Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(15):4338-4345. doi:10.7150/jca.123558 This issue Cite
Research Paper
Genetic association of NEAT1 gene polymorphism with the progression of colorectal cancer
1. School of Medicine, Chung Shan Medical University, Taichung, Taiwan.
2. Department of Surgery, Chung Shan Medical University Hospital, Taichung, Taiwan.
3. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan.
4. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan.
5. Department of Pharmacology, School of Medicine, China Medical University, Taichung, Taiwan.
6. Department of Medical Laboratory Science and Biotechnology, Asia University, Taichung, Taiwan.
7. Chinese Medicine Research Center, China Medical University, Taichung, Taiwan.
8. Department of Mathematics and Statistics, Florida Atlantic University, Florida, USA.
9. Whole-Genome Research Core Laboratory of Human Diseases, Chang Gung Memorial Hospital, Keelung, Taiwan.
10. Department of Medical Biotechnology and Laboratory Science, College of Medicine, Chang Gung University, Taoyuan, Taiwan.
Received 2025-8-11; Accepted 2025-10-16; Published 2025-10-24
Abstract
Colorectal cancer (CRC) is a globally common malignancy, whose complex disease etiology involves a genetic element. Nuclear enriched abundant transcript 1 (NEAT1), a long noncoding RNA (lncRNA) gene, has been demonstrated to play a key role in cancer development. To clarify the potential effect of NEAT1 gene polymorphisms on CRC susceptibility, three NEAT1 single-nucleotide polymorphisms (SNPs), including rs3825071, rs3741384, and rs512715, were assessed in 485 CRC patients and 485 sex- and age-matched non-cancer controls. We did not detect any significant association of these SNPs with the occurrence and clinicopathological features of CRC. Nevertheless, one SNP of NEAT1 gene, rs3825071, was found in association with the distant metastasis (CT+TT: CC, OR, 2.644; 95% CI, 1.328-5.263; p=0.005) among relatively younger patients (< 65 years old), indicating an age-specific effect of NEAT1 gene polymorphisms on the spread of CRC. Our stratification analysis revealed that the association of rs3825071 with CRC metastasis is anatomical site-specific, as cases of colon tumors but not of rectal tumors who bear at least one polymorphic allele of rs3825071 more commonly developed metastasis. Further exploration using the datasets from the Genotype-Tissue Expression (GTEx) Portal and The Cancer Genome Atlas (TCGA) showed that rs3825071 genotypes affected NEAT1 expression in the colon tissues, and elevated NEAT1 levels were associated with a worse survival rate in relatively younger patients (< 65 years old) with colon adenocarcinoma. These data suggest that altered expression levels of NEAT1 due to genetic polymorphisms may influence the progression of colon cancer.
Keywords: Colorectal cancer, nuclear enriched abundant transcript 1, single-nucleotide polymorphism, metastasis
Introduction
Colorectal cancer (CRC) is one of the most common cancers globally [1]. It remains the most prevalent malignancy among men and the second among women in Taiwan, representing a leading cause of deaths due to cancer [2]. In spite of the recent progress on surgical procedures and other therapeutic strategies, the survival rate of CRC in Taiwan is lower than that in US [2], with the age-standardized death rate of CRC increasing over the years [3]. Such high occurrence and mortality rates are mainly owing to the nature of its heterogeneous risk factors. Diet and long-lasting exposure of tumor-causing constituents (e.g., alcohol and cigarette) have been known as key environmental parameters that promote colorectal tumorigenesis [4]. In addition, various genetic aberrations that affect proteolytic reactions, angiogenic responses, and cellular adhesion have been shown to orchestrate the development and progression of CRC [5]. Recently, apart from host factors, disturbance in intestinal microbiota lying at the junction of those etiological parameters described above, has emerged as a major contributor of CRC [6]. Taking the complexity of CRC pathogenesis into consideration, all these risks tend to be mutually interlinked and imperative to evaluate the disease prognosis.
Current advances on the exploration of long non-coding RNA (lncRNA) biology have led to a huge paradigm shift in functional genetics [7-9]. To date, a great variety of lncRNAs are identified to play major roles in cancer [10, 11]. One such lncRNA, nuclear enriched abundant transcript 1 (NEAT1), is bound by multiple RNA-interacting proteins to form nuclear paraspeckles, a group of highly dynamic nuclear subdomains that act as gene regulatory condensates in many healthy and disease settings, including malignancies [12]. NEAT1 is functionally involved in tumorigenesis in many ways, comprising physical binding to microRNAs (miRNAs), orchestration of gene articulation, modulation of epigenetics, and engagement in signaling cascades [13]. Moreover, the intricacy of NEAT1's functions in cancer development is amplified by its participation in regulating cancer cell stemness and metabolism [14]. Recently, its functional association with autophagy further intensifies the complexity of NEAT1's roles in cancer biology [15]. In CRC, NEAT1 is upregulated and induces colorectal cell carcinogenesis by targeting several miRNA/transcription factor axes, such as miR-34a/SIRT1/Wnt/β-catenin, miR-185-5p/IGF2, miR-495-3p/CDK6, miR-150-5p/CPSF4, and so on [16]. Additionally, this RNA can interact with enhancer of zeste homolog 2 (EZH2), affecting the expression of genes involved in epithelial-to-mesenchymal transition (EMT) and tumor metastasis [17]. Furthermore, serum NEAT1 levels are significantly elevated in patients with CRC compared to healthy controls [18]. These findings suggest that NEAT1, functioning as a scaffold for RNA and protein molecules, is capable of governing the development and progression of CRC.
