3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(11):3296-3313. doi:10.7150/jca.113046 This issue Cite
Research Paper
Investigating the Role of Lactate-Related Genes in Radiotherapy Resistance of Lung Cancer by Integrated Bioinformatics and Experiment Validation
1. Department of Radiation Oncology, Nantong Tumor Hospital, Affiliated Tumor Hospital of Nantong University, Nantong 226361, China.
2. Department of Oncology, Haimen People's Hospital, Nantong 226100, China.
3. Department of Radiation Oncology, The Fourth Affiliated Hospital of Soochow University, Suzhou, Dushu Lake Hospital, Medical Center of Soochow University, Suzhou 215000, China.
* Equal contribution and share first authorship.
Received 2025-3-1; Accepted 2025-6-4; Published 2025-7-24
Abstract
Radiotherapy is a standard treatment for advanced lung cancer, but resistance remains a significant cause of treatment failure. This study aimed to investigate lactate-associated genes to identify patients likely to benefit from radiotherapy. RNA-seq data from 99 patients with lung cancer who underwent radiotherapy were analyzed to identify differentially expressed genes (DEGs) between resistant and sensitive cases. Bioinformatics tools were used to assess the prognostic relevance of lactate-related genes, and a risk score model was develpoed based on these genes. Dysregulation of these genes in patients with lung cancer undergoing radiotherapy was validated through in vitro experiments. Molecular docking was used to explore potential radiosensitizers. The analysis identified 1482 DEGs, with enrichment analysis highlighting lactate metabolism pathways. A risk score model was constructed using the lactate-related genes ADAMTS3, FADS2, and RTBDN to classify patients into high- and low-risk subgroups. Functional enrichment analysis revealed the model's impact on DNA repair and tumor immunity. A nomogram was developed for clinical implementation. Wet lab experiments further confirmed these findings. In conclusion, a novel risk score model based on lactate-related genes was developed to predict radiotherapy outcomes in lung cancer. FADS2 was identified as a potential biomarker for predicting resistance to radiotherapy. This study is the first to examine the predictive value of lactate-related genes for radiotherapy efficacy in lung cancer, offering valuable insights for personalized treatment strategies to improve therapeutic outcomes.
Keywords: lung cancer, radiotherapy resistance, lactate regulator, risk score, FADS2
Introduction
Lung cancer is the most prevalent and fatal malignancy worldwide, with a five-year overall survival (OS) rate of only 19%[1, 2]. Due to its subtle symptoms, most patients are diagnosed at advanced stages, resulting in poor prognoses[3]. Consequently, for advanced lung cancer, the primary treatment options include radiotherapy in combination with targeted therapy, chemotherapy, and immunotherapy, all of which have significantly improved survival rates[4, 5]. However, radiotherapy resistance remains the predominant cause of treatment failure. Thus, understanding the molecular mechanisms underlying radiotherapy resistance is crucial for developing more effective strategies to improve patient outcomes.
Radiotherapy kills tumor cells primarily through the generation of reactive oxygen species (ROS) and DNA breakage[6]. However, tumor heterogeneity leads to the survival of residual tumor cells, which can repair DNA, activate immune responses, promote cancer stem cells, and suppress various forms of cell death, all of which contribute to radiotherapy resistance[6-8]. Notably, all these processes require energy, which is often provided through altered metabolism. The Warburg effect, a hallmark of tumor metabolism, promotes rapid energy production via aerobic glycolysis, resulting in a lactic acid microenvironment[9]. Glycolysis has been linked to tumor metastasis and resistance to both chemotherapy and radiotherapy[10-13]. Lactate plays a pivotal role in delivering oxidative and gluconeogenic substrates and transducing cellular signaling in these processes[14, 15]. In lung cancer, lactate-related genes have been shown to influence prognosis and immune response[16-18]. However, studies investigating the regulation of lactate-related genes in the context of radiotherapy resistance in lung cancer remain scarce.
This study identified three prognostic lactate-related differentially expressed genes (LRDs) associated with radiotherapy resistance in patients with lung cancer from TCGA data. A risk score model based on these LRDs was developed and validated for clinical application, and the related mechanisms were further explored through bioinformatics analysis and in vitro experiments. Our findings suggest that lactate regulators could serve as biomarkers for predicting radiotherapy resistance in patients with lung cancer, providing clinicians with valuable tools to tailor personalized treatment strategies and improve therapeutic outcomes.
