3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(13):4037-4046. doi:10.7150/jca.119306 This issue Cite
Review
Epigenetic regulation espeically histone modifications in breast cancer: A viable and emerging targeted therapeutic strategy
1. Central Laboratory, Department of Scientific Research, Changshu Hospital Affiliated to Soochow University, Changshu No.1 People's Hospital, Changshu, 215500, Jiangsu Province, China.
2. General Surgery, Cancer Center, Department of Breast Surgery, Zhejiang Provincial People's Hospital (Affiliated People's Hospital), Hangzhou Medical College, Hangzhou, 310014, Zhejiang Province, China.
3. Department of Pharmacy, The Third Affiliated Hospital (The Affiliated Luohu Hospital) of Shenzhen University, Shenzhen, 518001, China.
4. Department of Pharmacy, Changshu Hospital Affiliated to Soochow University, Changshu No. 1 People's Hospital, Changshu, 215500, Jiangsu Province, China.
†Yibing Zhou, Haotian Liu and Weimin Hong contributed equally to this work and share first authorship.
Received 2025-6-10; Accepted 2025-9-7; Published 2025-9-22
Abstract
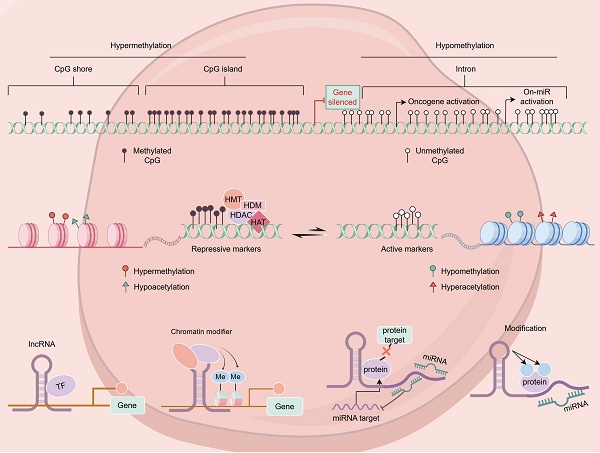
Epigenetic regulation, encompassing DNA methylation, histone modifications, and non-coding RNA activities, is a crucial mechanism through which gene expression is modified without corresponding changes in genomic DNA sequences. Dysregulation of these mechanisms can lead to histone and DNA modifications that either suppress or enhance the expression of disease progression-related genes. Among these regulatory processes, histone modifications are particularly significant, as they contribute to genomic stability, DNA repair, and chromatin dynamics, all of which influence breast cancer initiation and progression. This review offers a detailed analysis of the current state of research centered on epigenetic regulatory factors, with a particular focus on the role that histone modifications play in the treatment of breast cancer. It also examines the interplay between epigenetic modifications and the effectiveness of radiotherapy and chemotherapy when treating breast cancer. Lastly, this article explores the potential of epigenetic regulatory factors as viable targets for the future design of new anticancer therapies.
Keywords: breast cancer, epigenetic regulation, histone modification, histone target inhibitors, DNA methylation inhibitors
1. Introduction
Breast cancer is among the most widely diagnosed malignancies and remains a prominent driver of global cancer-related mortality in women [1]. GLOBOCAN 2022 indicates that 2.31 million new breast cancer diagnoses throughout the world in 2022, comprising 11.6% of all new cancer diagnoses. Over this same period, breast cancer was responsible for 666,000 deaths, comprising 6.9% of all cancer-related fatalities worldwide [2, 3]. Breast cancer subtypes are defined by the expression of the major biomarkers estrogen receptor (ER), progesterone receptor, human epidermal growth factor receptor 2 (HER2), and Ki67 [4]. The multidisciplinary treatment approach for operable breast cancer integrates local interventions, including surgery and radiotherapy, with systemic therapies, which include hormonal treatments, chemotherapy, and molecularly targeted agents. These systemic therapies can be administered as monotherapies or in combination regimens to enhance efficacy [5]. Despite advances in treatment, breast cancer remains a complex disease, often characterized by drug resistance and severe adverse effects associated with conventional therapies, which contribute to suboptimal patient outcomes [6]. The integration of epigenetic-targeting agents, such as inhibitors of histone modification, with existing therapeutic modalities offers a promising strategy for improving treatment efficacy and overcoming drug resistance [7].
2. Epigenetic Regulatory Mechanisms
Epigenetics consists of a class of heritable, reversible modifications capable of regulating gene expression without affecting the sequence of the underlying DNA. These mechanisms can entail DNA methylation, histone modifications, non-coding RNA activities, the remodeling of the chromatin, and nucleosome positioning [8]. In breast cancer, abnormal epigenetic modifications can result in tumor suppressor gene silencing and oncogene activation, thereby promoting tumor onset and progression [9] (Figure 1).
2.1 Breast cancer-related histone modifications
Histone modifications entail covalent chemical changes at the N-terminal histone tails, and can consist of phosphorylation, methylation, acetylation, and ubiquitination, among other post-translational modifications [10]. The resultant effects modulate chromatin structure and transcriptional activity, ultimately affecting gene expression. Dysregulated histone modifications contribute to aberrant gene expression in breast cancer, thereby facilitating tumor progression [11].
2.1.1 Histone acetylation
Histone acetylation is a process through which acetyl groups (-COCH₃) are enzymatically added to lysine residues, primarily on histones H3 and H4. Key acetylation sites can include H3K9, H3K14, H3K27, H4K5, H4K8, H4K12, and H4K16 [12]. This process is dynamically regulated by histone acetyltransferases (HATs), which promote acetylation, and histone deacetylases (HDACs), which remove acetyl groups [13]. Acetylation relaxes chromatin structure, enabling RNA polymerase and transcription factors to bind the DNA, leading to enhanced gene transcription, whereas deacetylation is related to gene silencing [14].
Various epigenetic alterations in breast cancer.
In breast cancer, histone acetylation is a major driver of tumor development through its ability to modulate gene expression. A notable observation in cancer is the global reduction of H4K16 acetylation, which is thought to occur early in breast tumorigenesis [15]. Suzuki et al. reported significantly lower ac-H4 and ac-H4K12 levels in ductal carcinoma in situ and invasive ductal carcinoma as compared to the normal mammary epithelium [16]. Additionally, H3K4ac has been linked to both early- and late-stage breast cancer, as it is highly enriched at gene promoters involved in cancer-related pathways, including estrogen response and epithelial-mesenchymal transition [17]. Beyond regulating oncogenes or tumor suppressor genes, abnormal histone acetylation is implicated in other biological processes relevant to breast cancer, such as DNA repair, metastasis, apoptotic death, metabolic activity, cellular homeostasis, cell cycle regulation, cell adhesion, and cellular proliferation [18].
