Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(13):3917-3927. doi:10.7150/jca.115044 This issue Cite
Research Paper
Identification of Plasma Protein Biomarkers and Drug Targets for Hematologic Malignancies by Proteome-wide Mendelian Randomization
1. Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, 100142, China.
2. Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Laboratory of Lymphoma Translational Research, Peking University Cancer Hospital & Institute, Beijing, 100142, China.
Received 2025-4-3; Accepted 2025-7-8; Published 2025-9-3
Abstract
Background: It has been reported that the proteome in blood was an important source for biomarker and therapeutic target discovery. However, up to now, few proteomes have been identified with the risk of hematologic malignancies.
Methods: Genome-wide association studies (GWASs) including 3,083 plasma proteins are based on data from 54,219 people in the UK Biobank Pharma Proteomics Project (UKB-PPP) and 35,559 individuals from Iceland (deCODE). Genetic correlations with 33 hematologic malignancies were derived from the FinnGen cohort and the UK Biobank Data. Further studies, including Bayesian colocalization, protein-protein interaction assessment, pathway enrichment analysis, and drug target evaluation, were performed to enhance knowledge and identify prospective therapeutic targets for 33 hematologic cancers.
Results: Our study indicated that 86 potential plasma proteins may have a substantial causal association with the incidence of 33 hematological tumors, such as BCL2, NFKB1, PARP1, and TNFRSF14. There are 18 proteins with strong evidence of genetic co-localization and 9 proteins with moderate support from colocalization analysis. Out of the 86 proteins, 51 have druggable targets, and 26 were identified as targets for current or prospective pharmaceuticals.
Conclusion: Our research revealed numerous significant proteins linked to the likelihood of hematologic malignancies. It may elucidate protein-mediated processes of hematological tumors and provide prospective treatment options for individuals with these conditions.
Keywords: hematologic malignancies, plasma protein, Mendelian randomization, drug target
Introduction
Hematologic malignancies, including conditions such as leukemia, lymphoma, and myeloma, arise from the alteration of hematopoietic cells [1]. They are characterized by the unregulated expansion of hematopoietic cells, resulting in significant disturbances in hematopoietic and immunological processes, greatly impacting the patient's quality of life and overall survival rate [2]. The primary treatment modalities for hematological tumors consist of chemotherapy, radiation, and stem cell transplantation [3]. Despite advancements in patient prognosis via these treatments, they often present limits, including restricted effectiveness, considerable adverse effects, and the development of resistance. Consequently, investigating the molecular causes of hematological malignancies is essential for improving patient prognosis. Recent advancements in comprehending the molecular foundation of hematological tumors have generated interest in discovering biomarkers that may function as therapeutic targets, with several plasma proteins already recognized as prospective biomarkers and therapeutic argots [4]. The identification of atypical protein expression patterns in the plasma of individuals with malignant hematological disorders has initiated new research opportunities. These proteins provide insights into illness development and prognosis and may also serve as effective targets for innovative therapeutic approaches, possibly enhancing clinical tomes [5]. Through the use of genetic variation as an instrumental variable, Mendelian randomization (MR) provides a robust framework for conducting an investigation into the causal relationship that exists between plasma proteins and the consequences of sickness [6]. This approach allows researchers to alleviate confounding biases and reverse causality problems often identified in observational studies [7].
The primary aim of this project is to comprehensively discover and assess plasma protein biomarkers linked to hematological malignancies via a Mendelian randomization framework. GWAS of levels of circulating proteins could identify protein quantitative trait loci (pQTLs). Combining pQTLs and disease-variant associations can examine the causative effects of the proteins on the disease through Mendelian randomisation. This work is important due to recent improvements in proteomics technology that facilitate the detection of low-abundance proteins in plasma. These improvements may provide new insights into the diagnostic and prognostic capabilities of plasma proteins, hence facilitating the development of precision medicine techniques for managing hematological tumors. This work included the collection of data from two extensive proteome-wide GWAS and 33 blood malignancy GWAS, followed by a proteome-wide MR analysis to comprehensively identify circulating protein biomarkers linked to the risk of blood malignancies. Given that MR alone may be inadequate for identifying genuine proteins associated with cancer causative pathways, a colocalization analysis was then performed. A pharmacological study was performed to investigate their potential as therapeutic targets for hematological malignancies.
Methods
Data sources
We identified cis-single-nucleotide polymorphisms (cis-SNPs) linked to plasma protein levels from two extensive GWAS, the UK Biobank Pharma Proteomics Project (UKB-PPP) [8] and the deCODE Health Study [9]. The genetic variation data pertaining to 27 hematological malignancies was acquired through the FinnGen consortium (https://www.finngen.fi/en/access_results). The genome-wide association study on six hematological malignancies was derived from the UK Biobank study (http://www.nealelab.is/uk-biobank/).
Selection of instrumental variables (IVs)
The plasma protein SNPs that satisfied the GWAS testing P value threshold (< 1×10-5) were identified.[10] All the IVs were categorized according to a linkage disequilibrium (LD) threshold (r2 < 0.001) within a distance of 10,000 kilobases (kb), utilizing the 1000 Genomes European reference panel on an individual basis. In instances of palindrome SNPs, the forward allele was established utilizing allele frequency data [11]. GWAS of levels of circulating proteins could identify protein quantitative trait loci (pQTLs). Cis-pQTLs were characterized as SNPs located within 1 MB of the gene that encodes the protein, with linkage disequilibrium assessed using the 1000 Genomes European panel [11]. To reduce the influence of weak instrumental bias, we calculated the F statistic for each SNP and excluded those with a F statistic lower than 10 [12]. SNPs with minor allele frequency (MAF)≤0.01 were removed. With the use of the Olink technology, the UKB-PPP was able to perform proteome profiling on blood plasma samples from 54,306 individuals, which resulted in the collection of information for 2,923 proteins [8]. During the Mendelian randomization investigation with two samples, we used index cis-SNPs as instrumental variables for a total of 2,112 proteins [8]. The deCODE Health project evaluated 4,907 aptamers in a group of 35,559 Icelanders using the SomaScan platform [9]. The results of this evaluation were used to produce index cis-SNPs for 1,970 plasma proteins. Each of the two examinations revealed that a total of 999 proteins had index cis-SNPs that overlapped with one another.