Materials and Methods
Data collection and sample preprocessing for radiotherapy patients with lung cancer
RNA sequencing, survival data, and clinical phenotypes were obtained from The Cancer Genome Atlas (TCGA) database. A total of 522 primary lung adenocarcinoma patients were obtained and underwent screening. The inclusion criteria were listed as following conditions: (a) patients with postoperative radiotherapy; (b) patients who have an evaluation of the efficacy of radiotherapy or information about their progress after radiotherapy; (c) patients who have a clear state of alive or death and a definite survival time. The exclusion criteria were listed as following conditions: (a) the patients whose number in the expression profile didn't match with the patient number corresponding to the clinical information (n=9); (b) patients with new primary tumor(n=12); (c) patients without postoperative radiotherapy or missing evaluation of the efficacy of radiotherapy or information about their progress after radiotherapy (n=389); (d) patients missing a clear state of alive or death, or a definite survival time (n=13). Finally, a cohort of 99 patients was extracted. Among them, 35 patients exhibited a complete response or partial response to radiotherapy, were categorized as the radiosensitive group. While 64 patients, exhibited progressive disease or stable disease or recurrence after radiotherapy, were included in the radioresistant group. Single cell sequencing analysis was performed on the lung cancer tissues in GSE179373.
Identification of differentially expressed genes (DEGs) and functional enrichment analysis
The threshold value for screening DEGs was set to |FC| > 1.2 and P < 0.05[19]. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of DEGs were analyzed to determine their potential functions and pathways. GO analysis, including biological processes, cellular components, and molecular functions, helped to understand the biological functions as well as positioning of DEGs. KEGG pathway analysis clarified the signaling pathways that DEGs may focus on. P < 0.05 was considered to be statistically significant. Gene set enrichment analysis (GSEA), and gene set variation analysis (GSVA) were conducted among DEGs using the R packages.
Construction of lactate-related risk score model
After lactate-related genes were downloaded from genecards database and were intersected with DEGs, LRDs were analyzed by Univariate COX regression analysis. Then, least absolute shrinkage and selection operator (LASSO) algorithm was performed on the significant LRDs in radiotherapy patients with lung cancer in TCGA cohort. Finally, the lactate related risk score model was constructed by the stepwise Cox regression algorithm. Radiotherapy patients with risk-score above the median were categorized as the high-risk subgroup, and the rest were included in the low-risk subgroup. For model, the time dependent receiver operating characteristic (ROC) was calculated. Survival analysis was used “survival”, “survminer” and “timeROC” packages, and the nomogram was used “rms” package.
Immune infiltration analysis
Cibersort algorithm analyzed the proportion of 22 immune cells in high-risk and low-risk subgroups. ssGSEA, xcell, ESTIMATE detected the different distribution of the immunity cells between the high-risk subgroup and low-risk subgroup using the SangerBox 3.0 platform.
Methylation and mutation analysis
The assessment of methylation status in ADAMTS3, FADS2, RTBDN between tumor and their corresponding para-cancerous tissues was conducted by the UALCAN database. The maftools package in R software was taken to download and extract mutation data, and the somatic mutation data of patients in high-risk subgroup and low-risk subgroup were then visually analyzed.
Immunohistochemical (IHC) analysis
The study was approved by the Research Ethics Board of the Tumor Hospital Affiliated with Nantong University (No.2023-053). Tissues slice was incubated with a primary antibody against a 1:100 dilution of FADS2 (Rabbit-anti-human, Proteintech) overnight at 4° C after the dewaxing and blocking of endogenous peroxides of the tissues. Visualisation of the antibody complex was achieved through a diaminobenzidine reaction. Tissue was counterstained by Meyer's haematoxylin. Based on the intensity of staining in the tumor from three hot spots of each tissue, IHC staining was analyzed by pathologists. Image J was used to score the intensity of staining.
Quantitative real-time PCR (qPCR)
RNA was isolated from H1299 radioresistant cell and radiosensitive cell A549[20, 21] by Trizol. Reverse transcription and quantitative PCR were carried out by using a two-step Prime Script TM RT reagent kit (TAKARA). Primers for the genes were synthesized and obtained from Thermo Fisher Scientific. The primer sequences are presented in Supplementary Table 1.
Cell culture and clonogenicity assay
The human lung cancer cell lines H1299, A549 were purchased from ATCC. Cell lines were maintained under a humidified atmosphere of 5% CO2 in air at 37℃ in RPMI 1640 medium supplemented with 10% FBS (Gibco BRL, USA) and 1% penicillin-streptomycin. H1299 or H1299 with lentivirus-mediated FADS2 overexpression were plated in 6-well plates prior to ionization radiation exposure (4 Gy, 8 Gy), and maintained in culture for 14 days. Afterward, when colonies were formed (defined as a bulk of at least 50 cells), cells were stained by crystal violet reagents.