2.1.2 Histone methylation
Histone methylation occurs on arginine, lysine, and histidine residue side chains, influencing gene expression through distinct methylation patterns. Lysine residues can undergo mono-, di-, or tri-methylation, while the asymmetric or symmetric methylation of arginine residues can take place. The types of histone methylation most commonly reported in the literature are H3K4, H3K9, H3K27, H3K36, H3K79, and H4K20 [19]. Different methylation states are associated with distinct genomic regions. For example, H3K4me2/3 is typically found at transcription start sites (TSS) of actively transcribed genes, while H3K4me1 is associated with active enhancer regions [20]. Consistently, H3K9me1 is related to active gene transcription, whereas H3K9me3 is linked to gene repression [21].
The tumor suppressor gene p53, which is frequently mutated or dysregulated in breast cancer, plays a crucial role in preventing oncogenesis [22]. H3K4 trimethylation at the p53 promoter region is related to its enhanced expression [23], while H3K27 trimethylation may play a role in its silencing [24]. These histone modifications can impair p53 function, promoting breast cancer progression. BRCA1, among the most well-established breast cancer susceptibility genes [25], is also regulated through histone methylation. H3K27me3-mediated methylation of the BRCA1 promoter is linked to its downregulation, increasing susceptibility to breast cancer development [26, 27].
2.1.3 Histone phosphorylation and ubiquitination
Beyond acetylation and methylation, histone phosphorylation and ubiquitination also play vital roles in the control of chromatin dynamics and the expression of genes. Histone H1 phosphorylation is closely linked with chromatin relaxation during interphase and its condensation in mitosis [28].
Histone ubiquitination, primarily facilitated by the Polycomb Repressive Complex 1 (PRC1), is integral to gene silencing in human cells [29]. PRT4165 is a potent inhibitor of PRC1-driven H2A ubiquitination, both in vivo and in vitro [30]. Functional assays of E3 ubiquitin ligase activity indicate that PRT4165 directly inhibits RNF2 and RING1A, key components responsible for the enzymatic activity within the PRC1 complex. Given its ability to suppress PRC1 function, PRT4165 presents a promising target for anticancer therapy [31].
2.2 Breast cancer-associated DNA methylation
DNA methylation is among the most widespread epigenetic modifications, typically occurring within CpG islands [32]. In normal cells, methylation patterns remain relatively stable; however, in breast cancer, genome-wide hypomethylation coupled with hypermethylation at specific promoter regions is frequently noted [33]. The aberrant hypermethylation of CpG islands in tumor suppressor gene promoters leads to transcriptional silencing, contributing to breast cancer initiation and progression [34].
In the context of breast cancer, multiple genes undergo significant methylation alterations [35]. Key tumor suppressor genes, including BRCA1, p16, and RASSF1A, often exhibit hypermethylation, leading to their silencing and subsequent loss of tumor-suppressive functions. In contrast, oncogenes such as c-Myc and cyclin D2 frequently undergo hypomethylation, leading to their overexpression and promoting uncontrolled cell proliferation and survival. Furthermore, the hypermethylation of DNA repair genes like MGMT and MLH1 exacerbates genomic instability [9].
DNA methylation markers provide several advantages over other types of tumor biomarkers in the context of breast cancer diagnostics, including improved sensitivity, non-invasiveness, and early detection [36]. Notably, aberrant methylation patterns can be identified in early-stage breast cancer and even in precancerous lesions [37]. For example, hypermethylation of APC and RASSF1A has been detected in breast cancer patient blood samples, highlighting their potential as non-invasive diagnostic markers [38, 39]. The integration of DNA methylation profiling with other omics data has led to significant improvements in early cancer screening accuracy [40].
The methylation status of specific genes serves as a valuable prognostic indicator for breast cancer patients. For instance, BRCA1 promoter hypermethylation is related to poor triple-negative breast cancer patient outcomes, while ESR1 gene methylation may predict responsiveness to endocrine therapy [41, 42]. These epigenetic markers hold great potential for guiding personalized treatment strategies.
2.3 miRNA expression in breast cancer
MicroRNAs (miRNAs) are small, non-coding RNAs, typically 21-23 nucleotides in length, that play vital roles in post-transcriptional gene regulation [43]. Some miRNAs are located within fragile genomic regions and are frequently subject to deletions or amplifications, resulting in their abnormal expression in various cancers, including breast cancer [44]. Depending on their function, miRNAs can function as tumor suppressors or oncogenic miRNAs (oncomiRs), influencing key pathways involved in cancer progression [45].
The first established link between miRNAs and cancer was demonstrated by Croce et al., who found that miR-15 and miR-16-1 can suppress tumor development via targeting the anti-apoptotic protein Bcl-2 [46]. In breast cancer, more than 40 miRNAs have been identified as key regulators, with the dysregulation of these miRNAs and their targets exhibiting either tumor-promoting or tumor-suppressing properties [47]. Notably, miR-126 and miR-335 suppress metastasis in breast cancer models in vivo [48].
miRNAs also hold promise as biomarkers for breast cancer subtyping, metastasis prediction, and therapy resistance assessment [49]. While chemotherapy remains a cornerstone of metastatic breast cancer treatment, a significant subset of patients fails to respond to standard chemotherapy or endocrine therapies [50]. Studies indicate that bone metastases have been associated with the presence of miR-10a and miR-10b in breast cancer [51]. Similarly, plasma levels of miR-210 that are elevated are related to trastuzumab sensitivity, tumor burden, and lymph node metastasis [52]. Identifying circulating miRNAs capable of early cancer detection or predicting therapeutic response could revolutionize breast cancer treatment, ultimately improving patient outcomes.
2.4 Chromatin remodeling and nucleosome positioning
Chromatin remodeling is driven by ATP-dependent chromatin remodeling complexes that modulate nucleosome positioning to regulate gene transcription. Nucleosomes, which serve as the fundamental chromatin units, are composed of DNA that wrap around histone octamers, facilitating DNA organization and protection within the nucleus. The positioning of nucleosomes determines the precise genomic arrangement of nucleosomes throughout chromatin landscapes [53]. The four major chromatin remodeling complex families in humans are SWI/SNF, ISWI, CHD, and INO80 [54]. Among these, BRG1, a core subunit of the SWI/SNF complex, has been implicated in breast cancer progression. Research has demonstrated the overexpression of BRG1 in invasive ductal carcinoma, with high BRG1 levels correlating with reduced overall survival and recurrence-free survival in breast cancer patients [55]. Despite these findings, research on the role of chromatin remodeling complexes in breast cancer remains limited, underscoring the need for further in-depth investigations.
3. Epigenetic Drugs in the Treatment of Breast Cancer
3.1 Histone target inhibitors
Histone targeting inhibitors represent a class of pharmacologically active compounds that modulate gene expression, chromatin architecture, and cellular differentiation through the regulation of histone function and post-translational modifications. These therapeutic agents exert their anti-neoplastic effects in breast cancer by specifically inhibiting various classes of histone-modifying enzymes (Figure 2).
