/C. Proc Natl Acad Sci U S A. 2012;109:18419-24
9. Domínguez A, Ramos-Morales F, Romero F, Rios RM, Dreyfus F, Tortolero M. et al. hpttg, a human homologue of rat pttg, is overexpressed in hematopoietic neoplasms. Evidence for a transcriptional activation function of hPTTG. Oncogene. 1998;17:2187-93
10. Liu J, Wang D, Li Y, Yao H, Zhang N, Zhang X. et al. Integrated in Silico-In Vitro Identification and Characterization of the SH3-Mediated Interaction between Human PTTG and its Cognate Partners in Medulloblastoma. Cell Biochem Biophys. 2018;76:83-90
11. Ochs F, Green C, Szczurek AT, Pytowski L, Kolesnikova S, Brown J. et al. Sister chromatid cohesion is mediated by individual cohesin complexes. Science. 2024;383:1122-30
12. Nishiyama T, Sykora MM, Huis in 't Veld PJ, Mechtler K, Peters JM. Aurora B and Cdk1 mediate Wapl activation and release of acetylated cohesin from chromosomes by phosphorylating Sororin. Proc Natl Acad Sci U S A. 2013;110:13404-9
13. Yuan X, Yan L, Chen Q, Zhu S, Zhou X, Zeng LH. et al. Molecular mechanism and functional significance of Wapl interaction with the Cohesin complex. Proc Natl Acad Sci U S A. 2024;121:e2405177121
14. Sun YX, Kucej M, Fan HY, Yu H, Sun QY, Zou H. Separase Is Recruited to Mitotic Chromosomes to Dissolve Sister Chromatid Cohesion in a DNA-Dependent Manner. Cell. 2009;137:123-32
15. Boland A, Martin TG, Zhang Z, Yang J, Bai XC, Chang L. et al. Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution. Nat Struct Mol Biol. 2017;24:414-8
16. Yu J, Raia P, Ghent CM, Raisch T, Sadian Y, Cavadini S. et al. Structural basis of human separase regulation by securin and CDK1-cyclin B1. Nature. 2021;596:138-42
17. Luo S, Tong L. Structural biology of the separase-securin complex with crucial roles in chromosome segregation. Curr Opin Struct Biol. 2018;49:114-22
18. Hornig NC, Knowles PP, McDonald NQ, Uhlmann F. The dual mechanism of separase regulation by securin. Curr Biol. 2002;12:973-82
19. Thomas C, Wetherall B, Levasseur MD, Harris RJ, Kerridge ST, Higgins JMG. et al. A prometaphase mechanism of securin destruction is essential for meiotic progression in mouse oocytes. Nat Commun. 2021;12:4322
20. Sorensen Turpin CG, Sloan D, LaForest M, Klebanow L, Mitchell D, Severson AF. et al. Securin regulates the spatiotemporal dynamics of separase. J Cell Biol. 2025;224:e202312099
21. Hellmuth S, Bottger F, Pan C, Mann M, Stemmann O. PP2A delays APC/C-dependent degradation of separase-associated but not free securin. EMBO J. 2014;33:1134-47
22. Wang T, Zou Y, Meng H, Zheng P, Teng J, Huang N. et al. Securin acetylation prevents precocious separase activation and premature sister chromatid separation. Curr Biol. 2024;34:1295-308 e5
23. Zhou CJ, Wang XY, Dong YH, Wang DH, Han Z, Zhang XJ. et al. CENP-F-dependent DRP1 function regulates APC/C activity during oocyte meiosis I. Nat Commun. 2022;13:7732
24. Sun SM, Zhao BW, Li YY, Liu HY, Xu YH, Yang XM. et al. Loss of UBE2S causes meiosis I arrest with normal spindle assembly checkpoint dynamics in mouse oocytes. Development. 2024;151:dev202285
25. Lin Y, Wei Z, Zhang L, Yao Y, Huang Y, Yao G. et al. Homozygous missense variations of APC12 cause meiotic metaphase I arrest in oocytes and female infertility. Am J Obstet Gynecol. 2025;232:547 e1- e17
26. Mora-Santos M, Limon-Mortes MC, Giraldez S, Herrero-Ruiz J, Saez C, Japon MA. et al. Glycogen synthase kinase-3beta (GSK3beta) negatively regulates PTTG1/human securin protein stability, and GSK3beta inactivation correlates with securin accumulation in breast tumors. J Biol Chem. 2011;286:30047-56