Univariate analysis of LRDs associated with OS of patients with lung cancer underlying radiotherapy in TCGA.
| Characteristics | Univariate analysis | |
|---|---|---|
| HR (95% CI) | P value | |
| FADS2 | 0.662 (0.537 - 0.816) | < 0.001 |
| PARP1 | 0.555 (0.369 - 0.834) | 0.005 |
| STC2 | 0.774 (0.648 - 0.924) | 0.005 |
| DARS2 | 0.592 (0.407 - 0.860) | 0.006 |
| LMNB1 | 0.675 (0.507 - 0.897) | 0.007 |
| ATP2A2 | 0.609 (0.424 - 0.873) | 0.007 |
| RCC2 | 0.598 (0.406 - 0.880) | 0.009 |
| PHGDH | 0.821 (0.708 - 0.952) | 0.009 |
| UPB1 | 1.246 (1.055 - 1.473) | 0.010 |
| ASPM | 0.773 (0.634 - 0.943) | 0.011 |
| PSTPIP1 | 1.382 (1.066 - 1.792) | 0.015 |
| KIF21A | 0.703 (0.528 - 0.935) | 0.016 |
| CAPN3 | 1.267 (1.040 - 1.543) | 0.019 |
| ALDH18A1 | 0.664 (0.472 - 0.934) | 0.019 |
| HNRNPU | 0.496 (0.275 - 0.895) | 0.020 |
| MKI67 | 0.787 (0.637 - 0.972) | 0.026 |
| VDAC1 | 0.625 (0.405 - 0.966) | 0.034 |
| RTBDN | 0.896 (0.809 - 0.993) | 0.036 |
| ADAMTS3 | 0.848 (0.726 - 0.991) | 0.038 |
| PITX3 | 0.881 (0.780 - 0.995) | 0.041 |
| ASIC2 | 0.852 (0.728 - 0.997) | 0.046 |
| ZMPSTE24 | 0.619 (0.385 - 0.993) | 0.047 |
| HSPA4 | 0.637 (0.406 - 0.998) | 0.049 |
Weighted gene co-expression network analysis (WGCNA)
The co-expression network of radiotherapy patients with lung cancer in TCGA was generated using WGCNA package. A suitable soft threshold β is calculated based on the criteria for scale-free networks. In the following step, the weighted adjacency matrix was converted into a topological overlap matrix (TOM), and the corresponding dissimilarity (1-TOM) was calculated. Module identification was conducted using the dynamic tree cutting approach. The modules most relevant to the clinical phenotype were selected for subsequent analysis.
Molecular docking
In order to estimate the likelihood of drug interactions, the DEGs was identified from the high- and low-risk subgroups. L1000FWD database was used to detect the signaling pathways affected by small molecule drugs. Drug-gene interactions were performed on the Network Analyst Database. Furthermore, Home for researches platform was used for visualizing the molecular docking.
Statistical analysis
SPSS 20.0 software was used for statistical analysis, and Graphpad Prism 9 software for drawing the survival curve and statistical chart of the difference between the two groups. The difference was tested by independent sample t-test. Cox proportional hazards regression model was taken for univariate and multivariate analysis, while Kaplan Meier method and Log-rank test for overall survival analysis. P < 0.05 was considered significant.
Results
Identification of the key biological processes and signaling pathways to radiotherapy resistance in lung cancer
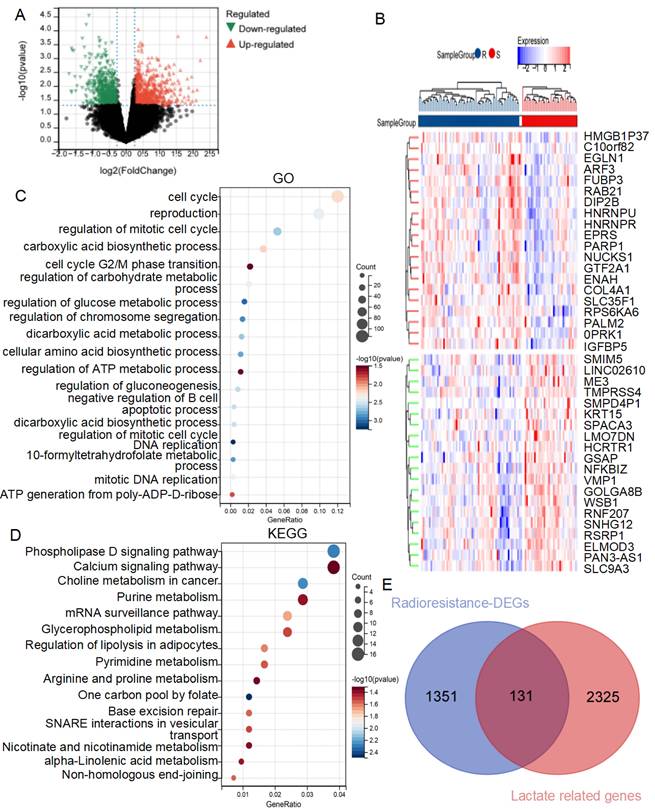
The workflow of this study is illustrated in Figure 1. To investigate the mechanisms of radiotherapy resistance in lung cancer, DEGs between radiotherapy-resistant and sensitive patients from the TCGA cohort were analyzed using limma. A total of 1482 DEGs (633 downregulated and 849 upregulated) as showed in volcano plot (Figure 2A), with the top 20 upregulated and downregulated DEGs displayed in the heatmap (Figure 2B). Further analysis of these DEGs using GO and KEGG enrichment revealed significant biological processes. GO analysis identified three primary categories: DNA damage repair (including the cell cycle, G2/M phase transition, and DNA replication)[22, 23], lactate metabolism (including regulation of carbohydrate metabolism, gluconeogenesis, and ATP generation from poly-ADP-D-ribose)[24, 25], and immune cell regulation (specifically the negative regulation of B cell apoptosis) (Figure 2C). KEGG analysis highlighted pathways associated with the Warburg effect, including phospholipase D signaling, glycerophospholipid metabolism, one-carbon pool by folate, and alpha-linolenic acid metabolism, as well as excision repair pathways[26-28] (Figure 2D). These findings highlight the critical roles of lactate metabolism, DNA repair, and immune response in radiotherapy resistance.