Lately, association studies using targeted gene approaches have unveiled a connection between single-nucleotide polymorphisms (SNPs) of NEAT1 gene and distinct types of cancers. Specifically, NEAT1 rs512715 was correlated with an increased risk of developing cervical cancer [19] and papillary thyroid carcinoma [20], as another SNP, rs2239895 polymorphisms, showed an association with the susceptibility to lung squamous cell carcinoma [21]. In addition, a link of NEAT1 rs3825071 with the risk of gastric cancer [22] and clinical stage and lymph node metastasis of tongue cancer [23] was recently detected. As emerging roles of NEAT1 in CRC biology were demonstrated, the influence of NEAT1 gene polymorphisms on the development of CRC remains unexplored. Here, we conducted a hypothesis-driven survey to clarify a genetic association of NEAT1 SNPs with colorectal tumorigenesis.
Materials and Methods
Study cohort
This study was approved by the institutional review board of Chung Shan Medical University Hospital, Taichung (CSMUH No: CS1-20111), Taiwan and recruited 485 CRC cases and 485 cancer-free controls between 2016 and 2023 to explore the impact of NEAT1 gene variations on the development of CRC. Informed written consent was provided by each subject at enrollment. Staging and grading of tumors were evaluated by a pathologist using the American Joint Committee on Cancer (AJCC) TNM staging system [24]. The control group encompassed subjects who did not report a history of cancer. Demographic data on age and gender were recorded from each participant.
Genotyping
The three selected NEAT1 genetic variants (rs3825071, rs3741384, and rs512715) were chosen based on prior evidence linking them to cancer susceptibility [19-23]. Specifically, rs3825071, rs3741384, and rs512715 have been associated with increased risks of cervical, thyroid, lung, gastric, and tongue cancers, highlighting their potential relevance to cancer development [19-23]. A QIAamp DNA blood mini kit (Qiagen, Valencia, CA, USA) was used to isolate genomic DNA from whole blood samples [25, 26]. Allelic discrimination for three NEAT1 SNPs was performed via the TaqMan assay with an ABI StepOne™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA).
Statistical analysis
The differences in age and gender between cases and controls were assessed by using Fisher's exact test. The Hardy-Weinberg equilibrium was tested with a chi-square goodness-of-fit test for biallelic markers in both study cohorts. The adjusted odds ratios (AORs) with their 95% confidence intervals (CIs) were calculated by multiple logistic regression models with adjustment for age and gender for determining the association of NEAT1 genotypes with the risk of CRC. Difference in NEAT1 expression levels among genotypic groups from the Genotype-Tissue Expression (GTEx) database [27] was calculated with one-way ANOVA. The association of NEAT1 levels with the survival of patients from the colon adenocarcinoma dataset of The Cancer Genome Atlas (TCGA) was estimated with a Kaplan-Meier plotter and compared by using the log-rank test. Data were analyzed by using SAS statistical software (Version 9.1, 2005; SAS Institute Inc., Cary, NC). A p value < 0.05 was considered significant.
Results
Demographic and clinical characteristics of study cohorts
In this association study, we have recruited 485 CRC patients to examine whether variants of NEAT1 SNPs contribute to CRC susceptibility. Considering that chronological age and gender represent tentative risks for colorectal neoplasms [28], 485 cancer-free subjects with matched age and gender were recruited as the control group for comparisons. The demographic and clinicopathologic properties of the case and control group were analyzed (Table 1). Several anatomical sites were represented among the cases, including right-sided colon (30.3%), left-sided colon (46.8%), and rectum (22.9%). Histological examination confirmed advanced stage III/IV in 51.8% of cases. Lymph node and distant metastasis occurred in 49.9.5% and 15.9% of our cases, respectively.
Age-specific effect of NEAT1 rs3825071 on CRC progression
To clarify whether NEAT1 gene polymorphisms are associated with the development and progression of CRC, rs3825071, rs3741384, and rs512715 were genotyped in this study. The genotypic frequencies of each SNP for both cohorts were examined (Table 2). Significant deviation from Hardy-Weinberg equilibrium in neither CRC nor control group was observed (p > 0.05) for three individual SNPs. Although none of these SNPs reached the threshold for significant associations, a marginal effect on CRC susceptibility was detected for specific genotypes of rs3741384 after the adjustment for potential confounders, age and gender (Table 2). Further assessment of NEAT1 association with clinicopathological characteristics of CRC patients also failed to detect any significant correlation of these NEAT1 variations with the progression of CRC (Table 3). Moreover, our stratification results demonstrated that the association of rs3825071 with distant metastasis of CRC (CT+TT: CC, metastatic tumor, OR, 2.644; 95% CI, 1.328-5.263; p=0.005) was exclusively detected in the younger age group (< 65 years old as the disease was diagnosed) (Table 4). Nevertheless, such genetic association with CRC metastasis was not seen in the more senior group (> 65 years old). These results indicate an age-specific effect of NEAT1 rs3825071 on the progression of CRC.