3.1.1 Histone deacetylase inhibitors (HDACi)
HDACs are often overexpressed in tumor cells, leading to excessive histone deacetylation, which in turn suppresses the transcription of essential housekeeping and tumor suppressor genes. This epigenetic silencing plays a role in malignant transformation [56]. HDAC inhibitors (HDACi) counteract this process by restoring tumor suppressor gene expression, resulting in the inhibition of tumor cell proliferation as well as the induction of apoptosis [57]. With over 50 HDAC inhibitors developed, they represent the largest class of epigenetic drugs and account for most FDA-approved epigenetic therapies for cancer [58]. Clinically, HDACi have demonstrated efficacy against various malignancies, including pancreatic, ovarian, breast, colon, prostate, and thyroid cancers [59-64].
HDACi are classified based on their chemical structure into short-chain fatty acids, benzamides, cyclic peptides, and hydroxamic acids [65]. A well-known example of a short-chain fatty acid inhibitor is butyrate, which, at higher doses, can suppress growth in multiple cancers, including colorectal, prostate, breast, endometrial, and cervical cancers [66]. Sodium butyrate has been found to exert both anti-proliferative and pro-apoptotic effects in breast cancer cells while modulating genotoxicity [67]. Entinostat, a synthetic benzamide-derived HDACi, enhances the expression of ER and aromatase, thereby restoring sensitivity to nonsteroidal aromatase inhibitors (NSAIs). A Phase 1 clinical trial found that the combination of entinostat with exemestane improved tolerability and antitumor efficacy in hormone receptor-positive (HR+) advanced breast cancer (ABC). The Phase 2 ENCORE301 trial, which included HR+ ABC patients who had progressed following endocrine therapy but had not received CDK4/6 inhibitors, revealed significantly improved progression-free and overall survival (PFS and OS) with the entinostat-exemestane combination compared to exemestane alone. Moreover, a multicenter Phase 3 clinical trial conducted in China demonstrated that entinostat significantly prolonged PFS in HR+/HER2- ABC patients following endocrine therapy failure [68]. Cyclic peptide HDAC inhibitors, such as Romidepsin (FK228, depsipeptide), have also been investigated in breast cancer. Romidepsin has completed a Phase 2 clinical trial for metastatic breast cancer (NCT02393794). In patients treated with Romidepsin combined with Cisplatin and the PD-1 inhibitor Nivolumab, the median progression-free survival (PFS) was 4.4 months, with a 1-year PFS rate of 23%. The median overall survival (OS) was 10.3 months, and the 1-year OS rate was 43%, showing significant efficacy [69, 70]. Studies indicate that FK228 and its analogs act as dual inhibitors of HDAC and PI3K, directly inhibiting PI3K activity while promoting apoptosis [71]. Hydroxamic acid-based HDACi, such as Vorinostat (SAHA) and Panobinostat, have been explored as ER+ breast cancer therapies. Vorinostat has demonstrated efficacy in enhancing anti-estrogen therapy and has shown promising outcomes in Phase 2 trials (NCT00365599) in patients resistant to tamoxifen or aromatase inhibitors. The efficacy is particularly more significant in patients with high histone acetylation or high HDAC2 expression. However, hematological toxicity requires close monitoring, and future efforts should focus on exploring biomarker-guided precision treatment strategies [72]. Panobinostat has been found to downregulate BRCA1 expression in breast cancer, thus sensitizing tumor cells to PARP inhibitors (PARPi) [73].
Histone targeting inhibitors agents exert their anti-neoplastic effects in breast cancer by specifically inhibiting various classes of histone-modifying enzymes.
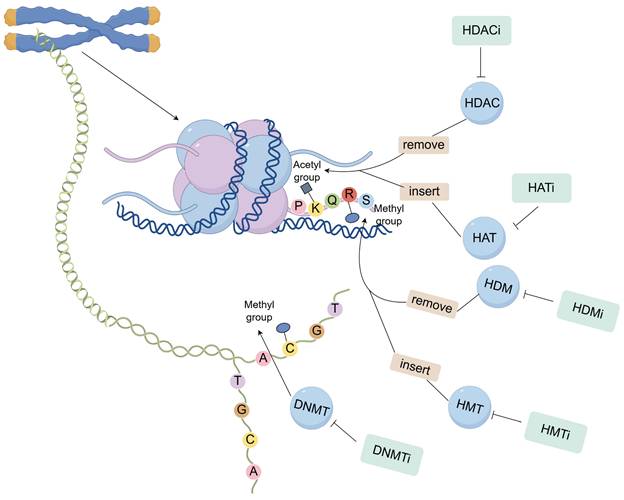
3.1.2 Histone methyltransferase inhibitors (HMTi)
Histone methyltransferases (HMTs) catalyze the methylation of specific histone residues, playing crucial roles in chromatin remodeling and gene expression regulation [74]. These enzymes are broadly classified into SET domain-containing and non-SET domain-containing families, with the former including EZH2, G9a, and SETD8, and the latter exemplified by DOT1L [75].
EZH2, the catalytic subunit of PRC2, mediates H3K27 trimethylation, leading to gene silencing. Silencing of PPP2R2B by EZH2 has been linked to poor prognosis in HER2-positive breast cancer and HER2-targeted therapy resistance, including trastuzumab and lapatinib. Inhibitors of EZH2, including GSK126 and EPZ-6438, have been shown to restore PPP2R2B expression, thereby mitigating resistance to HER2-targeted treatments [76, 77]. EPZ-6438 is currently being evaluated in a Phase 1/2 clinical trial (NCT01897571) for solid tumors and hematologic malignancies [78]. G9a, a major lysine methyltransferase, catalyzes the dimethylation of H3K9 and is implicated in multiple epigenetic modifications, including H3K27 methylation [79]. Inhibitors such as BIX-01294 have demonstrated the ability to promote the induction of apoptosis in breast cancer cells and reverse epithelial-mesenchymal transition, thereby reducing metastatic potential [80]. Similarly, the SETD8 inhibitor UNC0379 has been reported to inhibit breast cancer cell growth by inducing DNA damage and resulting in the arrest of the cell cycle [81]. DOT1L, a non-SET domain methyltransferase, regulates H3K79 methylation and is related to oncogenic gene expression. DOT1L inhibitors such as EPZ004777 have been shown to effectively suppress breast cancer growth and metastasis [82].
These HMTi not only exhibit standalone antitumor effects but also hold potential for combination therapies that enhance treatment efficacy. For example, combining EZH2 inhibitors with PARP inhibitors has been demonstrated to markedly enhance cytotoxicity in BRCA1-mutated breast cancer cells [83]. Furthermore, targeting HMTs may help overcome drug resistance, offering a promising avenue for improving breast cancer treatment outcomes [84].