27. Fuertes M, Sapochnik M, Tedesco L, Senin S, Attorresi A, Ajler P. et al. Protein stabilization by RSUME accounts for PTTG pituitary tumor abundance and oncogenicity. Endocr Relat Cancer. 2018;25:665-76
28. Wang Z, Melmed S. Pituitary tumor transforming gene (PTTG) transforming and transactivation activity. J Biol Chem. 2000;275:7459-61
29. Pei L. Identification of c-myc as a down-stream target for pituitary tumor-transforming gene. J Biol Chem. 2001;276:8484-91
30. McCabe CJ, Boelaert K, Tannahill LA, Heaney AP, Stratford AL, Khaira JS. et al. Vascular endothelial growth factor, its receptor KDR/Flk-1, and pituitary tumor transforming gene in pituitary tumors. J Clin Endocrinol Metab. 2002;87:4238-44
31. Chien W, Pei L. A novel binding factor facilitates nuclear translocation and transcriptional activation function of the pituitary tumor-transforming gene product. J Biol Chem. 2000;275:19422-7
32. Horwitz GA, Miklovsky I, Heaney AP, Ren SG, Melmed S. Human pituitary tumor-transforming gene (PTTG1) motif suppresses prolactin expression. Mol Endocrinol. 2003;17:600-9
33. Tong Y, Tan Y, Zhou C, Melmed S. Pituitary tumor transforming gene interacts with Sp1 to modulate G1/S cell phase transition. Oncogene. 2007;26:5596-605
34. Pei L. Activation of mitogen-activated protein kinase cascade regulates pituitary tumor-transforming gene transactivation function. J Biol Chem. 2000;275:31191-8
35. Romero F, Multon MC, Ramos-Morales F, Domínguez A, Bernal JA, Pintor-Toro JA. et al. Human securin, hPTTG, is associated with Ku heterodimer, the regulatory subunit of the DNA-dependent protein kinase. Nucleic Acids Res. 2001;29:1300-7
36. Bernal JA, Roche M, Mendez-Vidal C, Espina A, Tortolero M, Pintor-Toro JA. Proliferative potential after DNA damage and non-homologous end joining are affected by loss of securin. Cell Death Differ. 2008;15:202-12
37. Read ML, Modasia B, Fletcher A, Thompson RJ, Brookes K, Rae PC. et al. PTTG and PBF Functionally Interact with p53 and Predict Overall Survival in Head and Neck Cancer. Cancer Res. 2018;78:5863-76
38. Wu CC, Ekanem TI, Phan NN, Loan DTT, Hou SY, Lee KH. et al. Gene signatures and prognostic analyses of the Tob/BTG pituitary tumor-transforming gene (PTTG) family in clinical breast cancer patients. Int J Med Sci. 2020;17:3112-24
39. Hong Z, Wang Q, Hong C, Liu M, Qiu P, Lin R. et al. Identification of Seven Cell Cycle-Related Genes with Unfavorable Prognosis and Construction of their TF-miRNA-mRNA regulatory network in Breast Cancer. J Cancer. 2021;12:740-53
40. Yoon CH, Kim MJ, Lee H, Kim RK, Lim EJ, Yoo KC. et al. PTTG1 oncogene promotes tumor malignancy via epithelial to mesenchymal transition and expansion of cancer stem cell population. J Biol Chem. 2012;287:19516-27
41. Yin L, He Z, Yi B, Xue L, Sun J. Simvastatin Suppresses Human Breast Cancer Cell Invasion by Decreasing the Expression of Pituitary Tumor-Transforming Gene 1. Front Pharmacol. 2020;11:574068
42. Khazaei G, Shamsabadi FT, Yamchi A, Golalipour M, Jhingan GD, Shahbazi M. Proteomics evaluation of MDA-MB-231 breast cancer cells in response to RNAi-induced silencing of hPTTG. Life Sci. 2019;239:116873
43. Mei L. Multiple types of noncoding RNA are involved in potential modulation of PTTG1's expression and function in breast cancer. Genomics. 2022;114:110352