Construction of the lactate-related risk score model for radiotherapy patients with lung cancer
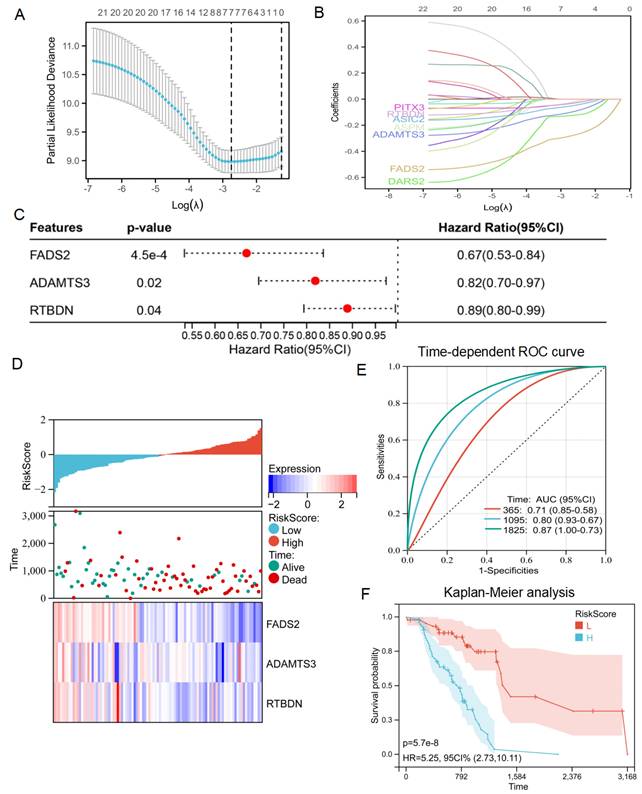
Based on these results, an intersection analysis of lactate-related genes and DEGs, identified 131 LRDs (Figure 2E). Subsequently, univariate Cox regression analysis was employed to construct a risk stratification system, with 23 LRDs found to be prognostically relevant for radiotherapy patients in the TCGA cohort (Table 1). To prevent overfitting of the prognostic signature, LASSO regression was applied to these 23 prognosis-related LRDs, resulting in the selection of seven LRDs based on the optimal λ value (Figures 3A, 3B). Stepwise regression further refined the model, extracting three key genes (Figure 3C). The risk score model was formulated using the following algorithm: risk score = (-0.257131817660543) × ADAMTS3 + (-0.500985047) × FADS2 + (-0.136319284) × RTBDN. The radiotherapy patients in the TCGA cohort were divided into high- and low-risk subgroups based on the median risk score for subsequent analysis. Comparisons of risk score distributions and survival statuses between these subgroups were performed (Figure 3D). The specificity and sensitivity of the signature were assessed by calculating the area under the curve (AUC) values for 1-, 3-, and 5-year survival, yielding AUCs of 0.71, 0.80, and 0.87, respectively, in the TCGA cohort (Figure 3E). Survival analysis revealed that patients in the high-risk subgroup had significantly poorer prognosis (P = 5.7e-8; Figure 3F).