The distri butions of demographic and clinicopathologic characteristics in 485 controls and 485 patients with CRC.
| Variable | Controls (N=485) n (%) | Patients (N=485) (%) | p value |
|---|---|---|---|
| Age (yrs) | 60.09 ± 8.81 | 63.16 ± 13.08 | |
| < 65 | 283 (58.4%) | 255 (52.6%) | 0.070 |
| ≥ 65 | 202 (41.6%) | 230 (47.4%) | |
| Gender | |||
| Male | 297 (61.2%) | 287 (59.2%) | 0.512 |
| Female | 188 (38.8%) | 198 (40.8%) | |
| Tumor location | |||
| Rectum | 111 (22.9%) | ||
| Left colon | 227 (46.8%) | ||
| Right colon | 147 (30.3%) | ||
| Stage | |||
| I+II | 234 (48.2%) | ||
| III+IV | 251 (51.8%) | ||
| Tumor T status | |||
| T1-T2 | 120 (24.7%) | ||
| T3-T4 | 365 (75.3%) | ||
| Lymph node status | |||
| N0 | 243 (50.1%) | ||
| N1+N2 | 242 (49.9%) | ||
| Metastasis | |||
| M0 | 408 (84.1%) | ||
| M1 | 77 (15.9%) | ||
| Lymphovascular invasion | |||
| No | 272 (56.1%) | ||
| Yes | 213 (43.9%) | ||
| Perineural invasion | |||
| No | 278 (57.3%) | ||
| Yes | 207 (42.7%) | ||
| Pathologic grading | |||
| Well | 7 (1.4%) | ||
| Moderately | 441 (90.9%) | ||
| Poorly | 37 (7.7%) |
rs3825071 was in association with metastasis of colon cancer but not that of rectal cancer
Given that rs3825071 was observed to be correlated with metastatic potential of CRC, we subsequently examined whether this genetic association was unique to the tumor location. We found that patients of colon cancer who carry at least one polymorphic allele of rs3825071 (CT and TT; OR, 1.919; 95% CI, 1.084-3.396; p = 0.024) more frequently developed distal metastasis (Table 5). Notably, this genetic association was exclusively observed in colon cancer but not in rectal cancer. These data indicate a promotive effect of NEAT1 gene polymorphisms on metastatic potential of colon adenocarcinoma but not on that of cancers in the rectum.
Functional and clinical relevance of rs3825071 in CRC
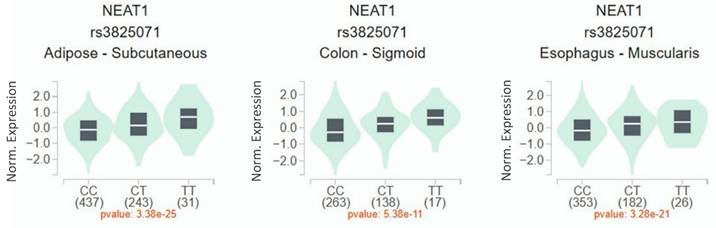
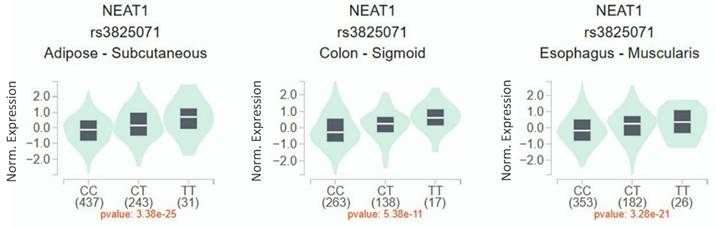
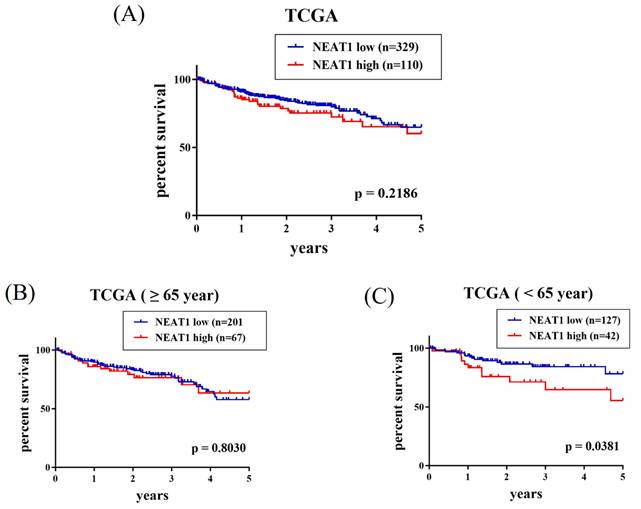
Since a connection of rs3825071 with CRC metastasis was identified, extra analyses employing public datasets were conducted to gain putative functional insights of this CRC-associated SNP. We found alterations of NEAT1 expression in the adipose (p = 3.38×10-25), colon tissues (p = 5.38×10-11) and esophagus tissue (p = 3.28×10-21) among subjects who carry different rs3825071 genotypes in the Genotype-Tissue Expression (GTEx) database (Figure 1). Moreover, further analysis of data from patients with colon adenocarcinoma in TCGA dataset revealed that cases of the younger age group (< 65 years old) with tumors expressing higher levels of NEAT1 exhibited a worse survival rate than those with tumors expressing lower levels of NEAT1 (Figure 2A-2C). These results support genetic associations detected in our study and suggest that altered expression levels of NEAT1 due to genetic polymorphisms may affect the progression and prognosis of colon cancer in relatively younger patients.