Mendelian randomization (MR)
Multiple MR techniques were used in order to assess the causal link that exists between the exposure and the result. The primary technique of MR analysis that was used was known as the inverse variance weighting (IVW)[13] approach. When only one SNP instrument was available, the method of causal estimation was the Wald method [14]. Following the use of Bonferroni adjustment for multiple testing, the combined relationship was shown to be statistically significant with a p value of less than 1.62×10-5 (0.05/3083 proteins). However, in order to explore more common plasma proteins of potential hematological malignancies, we set the P value at four levels. Level 1: P value is less than 1.62×10-5, denoted as “* * * *”; Level 2: P value ranges from 1.62×10-5 to 5×10-4, denoted as “* * *”; Level 3: P value ranges from 5×10-4 to 5×10-3, denoted as “* *”; Level 4: P value ranges from 5×10-3 to 5×10-2, denoted as “*”. Sensitivity analysis was employed to estimate pleiotropy and heterogeneity. We assessed heterogeneity using Cochrane's IVW Q statistics [15]. We evaluated vertical pleiotropy by examining the intercept obtained from the MR-Egger regression. The MR analyses were carried out using the “TwoSampleMR” software version 0.5.10[16] packages in R (version 4.3.2).
Colocalization analysis
We conducted colocalization analysis to see whether linkage disequilibrium accounted for the protein relationships reported with hematological malignancies. The study was conducted using a Bayesian framework, which took into consideration the evidence for five hypotheses that were incompatible with one another [17]. For every hypothesis test (H0, H1, H2, H3, and H4), a posterior probability is given to the hypothesis being tested. Under the condition that the posterior probability for shared causal changes (PH4) was more than 0.75, it was determined that two signals had strong evidence for colocalization. It was revealed that the medium colocalization indication lies between 0.5 and 0.75 for PH4. For the purpose of this study, the coloc tool in the R programming language (version 4.4.1) was used.
Protein-protein interaction (PPI) and functional enrichment analysis
Through the use of the Search Tool for the Retrieval of Interacting Genes (STRING, Version 11.5, https://string-db.org/), PPI networks were built in order to investigate the interactions that occurred between the MR-prioritized proteins. In addition, STRING was used to perform pathway analyses based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) in order to study the possibly enriched pathways that are associated with these proteins.
Mapping MR-prioritized proteins to drug targets
Plasma proteins are a rich therapeutic target reservoir. The list of druggable genes discovered by Finan et al. [18] was compared to the MR-prioritized proteins to see whether they overlap. Finan et al. [18] discovered 4,479 drugged or druggable genes and divided them into three groups by drug development level. Tier 1 has 1427 genes for effectiveness targets of licensed small compounds, biotherapeutic medicines, and clinical drug candidates. Tier 2 includes 682 genes with demonstrated bioactive drug-like small-molecule binding partners and ≥50% identity (over 75% sequencing) with authorized drug targets. Tier 3 contains 2,370 genes that encode secreted or extracellular proteins, are distantly related to known drug targets, and belong to key druggable gene families missing from tiers 1 or 2. Tier 3A prioritizes genes within ±50 kbp of GWAS SNPs and extracellular locations, whereas Tier 3B includes additional genes. MR-prioritized proteins were annotated as therapeutic targets by development phase using the Therapeutic Target Database (http://db.idrblab.net/ttd/). The database has 3,578 therapeutic targets, including 498 successful, 1,342 clinical trials, 185 preclinical/patented, and 1,553 research targets [19]. This analysis focuses on target type, drug connected to target, and illness treated by treatment.
Results
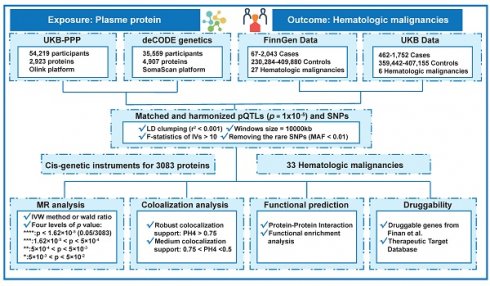
A summary of the conceptual framework of the research is shown in Figure 1. Each and every analysis made use of the data at the summary level that is shown in Table 1. The MR analysis included 1,970 proteins (Table S1) from the deCODE project and 2,112 proteins (Table S2) from the UKB-PPP collaboration; both sets of proteins were analyzed. Every single piece of outcome data showed that the least F statistic for the genetic instruments that were used was more than 10.
Estimating the effects of plasma proteins on 33 hematological tumors by using MR identified 86 plasma proteins
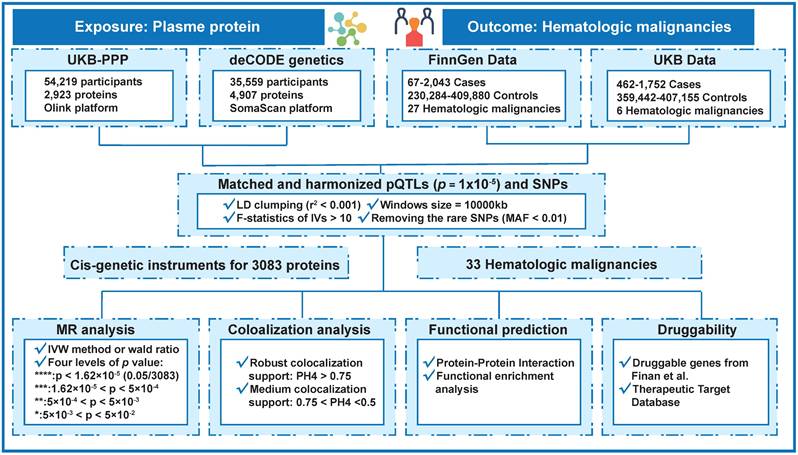
The overview of the findings from the examinations of thirty-three distinct hematologic cancers is shown in Figure 2 and Table S3. When the p-value of a protein is less than 5×10-4, we consider it to be a candidate plasma protein. This methodology allows us to investigate a greater number of prospective plasma proteins that are often associated with hematological malignancies. The genetically predicted amounts of 86 proteins were shown to have a substantial association with the probability of hematological tumors, as shown in Figure 2 of the combined analysis of two outcomes data. We found that some proteins have causal associations in multiple hematological tumors. Our findings indicate that certain plasma proteins exhibit causative roles in various hematological tumors, specifically ISOC1 in 14 tumors, ADK in 11 tumors, FKBPL in 10 tumors, and BCL2 in 9 tumors. In addition, we found that some plasma proteins play the same role in many blood tumors. ACTA2 serves as a risk factor across all seven types of blood tumors. CNTN1 serves as a protective factor in six types of hematological tumors (Figure 2 and Table S3).