Functional enrichment analysis of different risk score subgroups
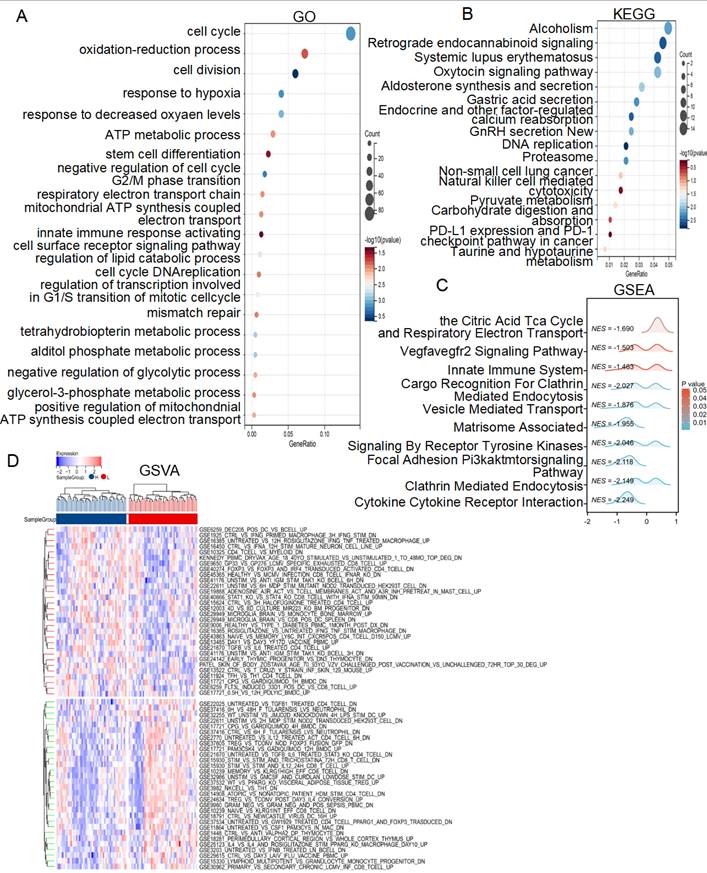
To investigate the molecular mechanisms underlying the different risk score subgroups, we performed a DEGs analysis of patients from the TCGA cohort and identified 1067 genes (|FC| > 1.2, P< 0.05). GO analysis revealed that the lactate-related risk score primarily impacted processes such as the cell cycle, DNA replication, oxidation-reduction processes, response to hypoxia, ATP metabolic process, mitochondrial ATP synthesis coupled electron transport, innate immune response activating cell surface receptor signaling pathway, regulation of lipid catabolic process, tetrahydrobiopterin metabolic process, alditol phosphate metabolic process, negative regulation of glycolytic process, glycerol-3-phosphate metabolic process (Figure 4A). KEGG analysis highlighted pathways involved in DNA replication, non-small cell lung cancer, natural killer cell mediated cytotoxicity, pyruvate metabolism, carbohydrate digestion and absorption-PD-L1 expression and PD-1 check point pathway in cancer (Figure 4B). Additionally, the GSEA algorithm explored the citric acid TCA cycle and respiratory electron transport, VEGFA-VEGFR2 signaling, innate immune system pathways, cargo recognition for clathrin mediated endocytosis, vesicle mediated transport, matrisome associated signaling by receptor tyrosine kinases were different between high- and low-risk score subgroups (Figure 4C). GSVA algorithm further revealed differences in multiple immune regulatory pathways between the subgroups (Figure 4D). These results suggest that lactate-related risk scores modulate radiotherapy outcomes through energy metabolism, immune infiltration, and DNA repair mechanisms.
Immune infiltration analysis for lactate-related risk score
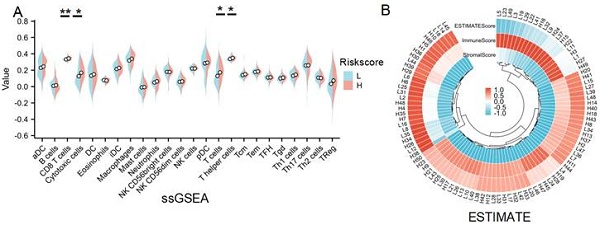
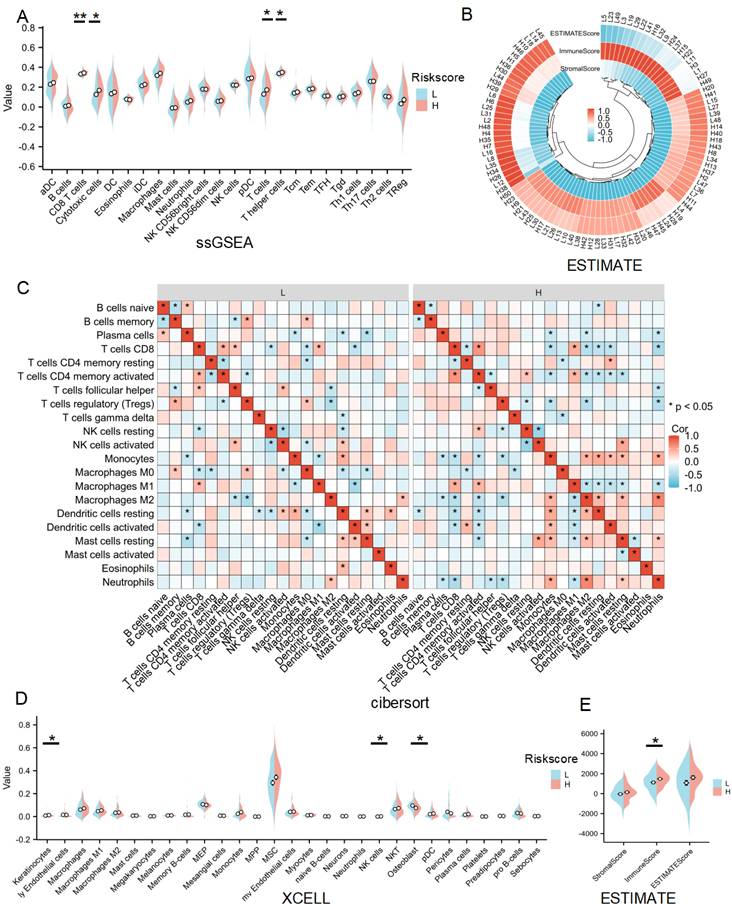
Given that GO, KEGG, GSEA and GSVA analyses all indicated a significant role of LRDs in immune infiltration, immune cell infiltration was assessed across 24 cell types using ssGSEA (Figure 5A). The results showed that CD8+ T cells, cytotoxic cells, T cells, and T helper cells were significantly more abundant in the high-risk score subgroup compared to the low-risk