3.1.3 Histone lysine demethylase inhibitors (HDMi)
Histone lysine demethylases are enzymes that remove methyl groups from specific histone lysine residues, thereby modulating gene expression. In accordance with their catalytic mechanisms, these enzymes are classified into lysine-specific demethylases (LSDs) and the Jumonji C (JmjC) domain-containing family [85]. LSD1 was the first histone demethylase to be identified and primarily mediates the demethylation of H3K4me1/2 and H3K9me1/2 [86]. The JmjC domain-containing family is further divided into multiple subgroups, including the KDM5 and KDM6 families, each targeting distinct histone methylation sites [87].
Among histone demethylases, LSD1 is particularly well-studied due to its frequent overexpression in breast cancer [88]. By demethylating H3K4me2, LSD1 downregulates tumor suppressor genes, while its activity on H3K9me2 promotes epithelial-mesenchymal transition (EMT), enhancing cancer cell proliferation, invasion, and metastasis [89]. Inhibitors targeting LSD1, such as Tranylcypromine (TCP) and ORY-1001, have demonstrated the ability to suppress ER+ breast cancer cell growth and increase sensitivity to endocrine therapy [90, 91]. The KDM5 family, including KDM5A and KDM5B, is also frequently overexpressed in breast cancer. These enzymes catalyze the removal of H3K4me3, thereby suppressing DNA damage repair and cell cycle regulation-related genes. This repression contributes to tumor progression and drug resistance [92]. Notably, KDM5 inhibitors such as CPI-455 and KDOAM-25 have shown potential in reversing resistance to chemotherapy, thereby enhancing treatment efficacy [93]. The role of KDM6 family members, UTX and JMJD3, in breast cancer is complex. While UTX serves as a tumor suppressor, with its inactivation leading to increased H3K27me3 levels that repress tumor suppressor genes, JMJD3 exhibits oncogenic potential by demethylating H3K27me3 and activating oncogene expression [94]. The KDM6 inhibitor GSK-J4 has been found to effectively inhibit the proliferation and invasiveness of triple-negative breast cancer (TNBC) cells [95].
3.1.4 Histone acetyltransferase inhibitors (HATi)
HATs play a dual role in cancer biology, functioning as both tumor suppressors and oncogenic drivers depending on the context [96]. These enzymes catalyze acetyl-CoA-derived acetyl group transfer to histone lysine residues, thus altering chromatin structure and gene expression. According to their sequence homology and substrate specificity, HATs are classified into the HAT1, GCN5/PCAF, MYST, p300/CBP, and Rtt109 subfamilies [97].
HATi are broadly divided into bisubstrate inhibitors and small-molecule inhibitors [96]. The clinical application of bisubstrate inhibitors is limited due to their poor metabolic stability and low cell permeability. Consequently, most currently available HATi are small-molecule compounds, many of which are derived from natural sources. For instance, anacardic acid, a naturally occurring HAT inhibitor, can markedly inhibit p300 and the p300/CBP-associated factor [98]. However, the phenolic structures in many natural HATi make them susceptible to oxidation, which limits their stability. Several synthetically designed HAT inhibitors have been developed to address these limitations [99]. A-485, a selective inhibitor targeting p300/CBP, has demonstrated potent anticancer activity against multiple malignancies, including colon, liver, and prostate cancers, as well as hematologic cancers [100]. Another notable HATi, C646, preferentially targets p300 and has been shown to suppress the survival and invasiveness of gastric cancer cells [101]. Additionally, Remodelin, an inhibitor of the acetyltransferase NAT10, has been reported to overcome doxorubicin resistance in breast cancer by reversing EMT, a key process in tumor progression [102].
3.2 DNA methylation inhibitors (DNMTi)
DNA methylation, a critical epigenetic modification, is primarily mediated by DNA methyltransferases (DNMTs) [103]. The first epigenetic drugs developed to treat cancer were DNMT inhibitors (DNMTi), which include nucleoside analogs and non-nucleoside inhibitors [104]. Nucleoside analogs function by incorporating into DNA, forming covalent complexes with DNMTs that trigger their degradation [105]. Among these, Azacitidine was the first FDA-approved epigenetic therapy for cancer. This cytidine analog irreversibly binds DNMTs, leading to DNA demethylation and reactivation of silenced tumor suppressor genes [106]. Another FDA-approved nucleoside analog, Decitabine, incorporates into DNA and induces hypomethylation, thereby disrupting DNA replication and promoting S-phase cell cycle arrest [107]. Both Azacitidine and Decitabine have received approval as treatments for acute myeloid leukemia, chronic myelomonocytic leukemia, and myelodysplastic syndromes [108, 109]. Zebularine, another cytidine analog, selectively inhibits DNMT1 and forms covalent complexes with DNMTs, effectively reversing gene silencing. Compared to other DNMTi, Zebularine exhibits greater chemical stability and lower toxicity. It has been shown to demethylate the p16 gene promoter, leading to the reactivation of the p16 tumor suppressor gene, which is frequently silenced in cancer [110]. Currently, Zebularine is a potential anticancer agent for breast cancer treatment, either as a monotherapy or in combination regimens [111]. Non-nucleoside DNMTi, in contrast, directly interact with the catalytic domain of DNMTs, rendering them inactive [112]. However, the clinical development of this class of inhibitors remains in the early research phase. Several promising compounds, including procainamide, SGI-1027, DC-05 analogs, and derivatives of quinazoline, propiophenone, and pyrrolopyridine, are being investigated for their potential as anticancer agents [113].
4. Summary and Future Perspectives
Epigenetic alterations, particularly aberrant histone modifications and DNA methylation, are central to breast cancer development and therapeutic resistance. HDACi have been widely explored, but their use as monotherapies has shown only limited efficacy in clinical trials. Consequently, current research is focused on their combination together with chemotherapy and targeted treatments to enhance therapeutic outcomes. A number of clinical trials are underway.
Currently, emerging epigenetic targets and innovative drugs mainly include KAT6A/6B inhibitors, NSD2/WHSC1-targeted therapies, and super-enhancer regulation. The first-in-class KAT6 inhibitor, PF-07248144, demonstrated promising and durable anti-tumor activity in later-line treatment of ER+/HER2- advanced breast cancer. When combined with fulvestrant, the objective response rate (ORR) reached 30.2%, with a median duration of response (DOR) of 9.2 months. The median progression-free survival (PFS) was 10.7 months—significantly longer than the 1-2 months observed with fulvestrant monotherapy—and efficacy was independent of ESR1 or PIK3CA mutations [114]. Histone methyltransferase NSD2/WHSC1 has been found to be highly expressed in triple-negative breast cancer (TNBC) and is associated with poor prognosis, as it accelerates tumor metastasis by promoting autophagy. Inhibition of NSD2 can block autophagic flux and significantly suppress TNBC metastasis in animal models. Small-molecule inhibitors targeting NSD2 are in the preclinical development stage [115]. The targets regulated by Super-Enhancers are FOXC1 and ANLN. CRISPR/Cas9-mediated knockout of the FOXC1 enhancer or targeted inhibition of the ANLN enhancer to reduce its expression can suppress tumor growth and metastasis [116].