Discussion
The development and progression of colorectal tumorigenesis are a series of intricate processes modulated by the combination of both acquired and genetic parameters. In this survey, we reported that NEAT1 gene polymorphism, rs3825071, conferred the metastatic potential of CRC in an age- and anatomical site-specific manner. Furthermore, rs3825071 may be involved in the regulation of NEAT1 gene expression, which is associated with the survival in colon cancer patients of the younger age group.
Genotype distribution of NEAT1 gene polymorphisms in 485 controls and 485 patients with CRC.
| Variable | Controls (N=485) n (%) | Patients (N=485) n (%) | AOR (95% CI) | p value |
|---|---|---|---|---|
| rs3825071 | ||||
| CC | 336 (69.3%) | 334 (68.9%) | 1.000 (reference) | |
| CT | 133 (27.4%) | 142 (29.3%) | 1.077 (0.813-1.427) | p=0.605 |
| TT | 16 (3.3%) | 9 (1.8%) | 0.600 (0.260-1.381) | p=0.230 |
| CT+TT | 149 (30.7%) | 151 (31.1%) | 1.027 (0.782-1.350) | p=0.846 |
| rs3741384 | ||||
| GG | 350 (72.2%) | 324 (66.8%) | 1.000 (reference) | |
| GA | 125 (25.8%) | 151 (31.1%) | 1.303 (0.983-1.728) | p=0.065 |
| AA | 10 (2.1%) | 10 (2.1%) | 1.099 (0.450-2.680) | p=0.836 |
| GA+AA | 135 (27.8%) | 161 (33.2%) | 1.288 (0.979-1.696) | p=0.071 |
| rs512715 | ||||
| GG | 257 (53.0%) | 254 (52.4%) | 1.000 (reference) | |
| GC | 186 (38.4%) | 196 (40.4%) | 1.063 (0.815-1.387) | p=0.651 |
| CC | 42 (8.6%) | 35 (7.2%) | 0.836 (0.516-1.354) | p=0.466 |
| GC+CC | 228 (47.0%) | 231 (47.6%) | 1.021 (0.793-1.315) | p=0.870 |
Association between the clinical status and NEAT1 genotypes in 485 CRC patients.
| Variable | rs3825071 | rs3741384 | rs512715 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CC (N=334) | CT + TT (N=151) | p value | GG (N=324) | GA + AA (N=161) | p value | GG (N=254) | GC + CC (N=231) | p value | |
| Stages | |||||||||
| I+II | 161 (48.2%) | 73 (48.3%) | p=0.977 | 154 (47.5%) | 80 (49.7%) | p=0.654 | 123 (48.4%) | 111 (48.1%) | p=0.935 |
| III+IV | 173 (51.8%) | 78 (51.7%) | 170 (52.5%) | 81 (50.3%) | 131 (51.6%) | 120 (51.9%) | |||
| Tumor T status | |||||||||
| T1+T2 | 80 (24.0%) | 40 (26.5%) | p=0.549 | 78 (24.1%) | 42 (26.1%) | p=0.629 | 62 (24.4%) | 58 (25.1%) | p=0.859 |
| T3+T4 | 254 (76.0%) | 111 (73.5%) | 246 (75.9%) | 119 (73.9%) | 192 (75.6%) | 173 (74.9%) | |||
| Lymph node status | |||||||||
| Negative | 168 (50.3%) | 75 (49.7%) | p=0.898 | 163 (50.3%) | 80 (49.7%) | p=0.898 | 130 (51.2%) | 113 (48.9%) | p=0.619 |
| Positive | 166 (49.7%) | 76 (50.3%) | 161 (49.7%) | 81 (50.3%) | 124 (48.8%) | 118 (51.1%) | |||
| Metastasis | |||||||||
| Negative | 288 (86.2%) | 120 (79.5%) | p=0.059 | 271 (83.6%) | 137 (85.1%) | p=0.680 | 217 (85.4%) | 191 (82.7%) | p=0.408 |
| Positive | 46 (13.8%) | 31 (20.5%) | 53 (16.4%) | 24 (14.9%) | 37 (14.6%) | 40 (17.3%) | |||
| Lymphovascular invasion | |||||||||
| No | 190 (56.9%) | 82 (54.3%) | p=0.596 | 178 (54.9%) | 94 (58.4%) | p=0.471 | 98 (54.1%) | 98 (52.1%) | p=0.408 |
| Yes | 144 (43.1%) | 69 (45.7%) | 146 (45.1%) | 67 (41.6%) | 83 (45.9%) | 90 (47.9%) | |||
| Perineural invasion | |||||||||
| No | 191 (57.2%) | 87 (57.6%) | p=0.929 | 178 (54.9%) | 100 (62.1%) | p=0.133 | 135 (53.1%) | 137 (59.3%) | p=0.172 |
| Yes | 143 (42.8%) | 64 (42.4%) | 146 (45.1%) | 61 (37.9%) | 119 (46.9%) | 94 (40.7%) | |||
| Cell differentiation | |||||||||
| Well/ Moderately | 306 (91.6%) | 142 (94.0%) | p=0.352 | 301 (92.9%) | 49 (91.3%) | p=0.533 | 139 (54.7%) | 139 (60.2%) | p=0.226 |
| Poorly | 28 (8.4%) | 9 (6.0%) | 23 (7.1%) | 14 (8.7%) | 115 (45.3%) | 92 (39.8%) | |||
Association between the clinical status and NEAT1 rs3825071 genotypes in 485 CRC patients of different age groups.