27 plasma proteins were verified by colocalization evidence
27 plasma proteins were verified by colocalization evidence. 18 proteins demonstrated strong support of colocalization analysis (PH4 > 0.75) out of 86 MR-identified proteins in connection to hematological tumors (P value of MR < 5×10-4) (Figure 3 and Table S4). Nine proteins obtained medium support of colocalization analysis (0.75 < PH4 < 0.5) (Figure 3 and Table S4). Some diseases have colocalization with multiple other diseases. For example, myeloproliferative diseases (excluding chronic myeloid leukemia, CML) have the most colocalized genes, including RPN1, BRAP, PPP1CC, ERP29, and PARP1. Moreover, myeloproliferative diseases (excluding CML) and essential (haemorrhagic) thrombocythaemia have three identical colocalized genes, namely BRAP, PPP1CC, and ERP29. In addition, certain genes have colocalization with multiple diseases. For instance, HSD17B8 has colocalization with lymphoid leukemia and chronic lymphocytic leukemia; RPN1 has colocalization with myeloproliferative diseases (excluding CML) and polycythaemia vera; and PARP1 has colocalization with myeloproliferative diseases (excluding CML) and polycythaemia vera.
PPI networks and KEGG pathway of the 86 MR-prioritized proteins
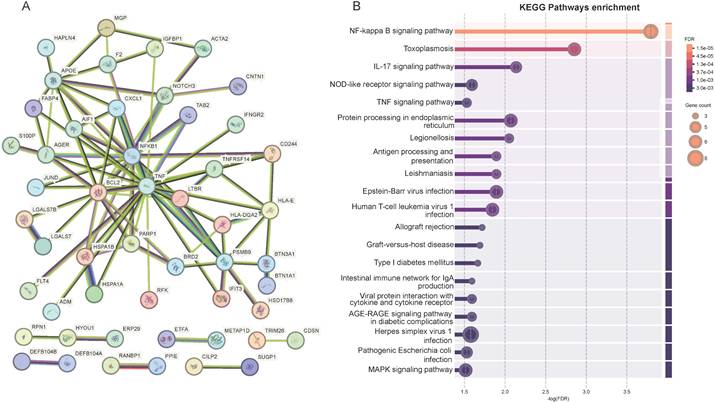
PPI and pathway studies were carried out in order to get a better knowledge of the etiology of hematological tumors and to gain a better understanding of the association between 86 MR-prioritized proteins and their enriched activities (Figure 4A, Table S5). The PPI network comprised 87 nodes and produced 92 edges, significantly exceeding the anticipated 58 edges based on an interaction score threshold of 0.4 (medium confidence) (enrichment p-value: 8.86×10-5) (Figure 4A, Table S5). By KEGG enrichment analysis, 86 proteins were mainly enriched in the NF-kappa B signaling pathway, the IL-17 signaling pathway, the NOD-like receptor signaling pathway, the TNF signaling pathway, and so on (Figure 4B, Table S6).
Flowchart of the MR-based analytical framework for evaluating the effect of plasma proteome on 33 hematologic malignancies. UKB-PPP, UK Biobank Pharma Proteomics Project; pQTLs, protein quantitative trait loci; SNP, single-nucleotide polymorphisms; LD, linkage disequilibrium; MAF, minor allele frequency; MR, Mendelian randomization; IVW, inverse variance weighting.
Characteristics of exposures and outcome.
| Variable | Source | Cases | Controls |
|---|---|---|---|
| Exposure | |||
| 4,907 plasma protein (pQTLs) | deCODE Health study | 35,559 | / |
| 2,923 plasma protein (pQTLs) | UK Biobank Pharma Proteomics Project (UKB-PPP) | 54,219 | / |
| Outcome | |||
| Hodgkin lymphoma | FinnGen data CD2_HODGKIN_LYMPHOMA_EXALLC | 846 | 324,650 |
| Diffuse large B-cell lymphoma | FinnGen data C3_DLBCL_EXALLC | 1,050 | 314,193 |
| Follicular lymphoma | FinnGen data CD2_FOLLICULAR_LYMPHOMA_EXALLC | 1,181 | 324,650 |
| Mantle cell lymphoma | FinnGen data C3_MANTLE_CELL_LYMPHOMA_EXALLC | 210 | 314,193 |
| Marginal zone B-cell lymphoma | FinnGen data C3_MARGINAL_ZONE_LYMPHOMA_EXALLC | 202 | 314,193 |
| Mature T/NK-cell lymphomas | FinnGen data CD2_TNK_LYMPHOMA_EXALLC | 363 | 324,650 |
| Waldenstrom macroglobulinemia, lymphoplasmacytic lymphoma | FinnGen data C3_MACROGLOBULINEMIA_EXALLC | 88 | 314,193 |
| Other and unspecified types of non Hodgkin lymphoma | FinnGen data CD2_NONHODGKIN_NAS_EXALLC | 1,171 | 324,650 |
| Non-Hodgkin lymphoma | FinnGen data C3_NONHODGKIN_EXALLC | 1,072 | 314,193 |
| Lymphoid leukaemia | FinnGen data CD2_LYMPHOID_LEUKAEMIA_EXALLC | 1,617 | 324,650 |
| Acute lymphocytic leukaemia | FinnGen data C3_ALL_EXALLC | 197 | 314,192 |
| Chronic lymphocytic leukaemia | FinnGen data C3_CLL_EXALLC | 668 | 314,189 |
| Myeloid leukaemia | FinnGen data CD2_MYELOID_LEUKAEMIA_EXALLC | 734 | 324,650 |
| Acute myeloid leukaemia | FinnGen data C3_AML_EXALLC | 244 | 314,192 |