Despite the promise of histone modification-based therapies, a major challenge lies in their broad-spectrum activity, which can lead to off-target effects and toxicity as a result of the disruption of gene expression within healthy cells. Therefore, improving the selectivity of epigenetic drugs remains a key priority in drug development. Precision medicine demands the multi-dimensional integration of molecular data to shift from "one drug for one disease" to "one strategy for one patient." PIK3CA is one of the most prevalent mutated genes in breast cancer. For patients with hormone receptor-positive, HER2-negative locally advanced or metastatic breast cancer, PIK3CA testing should be routinely performed before first-line treatment. If mutations are detected, patients may benefit from treatment with PI3K inhibitors such as inavolisib and alpelisib [117]. Future advancements in precision medicine may enable single-gene epigenetic editing approaches, allowing for highly targeted regulation of gene expression. This strategy could facilitate the development of more personalized treatments, minimizing adverse effects while maximizing therapeutic efficacy. As studies in the field of epigenetics continue to expand, the discovery of new histone-modifying enzymes and regulatory pathways will provide further opportunities for the development of targeted epigenetic therapies. These advancements hold great potential for improving cancer treatment, particularly for breast cancer, by offering novel therapeutic approaches to overcome drug resistance and enhance patient outcomes.
Acknowledgements
We thank the State Administration of Traditional Chinese Medicine, Zhejiang Provincial Key Laboratory, Key Laboratory for diagnosis and treatment of upper limb edema and stasis of breast cancer and figure support was provided by Figdraw.
Funding
This research was supported by the National Natural Science Foundation of China (Grant No. 82404685), Research Project of Jiangsu Association of Chinese Medicine (CYTF2024050), Suzhou Science and Technology Development Program (SKYD2023087), Changshu Medical Key Subjects (CSZDXK202301, CSZDXK202302), and Changshu Health Commission Science and Technology Program (CSWS202208、CSWS 202211).
Author contributions
Yibing Zhou: Writing - original draft, Writing - review & editing. Haotian Liu: Writing - review & editing. Weimin Hong: Writing - review & editing. Haotian Su: Writing - review & editing. Yuxiao Mu: Writing - review & editing. Yijie Cheng: Writing - review & editing. Chaoshen Wu: Writing - review & editing. Xuli Meng: Funding acquisition, Supervision. Da Qian: Writing - review & editing.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Akram M, Iqbal M, Daniyal M. et al. Awareness and current knowledge of breast cancer. Biol Res. 2017;50(1):33
2. Bray F, Laversanne M, Sung H. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229-263
3. Qian D, Hong W, Li S. et al. Trends in the global, national, and regional burden of breast cancer among adolescents and young adults from 1990 to 2021: Analyses of the 2021 global burden of disease study. Breast. 2025;82:104486
4. Shien T, Iwata H. Adjuvant and neoadjuvant therapy for breast cancer. Jpn J Clin Oncol. 2020;50(3):225-229
5. He M, Jiang Y Z, Gong Y. et al. Intensive chemotherapy versus standard chemotherapy among patients with high risk, operable, triple negative breast cancer based on integrated mRNA-lncRNA signature (BCTOP-T-A01): randomised, multicentre, phase 3 trial. Bmj. 2024;387:e079603
6. Lin Y, Zhu S, Hu C. et al. Cross-Species Suppression of Hepatoma Cell Growth and Migration by a Schistosoma japonicum MicroRNA. Mol Ther Nucleic Acids. 2019;18:400-412
7. Joshi DC, Sharma A, Prasad S. et al. Novel therapeutic agents in clinical trials: emerging approaches in cancer therapy. Discov Oncol. 2024;15(1):342
8. Zhang L, Lu Q, Chang C. Epigenetics in Health and Disease. Adv Exp Med Biol. 2020;1253:3-55
9. Mathur R, Jha NK, Saini G. et al. Epigenetic factors in breast cancer therapy. Front Genet. 2022;13:886487
10. Chen W, Dong L, Wei C. et al. Role of epigenetic regulation in diminished ovarian reserve. J Assist Reprod Genet. 2025;42(2):389-403
11. Li W, Wu H, Sui S. et al. Targeting Histone Modifications in Breast Cancer: A Precise Weapon on the Way. Front Cell Dev Biol. 2021;9:736935
12. Thakur C, Qiu Y, Fu Y. et al. Epigenetics and environment in breast cancer: New paradigms for anti-cancer therapies. Front Oncol. 2022;12:971288
13. Yao W, Wang T, Huang F. p300/CBP as a Key Nutritional Sensor for Hepatic Energy Homeostasis and Liver Fibrosis. Biomed Res Int. 2018;2018:8168791
14. Abballe L, Miele E. Epigenetic modulators for brain cancer stem cells: Implications for anticancer treatment. World J Stem Cells. 2021;13(7):670-684
15. Kumar R, Paul A M, Rameshwar P. et al. Epigenetic Dysregulation at the Crossroad of Women's Cancer. Cancers (Basel). 2019;11(8):1193
16. Pramanik SD, Kumar Halder A, Mukherjee U. et al. Potential of histone deacetylase inhibitors in the control and regulation of prostate, breast and ovarian cancer. Front Chem. 2022;10:948217
17. Messier T L, Gordon J A, Boyd J R. et al. Histone H3 lysine 4 acetylation and methylation dynamics define breast cancer subtypes. Oncotarget. 2016;7(5):5094-109
18. Guo P, Chen W, Li H. et al. The Histone Acetylation Modifications of Breast Cancer and their Therapeutic Implications. Pathol Oncol Res. 2018;24(4):807-813
19. Ning B, Li W, Zhao W. et al. Targeting epigenetic regulations in cancer. Acta Biochim Biophys Sin (Shanghai). 2016;48(1):97-109
20. McCornack C, Woodiwiss T, Hardi A. et al. The function of histone methylation and acetylation regulators in GBM pathophysiology. Front Oncol. 2023;13:1144184