| Variable | Aged < 65 (N=255) | Aged ≥ 65 (N=230) | ||||||
|---|---|---|---|---|---|---|---|---|
| CC (N=176) | CT + TT (N=79) | OR (95% CI) | p value | CC (N=158) | CT + TT (N=72) | OR (95% CI) | p value | |
| Stages | ||||||||
| I+II | 87 (49.4%) | 38 (48.1%) | 1.000 | p=0.844 | 74 (46.8%) | 35 (48.6%) | 1.000 | p=0.803 |
| III+IV | 89 (50.6%) | 41 (51.9%) | 1.055 (0.620-1.794) | 84 (53.2%) | 37 (51.4%) | 0.931 (0.533-1.627) | ||
| Tumor T status | ||||||||
| T1+T2 | 45 (25.6%) | 22 (27.8%) | 1.000 | p=0.702 | 35 (22.2%) | 18 (25.0%) | 1.000 | p=0.634 |
| T3+T4 | 131 (74.4%) | 57 (72.2%) | 0.890 (0.490-1.617) | 123 (77.8%) | 54 (75.0%) | 0.854 (0.445-1.639) | ||
| Lymph node status | ||||||||
| Negative | 90 (51.1%) | 40 (50.6%) | 1.000 | p=0.941 | 78 (49.4%) | 35 (48.6%) | 1.000 | p=0.915 |
| Positive | 86 (48.9%) | 39 (49.4%) | 1.020 (0.600-1.735) | 80 (50.6%) | 37 (51.4%) | 1.031 (0.590-1.800) | ||
| Metastasis | ||||||||
| Negative | 156 (88.6%) | 59 (74.7%) | 1.000 | p=0.005 | 132 (83.5%) | 61 (84.7%) | 1.000 | p=0.822 |
| Positive | 20 (11.4%) | 20 (25.3%) | 2.644 (1.328-5.263) | 26 (16.5%) | 11 (15.3%) | 0.916 (0.425-1.972) | ||
| Lymphovascular invasion | ||||||||
| No | 106 (60.2%) | 43 (54.4%) | 1.000 | p=0.385 | 84 (53.2%) | 39 (54.2%) | 1.000 | p=0.888 |
| Yes | 70 (39.8%) | 36 (45.6%) | 1.268 (0.742-2.167) | 74 (46.8%) | 33 (45.8%) | 0.960 (0.549-1.680) | ||
| Perineural invasion | ||||||||
| No | 103 (58.5%) | 46 (58.2%) | 1.000 | p=0.965 | 88 (55.7%) | 41 (56.9%) | 1.000 | p=0.860 |
| Yes | 73 (41.5%) | 33 (41.8%) | 1.012 (0.591-1.734) | 70 (44.3%) | 31 (43.1%) | 0.951 (0.542-1.668) | ||
| Cell differentiation | ||||||||
| Well/ Moderately | 165 (93.8%) | 73 (92.4%) | 1.000 | p=0.691 | 141 (89.2%) | 69 (95.8%) | 1.000 | p=0.100 |
| Poorly | 11 (6.2%) | 6 (7.6%) | 1.233 (0.439-3.461) | 17 (10.8%) | 3 (4.2%) | 0.361 (0.102-1.272) | ||
Association between the clinical status and NEAT1 rs3825071 genotypes in 485 patients with tumors of different anatomical sites.