| Chronic myeloid leukaemia | FinnGen data C3_CML_EXALLC | 115 | 314,192 |
| Chronic myeloid leukaemia [CML] BCR/ABL+ | FinnGen data CML | 258 | 409,880 |
| Monocytic leukaemia | FinnGen data CD2_MONOCYTIC_LEUKAEMIA_EXALLC | 85 | 324,650 |
| Leukaemia of unspecified cell type | FinnGen data CD2_LEUKAEMIA_NAS_EXALLC | 239 | 324,650 |
| Other leukaemias of specified cell type | FinnGen data CD2_OTHER_LEUKAEMIA_SPECIFIED_EXALLC | 67 | 324,650 |
| Multiple myeloma and malignant plasma cell neoplasms | FinnGen data CD2_MULTIPLE_MYELOMA_PLASMA_CELL_EXALLC | 1,337 | 324,650 |
| Multiple myeloma | FinnGen data C3_MULT_MYELOMA_EXALLC | 623 | 314,185 |
| Myeloproliferative diseases (CML excluded) | FinnGen data MYELOPROF_NONCML | 2,043 | 409,880 |
| Chronic myeloproliferative disease | FinnGen data CHRONMYELOPRO | 354 | 409,880 |
| Essential (haemorrhagic) thrombocythaemia | FinnGen data THROMBOCYTAEMIA | 1,062 | 313,473 |
| Polycythaemia vera | FinnGen data POLYCYTVERA | 1,004 | 313,577 |
| Eosinophilic disease (BM) | FinnGen data ESOSINOPHIL_DISEASE | 441 | 230,284 |
| Malignant immunoproliferative diseases | FinnGen data CD2_IMMUNOPROLIFERATIVE_EXALLC | 250 | 324,650 |
| Lymphomas | UK Biobank data | 1,752 | 359,442 |
| Leukaemia | UK Biobank data | 1,260 | 372,016 |
| Lymphoid leukaemia | UK Biobank data | 760 | 372,016 |
| Myeloid leukaemia | UK Biobank data | 462 | 372,016 |
| Multiple myeloma | UK Biobank data | 601 | 372,016 |
| Myeloproliferative neoplasms | UK Biobank data | 1,086 | 407,155 |
Evaluating the drug targets of the 86 MR-prioritized proteins
The assessment of human proteins utilizing MR evidence concentrated on their viability as therapeutic targets and their potential for drug development. We first compared the MR-prioritized proteins with the druggable genes identified by Finan et al [18]. Among the 86 proteins analyzed, 51 exhibited druggable targets, categorized as follows: 9 in tier 1, 8 in tier 2, 18 in tier 3A, and 16 in tier 3B (Table 2). Utilizing the Therapeutic Target Database, a total of 26 proteins were identified as targets for existing or potential drugs. Among these, 6 were classified as successful targets, 13 as clinical trial targets, 6 as literature-reported targets, and 1 as a discontinued target (Table 2; Table S7).
Discussion
This MR study investigated the relationships between 3,083 plasma proteins and the risk of 33 hematological tumors, complemented by a colocalization analysis. We identified 86 plasma proteins potentially causally associated with 33 hematological tumors, of which 27 proteins demonstrated colocalization support. Function prediction associated with these proteins enriched several pathways, including the NF-kappa B signaling pathway, IL-17 signaling pathway, NOD-like receptor signaling pathway, and TNF signaling pathway. The therapeutic potential of 86 proteins and their pharmaceutical properties were assessed.
There are a few proteins that have been found to be related to hematologic cancers, and these proteins were prioritized by our MR analysis in our research that used cis-pQTL. For example, BCL2, a key regulator of apoptosis, is frequently overexpressed in hematological tumors, contributing to chemotherapy resistance and poor prognosis [20]. Targeting BCL2 has emerged as a promising therapeutic approach, with BH3 mimetics showing particular potential [21, 22].
MR analysis on the associations of plasma protein with 33 hematological tumors. The gray squares in indicate missing data. * * * *, p < 1.62×10-5; * * *, 1.62×10-5 < p < 5×10-4; * *, 5×10-4 < p < 5×10-3; *, 5×10-3 < p < 5×10-2; †, P value is less than 0.05 in more than or equal to 7 hematological tumors. ‡, In more than or equal to 5 hematological tumors, the P value is less than 0.05, and the β value is greater than 0 or less than 0 at the same time.
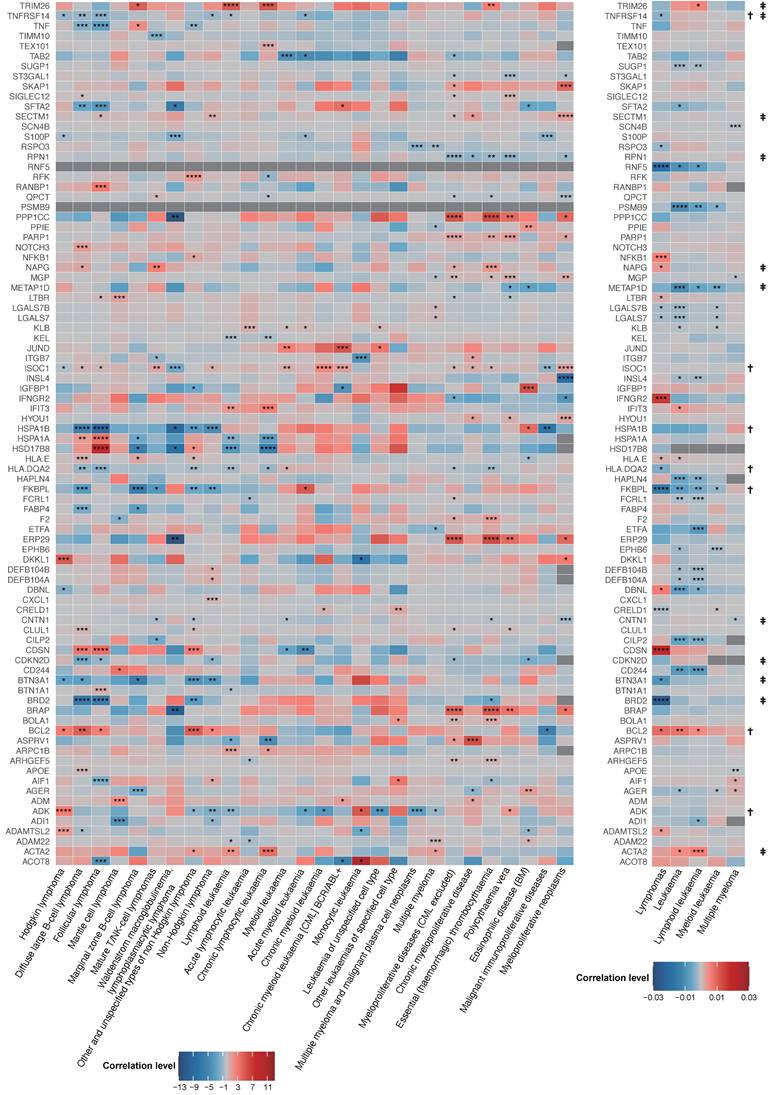
Colocalization analysis between plasma protein (P value of MR < 5×10-4) and hematologic malignancies.