21. Ohguchi H, Hideshima T, Anderson KC. The biological significance of histone modifiers in multiple myeloma: clinical applications. Blood Cancer J. 2018;8(9):83
22. Marvalim C, Datta A, Lee SC. Role of p53 in breast cancer progression: An insight into p53 targeted therapy. Theranostics. 2023;13(4):1421-1442
23. Majer AD, Hua X, Katona BW. Menin in Cancer. Genes (Basel). 2024;15(9):1231
24. Xu C, Xu Q, Yang H. H3K27 Acetylation-Activated GLDC Accelerated the Advancement of Oral Squamous Cell Carcinoma by Suppressing the p53 Signaling Pathway. Environ Toxicol. 2025;40(1):140-151
25. Tay JY, Ho JX, Cheo FF. et al. The Tumour Microenvironment and Epigenetic Regulation in BRCA1 Pathogenic Variant-Associated Breast Cancers. Cancers (Basel). 2024;16(23):3910
26. Gorodetska I, Lukiyanchuk V, Peitzsch C. et al. BRCA1 and EZH2 cooperate in regulation of prostate cancer stem cell phenotype. Int J Cancer. 2019;145(11):2974-2985
27. Judes G, Dagdemir A, Karsli-Ceppioglu S. et al. H3K4 acetylation, H3K9 acetylation and H3K27 methylation in breast tumor molecular subtypes. Epigenomics. 2016;8(7):909-24
28. Hu X, Chen LF. Pinning Down the Transcription: A Role for Peptidyl-Prolyl cis-trans Isomerase Pin1 in Gene Expression. Front Cell Dev Biol. 2020;8:179
29. Saygin C, Godley LA. Genetics of Myelodysplastic Syndromes. Cancers (Basel). 2021;13(14):3380
30. Tommasini D, Fox R, Ngo KJ. et al. Alterations in oligodendrocyte transcriptional networks reveal region-specific vulnerabilities to neurological disease. iScience. 2023;26(4):106358
31. Ismail IH, McDonald D, Strickfaden H. et al. A small molecule inhibitor of polycomb repressive complex 1 inhibits ubiquitin signaling at DNA double-strand breaks. J Biol Chem. 2013;288(37):26944-54
32. Hao X, Luo H, Krawczyk M. et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci U S A. 2017;114(28):7414-7419
33. Liu Y, Leng P, Liu Y. et al. Crosstalk between Methylation and ncRNAs in Breast Cancer: Therapeutic and Diagnostic Implications. Int J Mol Sci. 2022;23(24):15759
34. Schmitz RJ, Lewis ZA, Goll MG. DNA Methylation: Shared and Divergent Features across Eukaryotes. Trends Genet. 2019;35(11):818-827
35. Zhao J, Cheng G, Liu J. Combination of intensity modulated radiotherapy followed treatment with p38 MAPK activation inhibitor inhibits the proliferation of MCF-7 breast cancer cells. Saudi J Biol Sci. 2018;25(1):10-14
36. Hum M, Lee ASG. DNA methylation in breast cancer: early detection and biomarker discovery through current and emerging approaches. J Transl Med. 2025;23(1):465
37. Stastny I, Zubor P, Kajo K. et al. Aberrantly Methylated cfDNA in Body Fluids as a Promising Diagnostic Tool for Early Detection of Breast Cancer. Clin Breast Cancer. 2020;20(6):e711-e722
38. Khakpour G, Noruzinia M, Izadi P. et al. Methylomics of breast cancer: Seeking epimarkers in peripheral blood of young subjects. Tumour Biol. 2017;39(3):1010428317695040
39. Tang Q, Cheng J, Cao X. et al. Blood-based DNA methylation as biomarker for breast cancer: a systematic review. Clinical Epigenetics. 2016;8(1):115
40. Nguyen VTC, Nguyen TH, Doan NNT. et al. Multimodal analysis of methylomics and fragmentomics in plasma cell-free DNA for multi-cancer early detection and localization. Elife. 2023;12:RP89083
41. Lung DK, Reese RM, Alarid ET. Intrinsic and Extrinsic Factors Governing the Transcriptional Regulation of ESR1. Horm Cancer. 2020;11(3-4):129-147
42. Sheng X, Guo Y, Lu Y. Prognostic role of methylated GSTP1, p16, ESR1 and PITX2 in patients with breast cancer: A systematic meta-analysis under the guideline of PRISMA. Medicine (Baltimore). 2017;96(28):e7476
43. Treiber T, Treiber N, Meister G. Publisher Correction: Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20(5):321
44. Shah V, Shah J. Recent trends in targeting miRNAs for cancer therapy. J Pharm Pharmacol. 2020;72(12):1732-1749
45. Caetano S, Garcia AR, Figueira I. et al. MEF2C and miR-194-5p: New Players in Triple Negative Breast Cancer Tumorigenesis. Int J Mol Sci. 2023;24(18):14297
46. Abdi J, Jian H, Chang H. Role of micro-RNAs in drug resistance of multiple myeloma. Oncotarget. 2016;7(37):60723-60735
47. Bai W, Peng H, Zhang J. et al. LINC00589-dominated ceRNA networks regulate multiple chemoresistance and cancer stem cell-like properties in HER2(+) breast cancer. NPJ Breast Cancer. 2022;8(1):115
48. Shibuya N, Kakeji Y, Shimono Y. MicroRNA-93 targets WASF3 and functions as a metastasis suppressor in breast cancer. Cancer Sci. 2020;111(6):2093-2103
49. Ahmed R, Samanta S, Banerjee J. et al. Modulatory role of miRNAs in thyroid and breast cancer progression and insights into their therapeutic manipulation. Curr Res Pharmacol Drug Discov. 2022;3:100131
50. Mussell A, Shen H, Chen Y. et al. USP1 Regulates TAZ Protein Stability Through Ubiquitin Modifications in Breast Cancer. Cancers (Basel). 2020;12(11):3090
51. Zhang L, Lin Y, Zhang X. et al. Research progress of exosomes in orthopedics. Front Genet. 2022;13:915141
52. Cheng W, Xiao X, Liao Y. et al. Conducive target range of breast cancer: Hypoxic tumor microenvironment. Front Oncol. 2022;12:978276
53. Lai WKM, Pugh BF. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat Rev Mol Cell Biol. 2017;18(9):548-562
54. Nodelman IM, Bowman GD. Biophysics of Chromatin Remodeling. Annu Rev Biophys. 2021;50:73-93
55. Do SI, Yoon G, Kim HS. et al. Increased Brahma-related Gene 1 Expression Predicts Distant Metastasis and Shorter Survival in Patients with Invasive Ductal Carcinoma of the Breast. Anticancer Res. 2016;36(9):4873-82