| Variable | Rectum (N=111) | Colon (N=374) | ||||||
|---|---|---|---|---|---|---|---|---|
| CC (N=80) | CT + TT (N=31) | OR (95% CI) | p value | CC (N=254) | CT + TT (N=120) | OR (95% CI) | p value | |
| Stages | ||||||||
| I+II | 46 (57.5%) | 15 (48.4%) | 1.000 | p=0.387 | 115 (45.3%) | 58 (48.3%) | 1.000 | p=0.580 |
| III+IV | 34 (42.5%) | 16 (51.6%) | 1.443 (0.628-3.317) | 139 (54.7%) | 62 (51.7%) | 0.884 (0.572-1.366) | ||
| Tumor T status | ||||||||
| T1+T2 | 27 (33.8%) | 10 (32.3%) | 1.000 | p=0.881 | 53 (20.9%) | 30 (25.0%) | 1.000 | p=0.369 |
| T3+T4 | 53 (66.2%) | 21 (67.7%) | 1.070 (0.442-2.590) | 201 (79.1%) | 90 (75.0%) | 0.791 (0.474-1.320) | ||
| Lymph node status | ||||||||
| Negative | 47 (58.8%) | 16 (51.6%) | 1.000 | p=0.496 | 121 (47.6%) | 59 (49.2%) | 1.000 | p=0.782 |
| Positive | 33 (41.2%) | 15 (48.4%) | 1.335 (0.580-3.072) | 133 (52.4%) | 61 (50.8%) | 0.941 (0.609-1.452) | ||
| Metastasis | ||||||||
| Negative | 66 (82.5%) | 26 (83.9%) | 1.000 | p=0.863 | 222 (87.4%) | 94 (78.3%) | 1.000 | p=0.024 |
| Positive | 14 (17.5%) | 5 (16.1%) | 0.907 (0.297-2.771) | 32 (12.6%) | 26 (21.7%) | 1.919 (1.084-3.396) | ||
| Lymphovascular invasion | ||||||||
| No | 54 (67.5%) | 18 (58.1%) | 1.000 | p=0.350 | 136 (53.5%) | 64 (53.3%) | 1.000 | p=0.970 |
| Yes | 26 (32.5%) | 13 (41.9%) | 1.500 (0.639-3.520) | 118 (46.5%) | 56 (46.7%) | 1.008 (0.653-1.558) | ||
| Perineural invasion | ||||||||
| No | 53 (66.2%) | 21 (67.7%) | 1.000 | p=0.881 | 138 (54.3%) | 66 (55.0%) | 1.000 | p=0.903 |
| Yes | 27 (33.8%) | 10 (32.3%) | 0.935 (0.386-2.263) | 116 (45.7%) | 54 (45.0%) | 0.973 (0.629-1.506) | ||
| Cell differentiation | ||||||||
| Well/ Moderately | 80 (100.0%) | 30 (96.8%) | 1.000 | p=0.107 | 226 (89.0%) | 112 (93.3%) | 1.000 | p=0.182 |
| Poorly | 0 (0.0%) | 1 (3.2%) | ----- | 28 (11.0%) | 8 (6.7%) | 0.577 (0.255-1.306) | ||
Dysregulation of NEAT1 has been seen in various types of cancers, including CRC [16]. In accordance with our data that rs3825071 genotypes affected NEAT1 expression in the colon tissues, gastric cancer patients carrying the minor allele of rs3825071 (CT and TT) were shown to express a higher level of NEAT1 in the whole blood specimens as compared with homozygotes for the major allele (CC) [22]. These observations indicate a functional role of rs3825071 in acting as an expression quantitative trait locus (eQTL).
rs3825071 regulates the expression of NEAT1. eQTL analysis of rs3825071 in the adipose, colon tissues and esophagus tissue based on data from the GTEx portal. p value is calculated with one-way ANOVA.
Elevated NEAT1 expression levels are associated with a poor survival of patients with colon adenocarcinoma. Survival analysis of patients with colon adenocarcinoma of (A) All cases group, (B) older age group (> 65 years old), and (C) younger age group (< 65 years old) from The Cancer Genome Atlas (TCGA) database based on NEAT1 expression. p values were analyzed by log-rank test.
In addition to altered expression, functionality of rs3825071 might be also attuned based on the presence of polymorphic alleles. It has been predicted that the allelic change (C > T) of NEAT1 rs3825071 could give rise to corresponding alterations in the secondary structure of NEAT1 RNA transcripts, thereby abolishing the binding sites for hsa-miR-5092. Although the role of hsa-miR-5092 in CRC progression is yet mysterious, these findings collectively suggest that altered expression levels and sponging activities of NEAT1 rs3825071 attributed to its polymorphic alleles are likely implicated in governing the metastatic potential of CRC.
Moreover, it is striking that rs3825071 was merely linked to metastatic potentials of colon cancer whereas not to that of rectal cancer, indicating an anatomical site-specific influence of NEAT1 gene variations on the spread of colorectal neoplasms. Even though both rectal and colon malignancies arise in the large intestine and are usually viewed as a single tumor entity in most areas of clinical and basic research, considerable variations exist in molecular carcinogenesis, pathology, surgical topography and procedures, and multimodal treatment [29]. In terms of the carcinogenic mechanism, colon tumors often displayed increased expression levels of HOX gene family [30], higher pathway activities of MAPK cascades [31], and constitutive activation of KRAS [32] and BRAF [33, 34], as compared to rectal tumors. Besides distinctive modulation of numerous cancer hallmarks in cancers of the rectum and colon, these interrupted carcinogenic signaling events collectively induced a key transcriptional activator of NEAT1, hypoxia-inducible factor 2α (HIF-2α) [35], resulting in the discrepancies of local NEAT1 expression within CRC tumor compartments. These evidence, to some degree, accords with our finding that rs3825071 was in association with CRC in an anatomical site-specific manner.