Venetoclax, the first FDA-approved BCL2 inhibitor, has demonstrated significant efficacy in chronic lymphocytic leukemia and other hematologic cancers [23]. However, response to BCL2 inhibition varies across different malignancies, highlighting the need for biomarkers to predict treatment outcomes [23]. Ongoing research focuses on developing new BCL2 family inhibitors, optimizing combination therapies, and understanding resistance mechanisms [24]. As our understanding of BCL2 biology and apoptosis signaling improves, targeted therapies are expected to play an increasingly important role in treating hematological tumors [22, 25]. NF-κB, a crucial transcription factor family including NFKB1, plays a significant role in hematological tumors through its regulation of cell proliferation, apoptosis, and inflammation [26]. Constitutive NF-κB activation is common in these malignancies, contributing to enhanced cell survival and proliferation [27]. This activation can result from genetic alterations, microenvironmental factors, or chronic signaling [27]. NF-κB's pro-survival properties rely on the expression of anti-apoptotic molecules [28]. Targeting NF-κB and its regulators has emerged as a promising therapeutic approach for hematological tumors [29, 30]. PARP1 overexpression has been observed in various hematological tumors, suggesting its potential as a therapeutic target [31]. PARP inhibitors (PARPi) have shown promise in treating acute myeloid leukemia (AML) and other blood cancers, particularly in cases with specific genetic alterations such as RUNX1-RUNX1T1, PML-RARA, FLT3, and IDH1/2 mutations [32]. MYC-driven multiple myeloma cells exhibit increased sensitivity to PARPi [33]. Combining PARPi with other therapies, such as HDAC inhibitors or chemotherapeutic agents, has demonstrated synergistic effects [34]. TNFRSF14 mutations and 1p36 deletions are frequent genetic alterations in follicular lymphoma (FL), occurring in 18-40% of cases [35, 36]. These aberrations are associated with worse prognosis, increased risk of transformation, and inferior overall and disease-specific survival, particularly when both mutations and deletions are present [35, 37]. TNFRSF14 alterations reduce HVEM expression, enhancing the alloantigen-presenting capacity of lymphoma B cells and increasing the risk of acute graft-versus-host disease in allogeneic stem cell transplantation [38]. High TNFRSF14 expression correlates with poor outcomes and activation of the NF-kB pathway [37]. In contrast, high expression of BTLA, which interacts with TNFRSF14, is associated with better overall survival [37]. These findings highlight the importance of the BTLA-TNFRSF14 immune modulation pathway in FL pathobiology and prognosis. BRD2, a member of the bromodomain and extra-terminal (BET) family, plays a crucial role in hematological tumors. It promotes B-cell expansion and mitogenesis by regulating cyclin A expression [39]. Overexpression of BRD2 in transgenic mice leads to B-cell lymphoma and leukemia development [40]. BRD2 is also a critical mediator for STAT5 activity in leukemias and lymphomas [41]. BET inhibitors, such as JQ1, show promise in treating hematological tumors by targeting BRD proteins, including BRD2 and BRD4 [42]. In acute myeloid leukemia (AML), FABP4 promotes aggressiveness through a vicious loop with DNA methyltransferase 1 (DNMT1), enhancing aberrant DNA methylation [43, 44]. FABP4 inhibition suppresses AML progression and induces leukemia regression in mouse models [45]. Bone marrow adipocytes support AML proliferation through FABP4-mediated fatty acid transfer [45]. FCRL1, a member of the Fc receptor-like family, is predominantly expressed on B cells and plays a role in B cell regulation and malignancies [46]. It shows high expression in various B-cell non-Hodgkin lymphomas, chronic lymphocytic leukemia (CLL), and hairy cell leukemia [47]. FCRL1 has been identified as a potential target for immunotoxin therapy in these malignancies [48]. Its expression correlates with IGHV mutation status in CLL, making it a valuable prognostic marker [49]. FCRL1 promotes B cell proliferation and survival through the PI3K/AKT and NF-κB pathways [50]. Recent studies have highlighted the importance of IFIT family proteins in hematological tumors. IFIT1, IFIT2, IFIT3, and IFIT5 are overexpressed in acute myeloid leukemia (AML) patients, with higher levels of IFIT2, IFIT3, and IFIT5 predicting poor prognosis [51]. IFIT1 and IFIT3 have been implicated in proptosis induction in myeloma and leukemia cells [52].
Protein-protein interaction networks (A) and KEGG Pathways enrichment (B) of the 86 MR-prioritized proteins.
List of the 86 MR-prioritized proteins that were drug targets or to be druggable.