56. Vanaja GR, Ramulu HG, Kalle AM. Overexpressed HDAC8 in cervical cancer cells shows functional redundancy of tubulin deacetylation with HDAC6. Cell Commun Signal. 2018;16(1):20
57. Gu S, Hou Y, Dovat K. et al. Synergistic effect of HDAC inhibitor Chidamide with Cladribine on cell cycle arrest and apoptosis by targeting HDAC2/c-Myc/RCC1 axis in acute myeloid leukemia. Exp Hematol Oncol. 2023;12(1):23
58. Parveen R, Harihar D, Chatterji BP. Recent histone deacetylase inhibitors in cancer therapy. Cancer. 2023;129(21):3372-3380
59. Liu S, Zou Q, Chen JP. et al. Targeting enhancer reprogramming to mitigate MEK inhibitor resistance in preclinical models of advanced ovarian cancer. J Clin Invest. 2021;131(20):e145035
60. Song X, Wu X, Yang J. et al. Effect of histone deacetylases on pancreatic physiology and pathology: a review. Int J Biol Macromol. 2025;318(Pt 1):144837
61. Mustafa NM, Mustafa MT, Abushanab AK. et al. Efficacy and safety of entinostat plus exemestane in hormone receptor-positive breast cancer: a systematic review meta-analysis of randomized controlled trials. Breast Cancer Res Treat. 2025;212(3):417-426
62. Wang M, Chen Y, Tian L. et al. Vascular Normalization Augments the Anti-tumor Efficacy of Combined HDAC Inhibitor with Immunotherapy in Solid Tumors. Cancer Discovery. 2025;15(9):1883-1904
63. Shi Z, Liu M, Zhang X. et al. A novel selenium analog of HDACi-based twin drug induces apoptosis and cell cycle arrest via CDC25A to improve prostate cancer therapy. Theranostics. 2024;14(9):3565-3582
64. Cai X, Sun R, Yang L. et al. Proteomic analysis reveals modulation of key proteins in follicular thyroid cancer progression. Chin Med J (Engl). 2025
65. Xu X, Li X, Zhao Y. et al. Immunomodulatory Effects of Histone Deacetylation Inhibitors in Graft-vs.-Host Disease After Allogeneic Stem Cell Transplantation. Front Immunol. 2021;12:641910
66. Westheim AJF, Stoffels LM, Dubois LJ. et al. The Modulatory Effects of Fatty Acids on Cancer Progression. Biomedicines. 2023;11(2):280
67. Yuksel B, Deveci Ozkan A, Aydın D. et al. Evaluation of the antioxidative and genotoxic effects of sodium butyrate on breast cancer cells. Saudi J Biol Sci. 2022;29(3):1394-1401
68. Xu B, Zhang Q, Hu X. et al. Entinostat, a class I selective histone deacetylase inhibitor, plus exemestane for Chinese patients with hormone receptor-positive advanced breast cancer: A multicenter, randomized, double-blind, placebo-controlled, phase 3 trial. Acta Pharm Sin B. 2023;13(5):2250-2258
69. Das C, Bhattacharya A, Adhikari S. et al. A prismatic view of the epigenetic-metabolic regulatory axis in breast cancer therapy resistance. Oncogene. 2024;43(23):1727-1741
70. Sharma P, Abramson V G, O'Dea A. et al. Romidepsin (HDACi) plus cisplatin and nivolumab triplet combination in patients with metastatic triple negative breast cancer (mTNBC). Journal of Clinical Oncology. 2021;39(15_suppl):1076-1076
71. Pojani E, Barlocco D. Romidepsin (FK228), A Histone Deacetylase Inhibitor and its Analogues in Cancer Chemotherapy. Curr Med Chem. 2021;28(7):1290-1303
72. Thang NX, Yoo S, La H. et al. Epigenetic Factors as Etiological Agents, Diagnostic Markers, and Therapeutic Targets for Luminal Breast Cancer. Biomedicines. 2022;10(4):748
73. Sellars E, Savguira M, Wu J. et al. A high-throughput approach to identify BRCA1-downregulating compounds to enhance PARP inhibitor sensitivity. iScience. 2024;27(7):110180
74. Wang J, Shen F, Liu F. et al. Histone Modifications in Acute Kidney Injury. Kidney Dis (Basel). 2022;8(6):466-477
75. Kaur J, Daoud A, Eblen ST. Targeting Chromatin Remodeling for Cancer Therapy. Curr Mol Pharmacol. 2019;12(3):215-229
76. Elshazly AM, Gewirtz DA. An overview of resistance to Human epidermal growth factor receptor 2 (Her2) targeted therapies in breast cancer. Cancer Drug Resist. 2022;5(2):472-486
77. Bao Y, Oguz G, Lee W C. et al. EZH2-mediated PP2A inactivation confers resistance to HER2-targeted breast cancer therapy. Nat Commun. 2020;11(1):5878
78. Vougiouklakis T, Bernard BJ, Nigam N. et al. Clinicopathologic significance of protein lysine methyltransferases in cancer. Clin Epigenetics. 2020;12(1):146
79. Huang H, Li H, Zhao T. et al. TSPAN1-elevated FAM110A promotes pancreatic cancer progression by transcriptionally regulating HIST1H2BK. J Cancer. 2022;13(3):906-917
80. Zhao Z, Feng L, Peng X. et al. Role of histone methyltransferase SETDB1 in regulation of tumourigenesis and immune response. Front Pharmacol. 2022;13:1073713
81. Wang X, Cao C, Tan X. et al. SETD8, a frequently mutated gene in cervical cancer, enhances cisplatin sensitivity by impairing DNA repair. Cell Biosci. 2023;13(1):107
82. Neeli PK, Sahoo S, Karnewar S. et al. DOT1L regulates MTDH-mediated angiogenesis in triple-negative breast cancer: intermediacy of NF-κB-HIF1α axis. FEBS J. 2023;290(2):502-520
83. Chen MK. Efficacy of PARP inhibition combined with EZH2 inhibition depends on BRCA mutation status and microenvironment in breast cancer. FEBS J. 2021;288(9):2884-2887
84. Zhang L, Zhang X, Shi Y. et al. Role and potential therapeutic value of histone methyltransferases in drug resistance mechanisms in lung cancer. Front Oncol. 2024;14:1376916
85. Yang Y, Luan Y, Feng Q. et al. Epigenetics and Beyond: Targeting Histone Methylation to Treat Type 2 Diabetes Mellitus. Front Pharmacol. 2021;12:807413
86. Yang J, Talibov VO, Peintner S. et al. Macrocyclic Peptides Uncover a Novel Binding Mode for Reversible Inhibitors of LSD1. ACS Omega. 2020;5(8):3979-3995
87. Yin F, Hu Y, Cao X. et al. JmjC domain-containing histone demethylase gene family in Chinese cabbage: Genome-wide identification and expressional profiling. PLoS One. 2024;19(11):e0312798