Another intriguing finding detected here is that association of NEAT1 rs3825071 with CRC metastasis was merely observed in younger patients, revealing a role of chronological age in the genetic susceptibility to CRC. In some conditions, genetic etiologies have been found to display a stronger explanatory power in younger individuals, as compared with the older group [36]. This trend for genetic etiologies to lessen with increasing age has been reported in cancers [37, 38] and other conditions [39, 40]. Nevertheless, the relevance of genetic risk factors to human diseases is not equal among age contexts, though the rationales behind this phenomenon remain unclear. Through employing a proportional hazards model within an interval-based censoring approach on datasets from the UK Biobank, several facets regarding the correlation of genetic relative risks with age were posed [36]. Firstly, in a certain number of human disorders, statistical verification of a non-constant correlation between age and the impact of genetic etiologies was established. For such conditions, genetic etiologies mediated the largest influence at earlier ages, even though the magnitude and tendency of the drop-off varied across different diseases. Furthermore, the drop-off in genetic correlation with age cannot be attributed to latent variation in unmeasured covariates such as environmental parameters. These points of view underpin our observation that CRC is one of such illnesses affected by age-varying genetic risk profiles.
This investigation connected NEAT1 gene variants to the spread of colon cancer; however, additional efforts are necessary to deal with several study limitations. One caveat is the lack of data concerning the prolonged use of cigarettes and alcohol, potentially underestimating the impact of NEAT1 gene polymorphisms on the predisposition to CRC. Another issue is that the mechanistic role of rs3825071 in the promotion of colon cancer metastasis remains unanswered. How and to what extent the genetic polymorphism (C > T) affects its own expression or alters the interactions with its binding proteins or microRNAs needs further exploration. Moreover, in addition to rs3825071, another SNP (rs3741384) exhibited a marginal effect on CRC susceptibility. We acknowledge that many more significant associations are conceivably to be detected in a larger sample size. Lastly, the findings revealed in this study may be only applicable to particular ethnic groups and require replication experiments in different racial populations.
In conclusion, we detected a CRC-associated SNP in NEAT1 gene, rs3825071. Its connection with the metastatic responses of CRC was observed exclusively in younger patients (< 65 years old) with colon cancer. Induction of NEAT1 expression levels due to rs3825071 polymorphisms led to a poorer survival of younger patients with colon adenocarcinoma. These data unveil a novel link of NEAT1 variations to the progression and prognosis of CRC.
Acknowledgements
We would like to thank the Human Biobank of Chung Shan Medical University Hospital for providing the biological specimen and related clinical data for our research. This study was supported by Chung Shan Medical University Hospital (CSH-2024-C-027).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12-49
2. Kuo CN, Liao YM, Kuo LN, Tsai HJ, Chang WC, Yen Y. Cancers in Taiwan: Practical insight from epidemiology, treatments, biomarkers, and cost. J Formos Med Assoc. 2020;119:1731-41
3. Su SY, Huang JY, Jian ZH, Ho CC, Lung CC, Liaw YP. Mortality of colorectal cancer in Taiwan, 1971-2010: temporal changes and age-period-cohort analysis. Int J Colorectal Dis. 2012;27:1665-72
4. Hull R, Francies FZ, Oyomno M, Dlamini Z. Colorectal Cancer Genetics, Incidence and Risk Factors: In Search for Targeted Therapies. Cancer Manag Res. 2020;12:9869-82
5. Takayama T, Miyanishi K, Hayashi T, Sato Y, Niitsu Y. Colorectal cancer: genetics of development and metastasis. J Gastroenterol. 2006;41:185-92
6. Brennan CA, Garrett WS. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu Rev Microbiol. 2016;70:395-411
7. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A. et al. Landscape of transcription in human cells. Nature. 2012;489:101-8
8. Yeh JC, Chen YT, Chou YE, Su SC, Chang LC, Chen YL. et al. Interactive effects of CDKN2B-AS1 gene polymorphism and habitual risk factors on oral cancer. J Cell Mol Med. 2023;27:3395-403
9. Su SC, Hsieh MJ, Lin CW, Chuang CY, Liu YF, Yeh CM. et al. Impact of HOTAIR Gene Polymorphism and Environmental Risk on Oral Cancer. J Dent Res. 2018;97:717-24
10. Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965-81
11. Su SC, Yeh CM, Lin CW, Hsieh YH, Chuang CY, Tang CH. et al. A novel melatonin-regulated lncRNA suppresses TPA-induced oral cancer cell motility through replenishing PRUNE2 expression. J Pineal Res. 2021;71:e12760
12. Ingram HB, Fox AH. Unveiling the intricacies of paraspeckle formation and function. Curr Opin Cell Biol. 2024;90:102399