| Protein | Druggability tier | Target type | Protein | Druggability tier | Target type |
|---|---|---|---|---|---|
| ADK | Tier 2 | Clinical trial Target | HYOU1 | Tier 3B | / |
| ADM | Tier 3A | Clinical trial Target | IFNGR2 | Tier 1 | Successful Target |
| AGER | Tier 3A | Clinical trial Target | IGFBP1 | Tier 2 | Discontinued Target |
| AIF1 | / | Literature-reported target | INSL4 | Tier 3A | / |
| APOE | Tier 3A | Clinical trial Target | ITGB7 | Tier 1 | Successful Target |
| BCL2 | Tier 1 | Successful Target | KEL | Tier 3B | / |
| BOLA1 | Tier 3A | / | KLB | / | Clinical trial Target |
| BRD2 | Tier 2 | Clinical trial Target | LGALS7 | Tier 3B | / |
| BTN1A1 | Tier 3A | / | LGALS7B | Tier 3B | / |
| BTN3A1 | Tier 3B | / | LTBR | Tier 1 | Clinical trial Target |
| CD244 | Tier 3A | / | MGP | Tier 3B | / |
| CILP2 | Tier 3A | / | NFKB1 | Tier 1 | Successful Target |
| CLUL1 | Tier 3B | Literature-reported target | NOTCH3 | Tier 3B | Clinical trial Target |
| CNTN1 | Tier 3A | / | PARP1 | Tier 1 | Successful Target |
| CXCL1 | Tier 3B | Literature-reported target | PPIE | Tier 2 | / |
| DEFB104A | Tier 3B | / | PSMB9 | Tier 3B | Literature-reported target |
| DEFB104B | Tier 3B | / | QPCT | Tier 2 | Clinical trial Target |
| DKKL1 | Tier 3A | / | RPN1 | Tier 3A | / |
| EPHB6 | Tier 1 | Literature-reported target | RSPO3 | Tier 3A | Clinical trial Target |
| ERP29 | Tier 3A | / | SECTM1 | Tier 3B | / |
| F2 | Tier 1 | Successful Target | SFTA2 | Tier 3A | / |
| FABP4 | Tier 2 | / | SIGLEC12 | Tier 3B | / |
| FCRL1 | Tier 3A | / | ST3GAL1 | Tier 3B | / |
| HAPLN4 | Tier 3A | / | TEX101 | Tier 3B | / |
| HLA-DQA2 | Tier 3A | / | TNF | Tier 1 | Clinical trial Target |
| HSPA1A | Tier 2 | Clinical trial Target | TNFRSF14 | Tier 3A | Literature-reported target |
| HSPA1B | Tier 2 | Clinical trial Target |
In addition to some proteins that have been identified with hematological tumors, our research also found some plasma proteins that are related to most hematological tumors. For example, ISOC1 is causally associated with 14 hematological tumors. While ISOC1 promotes cell proliferation and tumor growth in pancreatic cancer [53] and lung cancer [54], it acts as a tumor suppressor in hepatocellular carcinoma [55]. ADK is causally associated with 14 hematological tumors. Dysregulation of ADK isoforms, particularly overexpression of the nuclear long isoform (ADK-L), has been observed in breast cancers, contributing to tumor growth and metastasis [56]. ADK deficiency increases susceptibility to carcinogens in the liver [57]. ADK's involvement in DNA methylation suggests an epigenetic role in cancer pathology [58]. FKBPL is causally associated with 11 hematological tumors. FKBPL, a novel member of the immunophilin family, has emerged as a potential cancer biomarker and therapeutic target. It plays crucial roles in steroid receptor signaling, particularly for estrogen, androgen, and glucocorticoid receptors [59]. High levels of FKBPL are associated with increased survival and improved response to endocrine therapy in breast cancer patients [60].
Our study highlights key targets like BCL2, NFKB1, and TNFRSF14, which regulate apoptosis, inflammation, and immune responses. While promising, their systemic inhibition poses toxicity risks. For example, BCL2 inhibitors (e.g., venetoclax) show efficacy in hematologic malignancies but cause severe neutropenia due to broad lymphoid dependency [61]. Similarly, NF-κB suppression may impair antimicrobial immunity [62], and PARP inhibitors induce hematologic toxicity from ubiquitous DNA repair roles [63]. Resistance remains a major hurdle. BCL2-targeted therapies fail via mutations (e.g., G101V) or compensatory MCL1 upregulation [64], while PARP inhibitor resistance arises through BRCA/PALB2 mutation reversal [65]. Combinatorial approaches (e.g., PARP + ATR inhibitors) could overcome escape mechanisms [66]. TNFRSF14 exemplifies context-dependency: loss drives lymphoma progression yet may enhance immunogenicity in specific subtypes, necessitating biomarker-driven stratification [38]. Emerging strategies (PROTACs, nanobodies) may bypass these limitations. Future work should: 1) Validate lead targets (PARP1, TNFRSF14) using PDX models and CRISPR screens; 2) Develop companion diagnostics via multi-omic profiling; 3) Optimize delivery systems (nanoparticles) to enhance specificity. Drug repurposing (e.g., NF-κB inhibitors) offers accelerated translation potential.
In conclusion, following the MR findings, we performed a series of subsequent analyses. Our study indicates that fewer than 50% of the associations demonstrate robust evidence of colocalization that supports causality. The assumption in Bayesian colocalization approaches that only one association signal exists per region may not accurately reflect reality, potentially leading to an underestimation of colocalization. Protein-protein interaction and pathway enrichment analyses were performed to investigate the relationships and functions of the identified proteins. Evaluation of drug targets was conducted for proteins with MR evidence, aiming to priorities drug discovery and facilitate the repurposing of existing drugs for hematological tumors.
Abbreviations
GWASs: Genome-wide association studies; UKB-PPP: UK Biobank Pharma Proteomics Project; MR: Mendelian randomization; IVs: instrumental variables; SNPs: single-nucleotide polymorphisms; LD: linkage disequilibrium; IVW: inverse variance weighting; KEGG: Kyoto Encyclopedia of Genes and Genomes; OR: Odds Ratio; PPI: Protein-protein interaction.
Supplementary Material
Supplementary tables.
Acknowledgements
Funding
This work was supported by the National Natural Science Foundation of China [grant numbers 82270153, 82270195, and 82070169]; Beijing Municipal General Fiscal Project (2025KT85); the Capital's Funds for Health Improvement and Research (Nos. 2022-1-2152 and 2024-1-2151); Beijing Natural Science Foundation (Nos. L244063 and L244025); Noncommunicable Chronic Diseases-National Science and Technology Major Project (2023ZD0500800); Beijing Research Ward Excellence Program (BRWEP2024W032150206, BRWEP2024 W032150205, BRWEP2024032150207, BRWEP2024W032150208, BRWEP2024W032150213) and the Peking University Cancer Hospital & Institute of Medicine Seed Fund for Interdisciplinary Research (JC202403).
Availability of data and materials
The GWAS Catalog summary statistics can be downloaded from the website https://www.ebi.ac.uk/gwas/. The FINNGEN summary data can be downloaded from the website https://www.finngen.fi/en/access_results. the UK Biobank study can be downloaded from the website http://www.nealelab.is/uk-biobank/.