44. Bashari N, Naghizadeh M, Chegini MK, Sadeghi ES, Zamani A, Mahdevar M. Therapeutic Potential of PLK1, KIF4A, CDCA5, UBE2C, CDT1, SKA3, AURKB, and PTTG1 Genes in Triple-Negative Breast Cancer: Correlating Their Expression with Sensitivity to GSK 461364 and IKK 16 Drugs. Biochem Genet. 2024
45. Talaat IM, Hamoudi RA, Yakout NM, Oweiss NY, Omar AM. Correlation of securin and Ki67 in invasive breast carcinoma. Histol Histopathol. 2019;34:697-709
46. Hatcher RJ, Dong J, Liu S, Bian GX, Contreras A, Wang T. et al. Pttg1/securin is required for the branching morphogenesis of the mammary gland and suppresses mammary tumorigenesis. Proc Natl Acad Sci U S A. 2014;111:1008-13
47. Wang ZY, Yu R, Melmed S. Mice lacking pituitary tumor transforming gene show testicular and splenic hypoplasia, thymic hyperplasia, thrombocytopenia, aberrant cell cycle progression, and premature centromere division. Mol Endocrinol. 2001;15:1870-9
48. Zhou C, Tong Y, Wawrowsky K, Bannykh S, Donangelo I, Melmed S. Oct-1 induces pituitary tumor transforming gene expression in endocrine tumors. Endocr Relat Cancer. 2008;15:817-31
49. Xing F, Zhao D, Wu SY, Tyagi A, Wu K, Sharma S. et al. Epigenetic and Posttranscriptional Modulation of SOS1 Can Promote Breast Cancer Metastasis through Obesity-Activated c-Met Signaling in African-American Women. Cancer Res. 2021;81:3008-21
50. Paizula X, Wulaying A, Chen D, Ou JH. KHSRP has oncogenic functions and regulates the expression and alternative splicing of DNA repair genes in breast cancer MDA-MB-231 cells. Sci Rep. 2024;14:14694
51. Meng CH, Zou Y, Hong WW, Bao CH, Jia XF. Estrogen-regulated PTTG1 promotes breast cancer progression by regulating cyclin kinase expression. Mol Med. 2020;26:33
52. Thompson AD, Kakar SS. Insulin and IGF-1 regulate the expression of the pituitary tumor transforming gene (PTTG) in breast tumor cells. FEBS Lett. 2005;579:3195-200
53. Scott OW, Tin Tin S, Cavadino A, Elwood JM. Statin use and breast cancer-specific mortality and recurrence: a systematic review and meta-analysis including the role of immortal time bias and tumour characteristics. Br J Cancer. 2025;133:539-54
54. Wang Z, Shi M, Liu B, Zhang X, Lin W, Yang Y. et al. Low-dose statins restore innate immune response in breast cancer cells via suppression of mutant p53. Front Pharmacol. 2025;16:1492305
55. Deng P, Tan M, Zhou W, Chen C, Xi Y, Gao P. et al. Bisphenol A promotes breast cancer cell proliferation by driving miR-381-3p-PTTG1-dependent cell cycle progression. Chemosphere. 2021;268:129221
56. Nguyen MP, Chen WC, Mirchia K, Choudhury A, Zakimi N, Nitturi V. et al. Pan-cancer copy number analysis identifies optimized size thresholds and co-occurrence models for individualized risk stratification. Nat Commun. 2025;16:6024
57. Lukow DA, Sausville EL, Suri P, Chunduri NK, Wieland A, Leu J. et al. Chromosomal instability accelerates the evolution of resistance to anti-cancer therapies. Dev Cell. 2021;56:2427-39 e4
58. Sala R, Farrell KC, Stearns T. Growth disadvantage associated with centrosome amplification drives population-level centriole number homeostasis. Mol Biol Cell. 2020;31:2646-56
59. Dias Louro MA, Bettencourt-Dias M, Bank C. Patterns of selection against centrosome amplification in human cell lines. PLoS Comput Biol. 2021;17:e1008765
60. Donepudi MS, Kondapalli K, Amos SJ, Venkanteshan P. Breast cancer statistics and markers. J Cancer Res Ther. 2014;10:506-11
61. Ellsworth RE, Ellsworth DL, Patney HL, Deyarmin B, Love B, Hooke JA. et al. Amplification of HER2 is a marker for global genomic instability. BMC Cancer. 2008 8