subgroup. ESTIMATE analysis showed the score of each sample (Figures 5B) and found immune score was higher in the high-risk subgroup (Figures 5C). CIBERSORT algorithms confirmed that CD4+ memory-activated T cells and M1 macrophages were inhibited, whereas M2 macrophages were significantly activated in the high-risk subgroup (Figures 5D). XCELL algorithm explored differences in Keratinocytes, NK cells and Osteoblast between the two subgroups (Figure 5D). Collectively, these results highlight the distinct patterns of immune cell infiltration between the lactate-related risk score subgroups, suggesting that immune modulation may influence the effectiveness of radiotherapy in patients with lung cancer.
The flow chart of the research. R: resistance, S: sensitive, DEGs: differentially expressed genes.
GO and KEGG analysis of DEGs from radioresistant patients comparing with sensitive ones with lung cancer in TCGA. (A) Volcano plot of the 1,482 DEGs. The red triangles represent the significantly up-regulated genes, and the blue triangles showed the significantly down-regulated genes. (B) Heat map showed the TOP20 up-regulated and down-regulated genes respectively. (C) GO enrichment analysis of the DEGs. (D) KEGG enrichment analysis of the DEGs. (E) The venn diagram of radiotherapy resistance DEGs and lactate-related genes.
Methylation and mutation analysis for lactate-related risk score
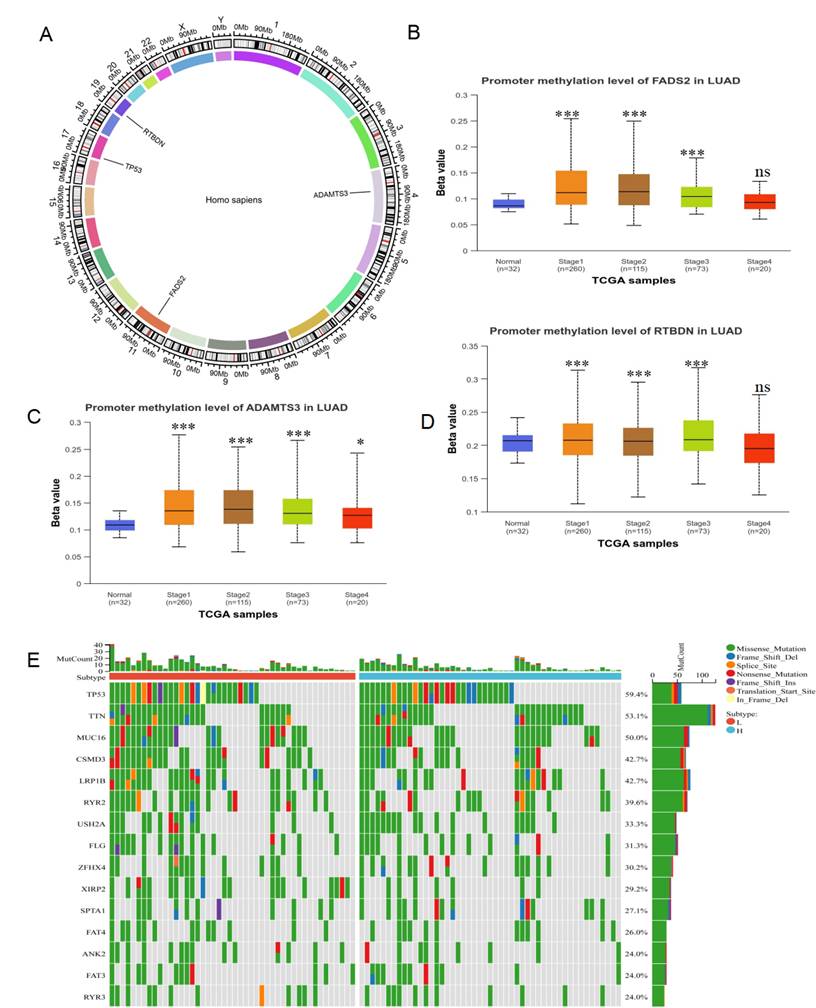
To explore the differences between lactate-related risk score subgroups, the chromosomal distribution of the three genes used to construct the lactate-related risk score was visualized through circos plots (Figure 6A). Given the role of aberrant DNA methylation in cancer-related dysregulation, the UALCAN database was utilized to investigate the relationship between the expression patterns of ADAMTS3, FADS2, and RTBDN and methylation levels in both normal and tumor tissues. The increased methylation of ADAMTS3, FADS2 and RTBDN was found in lung cancer tissues, potentially contributing to their downregulation (Figure 6B-6D). The gene mutation landscape revealed significant mutations in TP53, TTN, MUC16, USH2A, and ZFHX4 in the high-risk score subgroup, and in CSMD3, LRP1B, XIRP2, and RYR3 in the low-risk score subgroup (Figure 6E).