Author contributions
YQS and XMW conceived the project, supervised the research and revised the paper. TP analyzed most of the data. JYZ assisted with the data processing. TP and XMW wrote the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Abel GA, Klepin HD. Frailty and the management of hematologic malignancies. Blood. 2018;131:515-24
2. Choi H-S, Kim BS, Yoon S, Oh S-O, Lee D. Leukemic Stem Cells and Hematological Malignancies. Int J Mol Sci. 2024 25
3. Bair SM, Brandstadter JD, Ayers EC, Stadtmauer EA. Hematopoietic stem cell transplantation for blood cancers in the era of precision medicine and immunotherapy. Cancer. 2020;126:1837-55
4. Lou S, Jiang Z-L, Zhu Y-W, Zhang R-Y, Wang Y, Chu T. et al. Exploring the impact of hydrogen sulfide on hematologic malignancies: A review. Cell Signal. 2024;120:111236
5. Lu B, Shi J, Cheng T, Wang C, Xu M, Sun P. et al. Chemokine ligand 14 correlates with immune cell infiltration in the gastric cancer microenvironment in predicting unfavorable prognosis. Front Pharmacol. 2024;15:1397656
6. Sekula P, Del Greco M F, Pattaro C, Köttgen A. Mendelian Randomization as an Approach to Assess Causality Using Observational Data. J Am Soc Nephrol. 2016;27:3253-65
7. Birney E. Mendelian Randomization. Cold Spring Harb Perspect Med. 2022;12:4
8. Sun BB, Chiou J, Traylor M, Benner C, Hsu Y-H, Richardson TG. et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature. 2023;622:329-38
9. Ferkingstad E, Sulem P, Atlason BA, Sveinbjornsson G, Magnusson MI, Styrmisdottir EL. et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet. 2021;53:1712-21
10. Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Võsa U. et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. 2019;51:600-5
11. Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481-7
12. Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40:740-52
13. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658-65
14. Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26:2333-55
15. Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693-8
16. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408
17. Foley CN, Staley JR, Breen PG, Sun BB, Kirk PDW, Burgess S. et al. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat Commun. 2021;12:764
18. Finan C, Gaulton A, Kruger FA, Lumbers RT, Shah T, Engmann J. et al. The druggable genome and support for target identification and validation in drug development. Sci Transl Med. 2017;9:383
19. Zhou Y, Zhang Y, Lian X, Li F, Wang C, Zhu F. et al. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022;50:D1398-D407
20. Chanan-Khan A. Bcl-2 antisense therapy in hematologic malignancies. Curr Opin Oncol. 2004;16:581-5
21. Fowler-Shorten DJ, Hellmich C, Markham M, Bowles KM, Rushworth SA. BCL-2 inhibition in haematological malignancies: Clinical application and complications. Blood Rev. 2024;65:101195
22. Valentin R, Grabow S, Davids MS. The rise of apoptosis: targeting apoptosis in hematologic malignancies. Blood. 2018;132:1248-64
23. Vogler M, Walter HS, Dyer MJS. Targeting anti-apoptotic BCL2 family proteins in haematological malignancies - from pathogenesis to treatment. Br J Haematol. 2017;178:364-79
24. Wu H, Medeiros LJ, Young KH. Apoptosis signaling and BCL-2 pathways provide opportunities for novel targeted therapeutic strategies in hematologic malignances. Blood Rev. 2018;32:8-28
25. Anderson MA, Huang D, Roberts A. Targeting BCL2 for the treatment of lymphoid malignancies. Semin Hematol. 2014;51:219-27
26. Keutgens A, Robert I, Viatour P, Chariot A. Deregulated NF-kappaB activity in haematological malignancies. Biochem Pharmacol. 2006;72:1069-80
27. Imbert V, Peyron J-F. NF-κB in Hematological Malignancies. Biomedicines. 2017;5:27
28. Turco MC, Romano MF, Petrella A, Bisogni R, Tassone P, Venuta S. NF-kappaB/Rel-mediated regulation of apoptosis in hematologic malignancies and normal hematopoietic progenitors. Leukemia. 2004;18:11-7
29. Braun T, Carvalho G, Fabre C, Grosjean J, Fenaux P, Kroemer G. Targeting NF-kappaB in hematologic malignancies. Cell Death Differ. 2006;13:748-58
30. Cilloni D, Martinelli G, Messa F, Baccarani M, Saglio G. Nuclear factor kB as a target for new drug development in myeloid malignancies. Haematologica. 2007;92:1224-9
31. Machado CB, Da Silva EL, Dias Nogueira BM, De Moraes Filho MO, Montenegro RC, De Moraes MEA. et al. PARP1 Is Overexpressed in Hematological Malignant Cell Lines: A Framework for Experimental Oncology. Anticancer Res. 2021;41:2397-402
32. Kontandreopoulou C-N, Diamantopoulos PT, Tiblalexi D, Giannakopoulou N, Viniou N-A. PARP1 as a therapeutic target in acute myeloid leukemia and myelodysplastic syndrome. Blood Adv. 2021;5:4794-805
33. Caracciolo D, Scionti F, Juli G, Altomare E, Golino G, Todoerti K. et al. Exploiting MYC-induced PARPness to target genomic instability in multiple myeloma. Haematologica. 2021;106:185-95
34. Valdez BC, Nieto Y, Yuan B, Murray D, Andersson BS. HDAC inhibitors suppress protein poly(ADP-ribosyl)ation and DNA repair protein levels and phosphorylation status in hematologic cancer cells: implications for their use in combination with PARP inhibitors and chemotherapeutic drugs. Oncotarget. 2022;13:1122-35
35. Cheung KJJ, Johnson NA, Affleck JG, Severson T, Steidl C, Ben-Neriah S. et al. Acquired TNFRSF14 mutations in follicular lymphoma are associated with worse prognosis. Cancer Res. 2010;70:9166-74
36. Martin-Guerrero I, Salaverria I, Burkhardt B, Szczepanowski M, Baudis M, Bens S. et al. Recurrent loss of heterozygosity in 1p36 associated with TNFRSF14 mutations in IRF4 translocation negative pediatric follicular lymphomas. Haematologica. 2013;98:1237-41
37. Carreras J, Lopez-Guillermo A, Kikuti YY, Itoh J, Masashi M, Ikoma H. et al. High TNFRSF14 and low BTLA are associated with poor prognosis in Follicular Lymphoma and in Diffuse Large B-cell Lymphoma transformation. J Clin Exp Hematop. 2019;59:1-16