62. Smid M, Hoes M, Sieuwerts AM, Sleijfer S, Zhang Y, Wang Y. et al. Patterns and incidence of chromosomal instability and their prognostic relevance in breast cancer subtypes. Breast Cancer Res Treat. 2011;128:23-30
63. Birkbak NJ, Eklund AC, Li QY, McClelland SE, Endesfelder D, Tan P. et al. Paradoxical Relationship between Chromosomal Instability and Survival Outcome in Cancer. Cancer Res. 2011;71:3447-52
64. Bakhoum SF, Cantley LC. The Multifaceted Role of Chromosomal Instability in Cancer and Its Microenvironment. Cell. 2018;174:1347-60
65. Jamal-Hanjani M, A'Hern R, Birkbak NJ, Gorman P, Gronroos E, Ngang S. et al. Extreme chromosomal instability forecasts improved outcome in ER-negative breast cancer: a prospective validation cohort study from the TACT trial. Ann Oncol. 2015;26:1340-6
66. Ogbagabriel S, Fernando M, Waldman FM, Bose S, Heaney AP. Securin is overexpressed in breast cancer. Mod Pathol. 2005;18:985-90
67. Karra H, Pitkanen R, Nykanen M, Talvinen K, Kuopio T, Soderstrom M. et al. Securin predicts aneuploidy and survival in breast cancer. Histopathology. 2012;60:586-96
68. Karra H, Repo H, Ahonen I, Loyttyniemi E, Pitkanen R, Lintunen M. et al. Cdc20 and securin overexpression predict short-term breast cancer survival. Br J Cancer. 2014;110:2905-13
69. Talvinen K, Karra H, Pitkanen R, Ahonen I, Nykanen M, Lintunen M. et al. Low cdc27 and high securin expression predict short survival for breast cancer patients. APMIS. 2013;121:945-53
70. Pati D, Haddad BR, Haegele A, Thompson H, Kittrell FS, Shepard A. et al. Hormone-induced chromosomal instability in p53-null mammary epithelium. Cancer Res. 2004;64:5608-16
71. Yu R, Heaney AP, Lu W, Chen J, Melmed S. Pituitary tumor transforming gene causes aneuploidy and p53-dependent and p53-independent apoptosis. J Biol Chem. 2000;275:36502-5
72. Astrinidis A, Kim J, Kelly CM, Olofsson BA, Torabi B, Sorokina EM. et al. The Transcription Factor SP1 Regulates Centriole Function and Chromosomal Stability Through a Functional Interaction with the Mammalian Target of Rapamycin/Raptor Complex. Genes Chromosomes Cancer. 2010;49:282-97
73. Pecqueux C, Arslan A, Heller M, Falkenstein M, Kaczorowski A, Tolstov Y. et al. FGF-2 is a driving force for chromosomal instability and a stromal factor associated with adverse clinico-pathological features in prostate cancer. Urol Oncol. 2018;36:365.e15-e26
74. Bastianello G, Kidiyoor GR, Lowndes C, Li Q, Bonnal R, Godwin J. et al. Mechanical stress during confined migration causes aberrant mitoses and c-MYC amplification. Proc Natl Acad Sci U S A. 2024;121:e2404551121
75. Talvinen K, Tuikkala J, Nevalainen O, Rantanen A, Hirsimaki P, Sundstrom J. et al. Proliferation marker securin identifies favourable outcome in invasive ductal breast cancer. Br J Cancer. 2008;99:335-40
76. Talvinen K, Karra H, Hurme S, Nykanen M, Nieminen A, Anttinen J. et al. Securin promotes the identification of favourable outcome in invasive breast cancer. Br J Cancer. 2009;101:1005-10
77. Colak D, Nofal A, Albakheet A, Nirmal M, Jeprel H, Eldali A. et al. Age-specific gene expression signatures for breast tumors and cross-species conserved potential cancer progression markers in young women. PLoS ONE. 2013;8:e63204
78. Vihervuori H, Autere TA, Repo H, Kurki S, Kallio L, Lintunen MM. et al. Tumor-infiltrating lymphocytes and CD8(+) T cells predict survival of triple-negative breast cancer. J Cancer Res Clin Oncol. 2019;145:3105-14