88. Hu X, Xiang D, Xie Y. et al. LSD1 suppresses invasion, migration and metastasis of luminal breast cancer cells via activation of GATA3 and repression of TRIM37 expression. Oncogene. 2019;38(44):7017-7034
89. Liu J, Feng J, Li L. et al. Arginine methylation-dependent LSD1 stability promotes invasion and metastasis of breast cancer. EMBO Rep. 2020;21(2):e48597
90. Cuyàs E, Gumuzio J, Verdura S. et al. The LSD1 inhibitor iadademstat (ORY-1001) targets SOX2-driven breast cancer stem cells: a potential epigenetic therapy in luminal-B and HER2-positive breast cancer subtypes. Aging (Albany NY). 2020;12(6):4794-4814
91. Orouji E, Utikal J. Tackling malignant melanoma epigenetically: histone lysine methylation. Clin Epigenetics. 2018;10(1):145
92. McCann TS, Sobral LM, Self C. et al. Biology and targeting of the Jumonji-domain histone demethylase family in childhood neoplasia: a preclinical overview. Expert Opin Ther Targets. 2019;23(4):267-280
93. Li CY, Wang W, Leung CH. et al. KDM5 family as therapeutic targets in breast cancer: Pathogenesis and therapeutic opportunities and challenges. Mol Cancer. 2024;23(1):109
94. Das P, Taube JH. Regulating Methylation at H3K27: A Trick or Treat for Cancer Cell Plasticity. Cancers (Basel). 2020;12(10):2792
95. Taube JH, Sphyris N, Johnson KS. et al. The H3K27me3-demethylase KDM6A is suppressed in breast cancer stem-like cells, and enables the resolution of bivalency during the mesenchymal-epithelial transition. Oncotarget. 2017;8(39):65548-65565
96. Wapenaar H, Dekker FJ. Histone acetyltransferases: challenges in targeting bi-substrate enzymes. Clin Epigenetics. 2016;8:59
97. McCullough CE, Marmorstein R. Molecular Basis for Histone Acetyltransferase Regulation by Binding Partners, Associated Domains, and Autoacetylation. ACS Chem Biol. 2016;11(3):632-642
98. Li F, Yan W, Dong W. et al. PNSC928, a plant-derived compound, specifically disrupts CtBP2-p300 interaction and reduces inflammation in mice with acute respiratory distress syndrome. Biol Direct. 2024;19(1):48
99. Jiang Y, Han L, Yang J. et al. Identification of a novel immune checkpoint molecule V-set immunoglobulin domain-containing 4 that leads to impaired immunity infiltration in pancreatic ductal adenocarcinoma. Cancer Immunol Immunother. 2023;72(8):2701-2716
100. Kim E, Zucconi BE, Wu M. et al. MITF Expression Predicts Therapeutic Vulnerability to p300 Inhibition in Human Melanoma. Cancer Res. 2019;79(10):2649-2661
101. van den Bosch T, Boichenko A, Leus NGJ. et al. The histone acetyltransferase p300 inhibitor C646 reduces pro-inflammatory gene expression and inhibits histone deacetylases. Biochem Pharmacol. 2016;102:130-140
102. Wu J, Zhu H, Wu J. et al. Inhibition of N-acetyltransferase 10 using remodelin attenuates doxorubicin resistance by reversing the epithelial-mesenchymal transition in breast cancer. Am J Transl Res. 2018;10(1):256-264
103. Correa LO, Jordan MS, Carty SA. DNA Methylation in T-Cell Development and Differentiation. Crit Rev Immunol. 2020;40(2):135-156
104. Majchrzak-Celińska A, Warych A, Szoszkiewicz M. Novel Approaches to Epigenetic Therapies: From Drug Combinations to Epigenetic Editing. Genes (Basel). 2021;12(2):208
105. Fàbrega C, Clua A, Eritja R. et al. Oligonucleotides Carrying Nucleoside Antimetabolites as Potential Prodrugs. Curr Med Chem. 2023;30(11):1304-1319
106. Suraweera A, O'Byrne KJ, Richard DJ. Epigenetic drugs in cancer therapy. Cancer Metastasis Rev. 2025;44(1):37
107. Montalvo-Casimiro M, González-Barrios R, Meraz-Rodriguez MA. et al. Epidrug Repurposing: Discovering New Faces of Old Acquaintances in Cancer Therapy. Front Oncol. 2020;10:605386
108. Bataller A, Sasaki K, Urrutia S. et al. Oral decitabine cedazuridine with and without venetoclax in higher-risk myelodysplastic syndromes or chronic myelomonocytic leukemia: a propensity score-matched study. Blood Cancer J. 2025;15(1):50
109. Pasvolsky O, Saliba RM, Popat UR. et al. Azacitidine Post-transplant Maintenance Improves Disease Progression in High-Risk Acute Myeloid Leukemia and Myelodysplastic Syndrome. Clin Lymphoma Myeloma Leuk. 2024;24(5):e196-e204
110. Ben-Kasus T, Ben-Zvi Z, Marquez VE. et al. Metabolic activation of zebularine, a novel DNA methylation inhibitor, in human bladder carcinoma cells. Biochem Pharmacol. 2005;70(1):121-133
111. Yap ZH, Kong WY, Azeez AR. et al. Anti-Cancer Effects of Epigenetics Drugs Scriptaid and Zebularine in Human Breast Adenocarcinoma Cells. Anticancer Agents Med Chem. 2022;22(8):1582-1591
112. Zhang Z, Wang G, Li Y. et al. Recent progress in DNA methyltransferase inhibitors as anticancer agents. Front Pharmacol. 2022;13:1072651
113. Pechalrieu D, Etievant C, Arimondo PB. DNA methyltransferase inhibitors in cancer: From pharmacology to translational studies. Biochem Pharmacol. 2017;129:1-13
114. Mukohara T, Park YH, Sommerhalder D. et al. Inhibition of lysine acetyltransferase KAT6 in ER+HER2- metastatic breast cancer: a phase 1 trial. Nat Med. 2024;30(8):2242-2250
115. Chen D, Chen X, Yang M. et al. H3K36me2 methyltransferase NSD2/WHSC1 promotes triple-negative breast cancer metastasis via activation of ULK1-dependent autophagy. Autophagy. 2025;21(8):1824-1842
116. Huang H, Hu J, Maryam A. et al. Defining super-enhancer landscape in triple-negative breast cancer by multiomic profiling. Nat Commun. 2021;12(1):2242
117. Xiong X, Zheng L W, Ding Y. et al. Breast cancer: pathogenesis and treatments. Signal Transduct Target Ther. 2025;10(1):49
Author contact
![]() Corresponding authors: Chaoshen Wu, Chaoshen_Wucom. Xuli Meng, mxlmailcom. Da Qian, qianda0215edu.cn
Corresponding authors: Chaoshen Wu, Chaoshen_Wucom. Xuli Meng, mxlmailcom. Da Qian, qianda0215edu.cn

 Global reach, higher impact
Global reach, higher impact