13. Li K, Yao T, Zhang Y, Li W, Wang Z. NEAT1 as a competing endogenous RNA in tumorigenesis of various cancers: Role, mechanism and therapeutic potential. Int J Biol Sci. 2021;17:3428-40
14. Smith NE, Spencer-Merris P, Fox AH, Petersen J, Michael MZ. The Long and the Short of It: NEAT1 and Cancer Cell Metabolism. Cancers (Basel). 2022 14
15. Almujri SS, Almalki WH. The paradox of autophagy in cancer: NEAT1's role in tumorigenesis and therapeutic resistance. Pathol Res Pract. 2024;262:155523
16. Azizidoost S, Ghaedrahmati F, Anbiyaee O, Ahmad Ali R, Cheraghzadeh M, Farzaneh M. Emerging roles for lncRNA-NEAT1 in colorectal cancer. Cancer Cell Int. 2022;22:209
17. Alshahrani MY, Saleh RO, Hjazi A, Bansal P, Kaur H, Deorari M. et al. Molecular Mechanisms of Tumorgenesis and Metastasis of Long Non-coding RNA (lncRNA) NEAT1 in Human Solid Tumors; An Update. Cell Biochem Biophys. 2024;82:593-607
18. Mohamadnejad M, Firoozi MR, Hashemzadeh S, Zafari V, Zarredar H, Farzaneh R. et al. Serum Level of MVIH, HNF1A-AS1, and NEAT1 Long Noncoding RNAs: Potential Biomarkers for Colorectal Cancer. Asian Pac J Cancer Prev. 2025;26:1953-8
19. Fang J, Li Y, Zhang J, Yan M, Li J, Bao S. et al. Correlation between polymorphisms in microRNA-regulated genes and cervical cancer susceptibility in a Xinjiang Uygur population. Oncotarget. 2017;8:31758-64
20. Abdi Pastaki M, Salimi S, Heidari Z, Saravani M. An Association Between GAS5 rs145204276, NEAT1 rs512715, and MEG3 rs4081134 Gene Polymorphisms and Papillary Thyroid Carcinoma. Rep Biochem Mol Biol. 2023;12:487-94
21. Wang S, Cui Z, Li H, Li J, Lv X, Yang Z. et al. LncRNA NEAT1 polymorphisms and lung cancer susceptibility in a Chinese Northeast Han Population: A case-control study. Pathol Res Pract. 2019;215:152723
22. Ji X, Yan Y, Ma N, He G, Wang K, Zhang Y. et al. Variant of SNPs at lncRNA NEAT1 contributes to gastric cancer susceptibility in Chinese Han population. Int J Clin Oncol. 2021;26:694-700
23. Chen KJ, Chuang CY, Lien MY, Su CW, Chen MY, Tsai HC. et al. Genetic associations of Neat1 polymorphisms with clinicopathologic characteristics of tongue cancer. Int J Med Sci. 2025;22:1208-14
24. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Annals of surgical oncology. 2010;17:1471-4
25. Weng WC, Hsieh YH, Lin CY, Liu YF, Su SC, Wang SS. et al. Functional variants of the pentraxin 3 gene are associated with the metastasis and progression of prostate cancer. J Cell Mol Med. 2024;28:e70041
26. Chen YT, Lin CW, Chou YE, Su SC, Chang LC, Lee CY. et al. Potential impact of ADAM-10 genetic variants with the clinical features of oral squamous cell carcinoma. J Cell Mol Med. 2023;27:1144-52
27. Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013;45:580-5
28. Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14:89-103
29. Paschke S, Jafarov S, Staib L, Kreuser ED, Maulbecker-Armstrong C, Roitman M. et al. Are Colon and Rectal Cancer Two Different Tumor Entities? A Proposal to Abandon the Term Colorectal Cancer. Int J Mol Sci. 2018 19
30. Sanz-Pamplona R, Cordero D, Berenguer A, Lejbkowicz F, Rennert H, Salazar R. et al. Gene expression differences between colon and rectum tumors. Clin Cancer Res. 2011;17:7303-12
31. Iacopetta B. Are there two sides to colorectal cancer? Int J Cancer. 2002;101:403-8
32. Slattery ML, Curtin K, Wolff RK, Boucher KM, Sweeney C, Edwards S. et al. A comparison of colon and rectal somatic DNA alterations. Dis Colon Rectum. 2009;52:1304-11
33. Popovici V, Budinska E, Tejpar S, Weinrich S, Estrella H, Hodgson G. et al. Identification of a poor-prognosis BRAF-mutant-like population of patients with colon cancer. J Clin Oncol. 2012;30:1288-95
34. Budinska E, Popovici V, Tejpar S, D'Ario G, Lapique N, Sikora KO. et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol. 2013;231:63-76
35. Choudhry H, Albukhari A, Morotti M, Haider S, Moralli D, Smythies J. et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34:4482-90
36. Jiang X, Holmes C, McVean G. The impact of age on genetic risk for common diseases. PLoS Genet. 2021;17:e1009723
37. Conti DV, Darst BF, Moss LC, Saunders EJ, Sheng X, Chou A. et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat Genet. 2021;53:65-75
38. Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal L, Lee A. et al. Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am J Hum Genet. 2019;104:21-34
39. Isgut M, Sun J, Quyyumi AA, Gibson G. Highly elevated polygenic risk scores are better predictors of myocardial infarction risk early in life than later. Genome Med. 2021;13:13
40. Simino J, Shi G, Bis JC, Chasman DI, Ehret GB, Gu X. et al. Gene-age interactions in blood pressure regulation: a large-scale investigation with the CHARGE, Global BPgen, and ICBP Consortia. Am J Hum Genet. 2014;95:24-38
Author contact
![]() Corresponding authors: Shih-Chi Su, Email: ssu1org.tw; Shun-Fa Yang, Email: ysfedu.tw.
Corresponding authors: Shih-Chi Su, Email: ssu1org.tw; Shun-Fa Yang, Email: ysfedu.tw.

 Global reach, higher impact
Global reach, higher impact