Clinical applications for lactate-related risk score
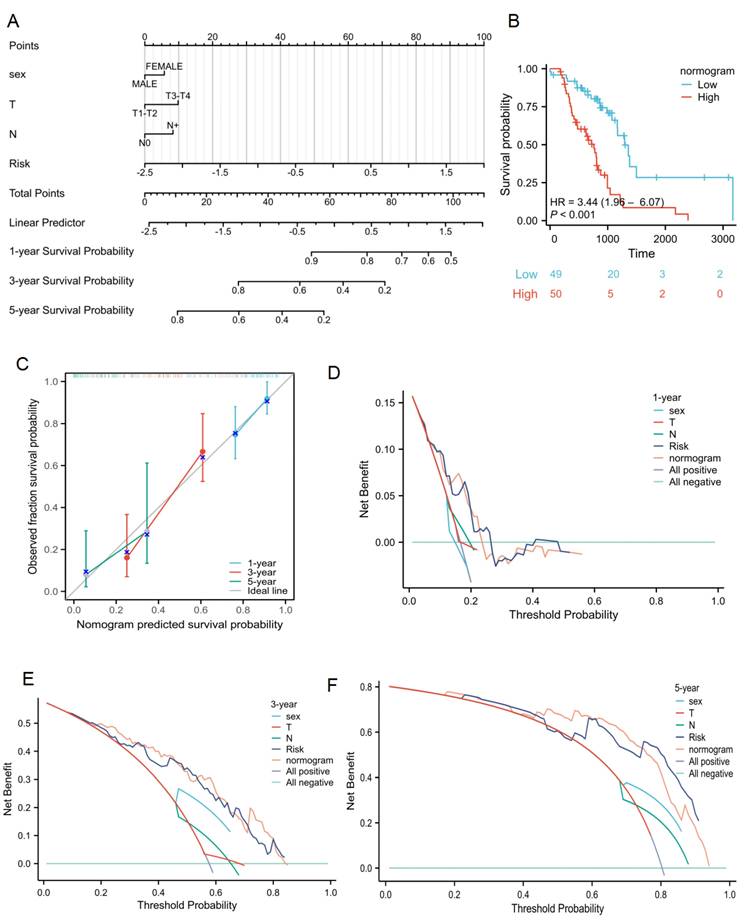
The relationship between the lactate-related risk score and clinical parameters was also assessed. The results showed significant correlations between the risk score and sex, radiotherapy response, T status, and N status (P < 0.001, Supplementary Table 2). To capture the complexity of the risk signature, a nomogram was developed incorporating both clinical information and the risk scores of radiotherapy patients with lung cancer from the TCGA cohort (Figure 7A). Patients with high risk scores had poorer survival compared to those with low scores (Figure 7B). Calibration plots for OS outcomes demonstrated strong concordance between predicted and observed OS at 1-, 3-, and 5-year intervals. The C-index for prediction was 0.701 (0.661-0.741), indicating a relatively good predictive performance (Figure 7C). Additionally, the nomogram model was evaluated using decision curve analysis (DCA) for radiotherapy patients with lung cancer in the TCGA cohort (Figure 7D-7F). Collectively, the lactate-related risk score emerged as an independent and robust prognostic indicator, offering enhanced predictive value when combined with clinical features for radiotherapy patients with lung cancer.