38. Kotsiou E, Okosun J, Besley C, Iqbal S, Matthews J, Fitzgibbon J. et al. TNFRSF14 aberrations in follicular lymphoma increase clinically significant allogeneic T-cell responses. Blood. 2016;128:72-81
39. Belkina AC, Blanton WP, Nikolajczyk BS, Denis GV. The double bromodomain protein Brd2 promotes B cell expansion and mitogenesis. J Leukoc Biol. 2014;95:451-60
40. Greenwald RJ, Tumang JR, Sinha A, Currier N, Cardiff RD, Rothstein TL. et al. E mu-BRD2 transgenic mice develop B-cell lymphoma and leukemia. Blood. 2004;103:1475-84
41. Liu S, Walker SR, Nelson EA, Cerulli R, Xiang M, Toniolo PA. et al. Targeting STAT5 in hematologic malignancies through inhibition of the bromodomain and extra-terminal (BET) bromodomain protein BRD2. Mol Cancer Ther. 2014;13:1194-205
42. Bhattacharya S, Piya S, Borthakur G. Bromodomain inhibitors: what does the future hold? Clin Adv Hematol Oncol. 2018;16:504-15
43. Yan F, Shen N, Pang JX, Zhao N, Zhang YW, Bode AM. et al. A vicious loop of fatty acid-binding protein 4 and DNA methyltransferase 1 promotes acute myeloid leukemia and acts as a therapeutic target. Leukemia. 2018;32:865-73
44. Yan F, Shen N, Pang JX, Zhang YW, Rao EY, Bode AM. et al. Fatty acid-binding protein FABP4 mechanistically links obesity with aggressive AML by enhancing aberrant DNA methylation in AML cells. Leukemia. 2017;31:1434-42
45. Shafat MS, Oellerich T, Mohr S, Robinson SD, Edwards DR, Marlein CR. et al. Leukemic blasts program bone marrow adipocytes to generate a protumoral microenvironment. Blood. 2017;129:1320-32
46. Mamidi MK, Huang J, Honjo K, Li R, Tabengwa EM, Neeli I. et al. FCRL1 immunoregulation in B cell development and malignancy. Front Immunol. 2023;14:1251127
47. Yousefi Z, Eskandari N. Prognostic significance of Fc receptor-like 1 in patients with chronic lymphocytic leukemia, hairy cell leukemia, and various B-cell non-Hodgkin's lymphoma. Leuk Res Rep. 2019;12:100181
48. Du X, Nagata S, Ise T, Stetler-Stevenson M, Pastan I. FCRL1 on chronic lymphocytic leukemia, hairy cell leukemia, and B-cell non-Hodgkin lymphoma as a target of immunotoxins. Blood. 2008;111:338-43
49. Li FJ, Ding S, Pan J, Shakhmatov MA, Kashentseva E, Wu J. et al. FCRL2 expression predicts IGHV mutation status and clinical progression in chronic lymphocytic leukemia. Blood. 2008;112:179-87
50. Yousefi Z, Sharifzadeh S, Yar-Ahmadi V, Andalib A, Eskandari N. Fc Receptor-Like 1 as a Promising Target for Immunotherapeutic Interventions of B-Cell-Related Disorders. Biomark Insights. 2019;14:1177271919882351
51. Zhao Y, Zhang Y, Lu W, Sun R, Guo R, Cao X. et al. The diagnostic/prognostic roles and biological function of the IFIT family members in acute myeloid leukemia. BMC Med Genomics. 2023;16:296
52. He Y, Jiang S, Cui Y, Liang J, Zhong Y, Sun Y. et al. Induction of IFIT1/IFIT3 and inhibition of Bcl-2 orchestrate the treatment of myeloma and leukemia via pyroptosis. Cancer Lett. 2024;588:216797
53. Cheng L, Zhao Y, Tang M, Luo Z, Wang X. Knockdown of ISOC1 suppresses cell proliferation in pancreatic cancer in vitro. Oncol Lett. 2019;17:4263-70
54. Shi J, Yang F, Zhou N, Jiang Y, Zhao Y, Zhu J. et al. Isochorismatase domain-containing protein 1 (ISOC1) participates in DNA damage repair and inflammation-related pathways to promote lung cancer development. Transl Lung Cancer Res. 2021;10:1444-56
55. Xiang J, Gao X-Q, Chen X-L, Lu Y-Y. ISOC1 is a novel potential tumor suppressor in hepatocellular carcinoma. Neoplasma. 2022;69:174-82
56. Shamloo B, Kumar N, Owen RH, Reemmer J, Ost J, Perkins RS. et al. Dysregulation of adenosine kinase isoforms in breast cancer. Oncotarget. 2019;10:7238-50
57. El-Kharrag R, Owen R, Boison D. Adenosine Kinase Deficiency Increases Susceptibility to a Carcinogen. J Caffeine Adenosine Res. 2019;9:4-11
58. Zhulai G, Oleinik E, Shibaev M, Ignatev K. Adenosine-Metabolizing Enzymes, Adenosine Kinase and Adenosine Deaminase, in Cancer. Biomolecules. 2022;12:418
59. McKeen HD, Brennan DJ, Hegarty S, Lanigan F, Jirstrom K, Byrne C. et al. The emerging role of FK506-binding proteins as cancer biomarkers: a focus on FKBPL. Biochem Soc Trans. 2011;39:663-8
60. McKeen HD, Byrne C, Jithesh PV, Donley C, Valentine A, Yakkundi A. et al. FKBPL regulates estrogen receptor signaling and determines response to endocrine therapy. Cancer Res. 2010;70:1090-100
61. Di Pasqua LG, Abdallah MM, Feletti F, Vairetti M, Ferrigno A. Venetoclax-Related Neutropenia in Leukemic Patients: A Comprehensive Review of the Underlying Causes, Risk Factors, and Management. Pharmaceuticals (Basel). 2024;17:484
62. Zhu M, Su F, Leng J, Jian S, Yi P, Wen C. et al. Two NF-κB subunits are associated with antimicrobial immunity in Hyriopsis cumingii. Dev Comp Immunol. 2022;129:104336
63. Li H, Liu Z-Y, Wu N, Chen Y-C, Cheng Q, Wang J. PARP inhibitor resistance: the underlying mechanisms and clinical implications. Mol Cancer. 2020;19:107
64. Khalsa JK, Cha J, Utro F, Naeem A, Murali I, Kuang Y. et al. Genetic events associated with venetoclax resistance in CLL identified by whole-exome sequencing of patient samples. Blood. 2023;142:421-33
65. Seeber A, Zimmer K, Kocher F, Puccini A, Xiu J, Nabhan C. et al. Molecular characteristics of BRCA1/2 and PALB2 mutations in pancreatic ductal adenocarcinoma. ESMO Open. 2020;5:e000942
66. Kim H, Xu H, George E, Hallberg D, Kumar S, Jagannathan V. et al. Combining PARP with ATR inhibition overcomes PARP inhibitor and platinum resistance in ovarian cancer models. Nat Commun. 2020;11:3726
Author contact
![]() Corresponding authors: Xiaomin Wang (Email: wangxiaominedu.cn) and Yuqin Song (Email: songyuqin622com).
Corresponding authors: Xiaomin Wang (Email: wangxiaominedu.cn) and Yuqin Song (Email: songyuqin622com).

 Global reach, higher impact
Global reach, higher impact