79. Hao M, Liu W, Ding C, Peng X, Zhang Y, Chen H. et al. Identification of hub genes and small molecule therapeutic drugs related to breast cancer with comprehensive bioinformatics analysis. PeerJ. 2020;8:e9946
80. Wei H, Ma Y, Chen S, Zou C, Wang L. Multi-omics analysis identifies PTTG1 as a prognostic biomarker associated with immunotherapy and chemotherapy resistance. BMC Cancer. 2024;24:1315
81. Wang L, Liu X. Multi-Omics Analysis of the Oncogenic Value of Pituitary Tumor-Transforming Gene 1 (PTTG1) in Human Cancers. Front Biosci (Landmark Ed). 2024;29:87
82. Kim S, Kon M, DeLisi C. Pathway-based classification of cancer subtypes. Biol Direct. 2012;7:21
83. Chen CC, Chang TW, Chen FM, Hou MF, Hung SY, Chong IW. et al. Combination of multiple mRNA markers (PTTG1, Survivin, UbcH10 and TK1) in the diagnosis of Taiwanese patients with breast cancer by membrane array. Oncology. 2006;70:438-46
84. Repo H, Loyttyniemi E, Kurki S, Kallio L, Kuopio T, Talvinen K. et al. A prognostic model based on cell-cycle control predicts outcome of breast cancer patients. BMC Cancer. 2020;20:558
85. Buus R, Sestak I, Barron S, Loughman T, Fender B, Ruiz CL. et al. Validation of the OncoMasTR Risk Score in Estrogen Receptor-Positive/HER2-Negative Patients: A TransATAC study. Clin Cancer Res. 2020;26:623-31
86. Abd El-Fattah AA, Sadik NAH, Shaker OG, Kamal AM. Single Nucleotide Polymorphism in SMAD7 and CHI3L1 and Colorectal Cancer Risk. Mediat Inflamm. 2018;2018:9853192
87. Barnekow E, Liu W, Franko MA, von Wachenfeldt A, Wendt C, Tham E. et al. Novel Shared Heritable Candidate Risk Loci of Breast and Endometrial Cancer-A Swedish Haplotype Genome-Wide Association Study. Int J Mol Sci. 2025;26:5461
88. Lo YL, Yu JC, Chen ST, Hsu GC, Mau YC, Yang SL. et al. Breast cancer risk associated with genotypic polymorphism of the mitotic checkpoint genes: a multigenic study on cancer susceptibility. Carcinogenesis. 2007;28:1079-86
89. Brendle A, Brandt A, Johansson R, Enquist K, Hallmans G, Hemminki K. et al. Single nucleotide polymorphisms in chromosomal instability genes and risk and clinical outcome of breast cancer: a Swedish prospective case-control study. Eur J Cancer. 2009;45:435-42
90. Gurvits N, Repo H, Loyttyniemi E, Nykanen M, Anttinen J, Kuopio T. et al. Prognostic implications of securin expression and sub-cellular localization in human breast cancer. Cell Oncol (Dordr). 2016;39:319-31
91. Repo H, Gurvits N, Loyttyniemi E, Nykanen M, Lintunen M, Karra H. et al. PTTG1-interacting protein (PTTG1IP/PBF) predicts breast cancer survival. BMC Cancer. 2017;17:705
92. Teveroni E, Di Nicuolo F, Vergani E, Oliva A, Vodola EP, Bianchetti G. et al. SPTBN1 Mediates the Cytoplasmic Constraint of PTTG1, Impairing Its Oncogenic Activity in Human Seminoma. Int J Mol Sci. 2023;24:16891
93. Wu H, Chen S, Liu C, Li J, Wei X, Jia M. et al. SPTBN1 inhibits growth and epithelial-mesenchymal transition in breast cancer by downregulating miR-21. Eur J Pharmacol. 2021;909:174401
94. Hum M, Lee ASG. DNA methylation in breast cancer: early detection and biomarker discovery through current and emerging approaches. J Transl Med. 2025;23:465
95. Qi L, Zhou B, Chen J, Hu W, Bai R, Ye C. et al. Significant prognostic values of differentially expressed-aberrantly methylated hub genes in breast cancer. J Cancer. 2019;10:6618-34
96. Xie Y, Wang R. Pttg1 Promotes Growth of Breast Cancer through P27 Nuclear Exclusion. Cell Physiol Biochem. 2016;38:393-400
97. Liao YC, Ruan JW, Lua I, Li MH, Chen WL, Wang JR. et al. Overexpressed hPTTG1 promotes breast cancer cell invasion and metastasis by regulating GEF-H1/RhoA signalling. Oncogene. 2012;31:3086-97
98. Li L, Liu J, Wang W, Fu Y, Deng Y, Li X. et al. Cancer stem cells promote lymph nodes metastasis of breast cancer by reprogramming tumor microenvironment. Transl Oncol. 2023;35:101733
99. Hamid T, Kakar SS. PTTG/securin activates expression of p53 and modulates its function. Mol Cancer. 2004;3:18
100. Ren JL, Pan JS, Lu YP, Sun P, Han J. Inflammatory signaling and cellular senescence. Cell Signal. 2009;21:378-83
101. Tschop K, Conery AR, Litovchick L, Decaprio JA, Settleman J, Harlow E. et al. A kinase shRNA screen links LATS2 and the pRB tumor suppressor. Genes Dev. 2011;25:814-30
102. Chen WS, Yu YC, Lee YJ, Chen JH, Hsu HY, Chiu SJ. Depletion of securin induces senescence after irradiation and enhances radiosensitivity in human cancer cells regardless of functional p53 expression. Int J Radiat Oncol Biol Phys. 2010;77:566-74
103. Yu YC, Yang PM, Chuah QY, Huang YH, Peng CW, Lee YJ. et al. Radiation-induced senescence in securin-deficient cancer cells promotes cell invasion involving the IL-6/STAT3 and PDGF-BB/PDGFR pathways. Sci Rep. 2013;3:1675
104. Ruan JW, Liao YC, Lua I, Li MH, Hsu CY, Chen JH. Human pituitary tumor-transforming gene 1 overexpression reinforces oncogene-induced senescence through CXCR2/p21 signaling in breast cancer cells. Breast Cancer Res. 2012;14:R106
105. Ghayad SE, Vendrell JA, Bieche I, Spyratos F, Dumontet C, Treilleux I. et al. Identification of TACC1, NOV, and PTTG1 as new candidate genes associated with endocrine therapy resistance in breast cancer. J Mol Endocrinol. 2009;42:87-103
106. Yu SY, Liu HF, Wang SP, Chang CC, Tsai CM, Chao JI. Evidence of securin-mediated resistance to gefitinib-induced apoptosis in human cancer cells. Chem Biol Interact. 2013;203:412-22
107. Suga T, Ishikawa A, Kohda M, Otsuka Y, Yamada S, Yamamoto N. et al. Haplotype-based analysis of genes associated with risk of adverse skin reactions after radiotherapy in breast cancer patients. Int J Radiat Oncol Biol Phys. 2007;69:685-93
108. Aguiar BRL, Ferreira EB, Normando AGC, Mazzeu JF, Assad DX, Guerra ENS. et al. Single nucleotide polymorphisms to predict acute radiation dermatitis in breast cancer patients: A systematic review and meta-analysis. Crit Rev Oncol Hematol. 2022;173:103651
109. Murray RJS, Tanteles GA, Mills J, Perry A, Peat I, Osman A. et al. Association between single nucleotide polymorphisms in the DNA repair gene and acute adverse skin reactions following radiotherapy. Radiother Oncol. 2011;99:231-4
110. Mumbrekar KD, Bola Sadashiva SR, Kabekkodu SP, Fernandes DJ, Vadhiraja BM, Suga T. et al. Genetic Variants in CD44 and MAT1A Confer Susceptibility to Acute Skin Reaction in Breast Cancer Patients Undergoing Radiation Therapy. Int J Radiat Oncol Biol Phys. 2017;97:118-27
111. Prabu SS, Ch'ng ES, Woon PY, Chen JH, Tang TH, Citartan M. Unravelling the diagnostic and therapeutic potentialities of a novel RNA aptamer isolated against human pituitary tumour transforming gene 1 (PTTG1) protein. Anal Chim Acta. 2020;1138:181-90
Author contact
![]() Corresponding authors: Tianning Wang: wangtianningedu.cn; Xianqiang Liu: arthur730318com; Huan Shi: 15553119187com.
Corresponding authors: Tianning Wang: wangtianningedu.cn; Xianqiang Liu: arthur730318com; Huan Shi: 15553119187com.

 Global reach, higher impact
Global reach, higher impact