Identification of lactate related signature via LASSO-stepwise algorithms. (A, B) LASSO analysis with minimal lambda value. (C) Three genes were screen out by stepwise Cox algorithm. (D) Lactate-related risk score model illustrating the distribution of survival status and the expression of the three candidate genes. (E) The time-dependent ROC curve for lactate-related risk score. (F) Kaplan-Meier analysis demonstrated the prognostic significance of the risk model in TCGA.
Enrichment analysis of the lactate-related risk score. GO (A), KEGG (B), GSEA (C) and GSVA (D) analysis of the DEGs and potential signaling pathways in different lactate-related risk score subgroups.
Expression and prognosis of LRDs in radiotherapy patients with lung cancer
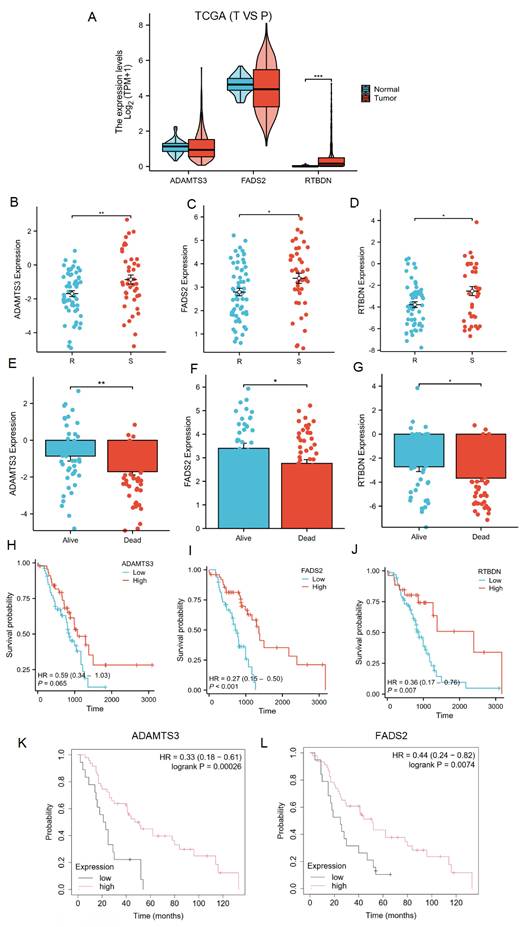
The lactate-related risk score, constructed using ADAMTS3, FADS2, and RTBDN, was further explored to assess their expression and prognostic value for radiotherapy patients with lung cancer across multiple databases. ADAMTS3 and FADS2 exhibited downregulation in tumor tissues compared to normal lung tissues, whereas RTBDN displayed an opposite trend (Figure 8A). Notably, the expression of these three genes was significantly downregulated in the radiotherapy-resistant group from the TCGA cohort (Figure 8B-8D), as well as in the deceased subgroup from the same cohort (Figure 8E-8G). Survival analysis revealed that radiotherapy patients with low FADS2 expression had a poorer prognosis compared to those with high expression of FADS2 in the TCGA cohort (P < 0.001). A similar trend was observed for ADAMTS3 (P = 0.065) and RTBDN (P = 0.007) (Figure 8H-8J). Moreover, validation in the KM-Plot database confirmed that radiotherapy patients with low expression of FADS2 and ADAMTS3 had better prognoses, whereas RTBDN did not show significant prognostic relevance (Figure 8K-8L). Based on these findings, FADS2 was prioritized for further validation.
Immune infiltration analysis of lactate-related risk score. (A) ssGSEA analysis detected immune cell expression between the high- and low-risk score subgroups. (B, E) The landscape and comparing of the immunoscore in high- and low-risk score subgroups by ESTIMATE algorithms. (C) CIBERSORT showed the different correlations between 22 immune cells in the high- and low-risk score subgroups, respectively. (D) Xcell algorithms detected immune cell expression between the high- and low-risk score subgroups. L:low-risk score subgroup, H: high-risk score subgroup, *P<0.05, **P<0.01.
Methylation and mutation analysis of LRDs. (A) The chromosomal distribution of LRDs by circos plots. Methylation analysis of FADS2 (B), ADAMTS3 (C) and RTBDN (D) in different stage of lung cancer and normal tissues was explored in the UALCAN database. (E) The top 15 mutation genes in high and low lactate-related risk score subgroups, respectively. ***:P < 0.001.
Clinical application of lactate-related risk score. (A) The nomogram of the risk score and clinical parameters for radiotherapy patients with lung cancer in TCGA cohort. (B) Kaplan-Meier plot analysis based on nomogram. (C) The calibration curves displayed the accuracy of the nomogram in the 1-, 3-, and 5-years. (D-F) DCA curves to assess the ability of sex, T stage, N stage, risk score, and their combination to predict 1-, 3-, 5-years overall survival of radiotherapy patients with lung cancer in TCGA cohort.
Validation of the LRDs in lung cancer tissues and radioresistant cells
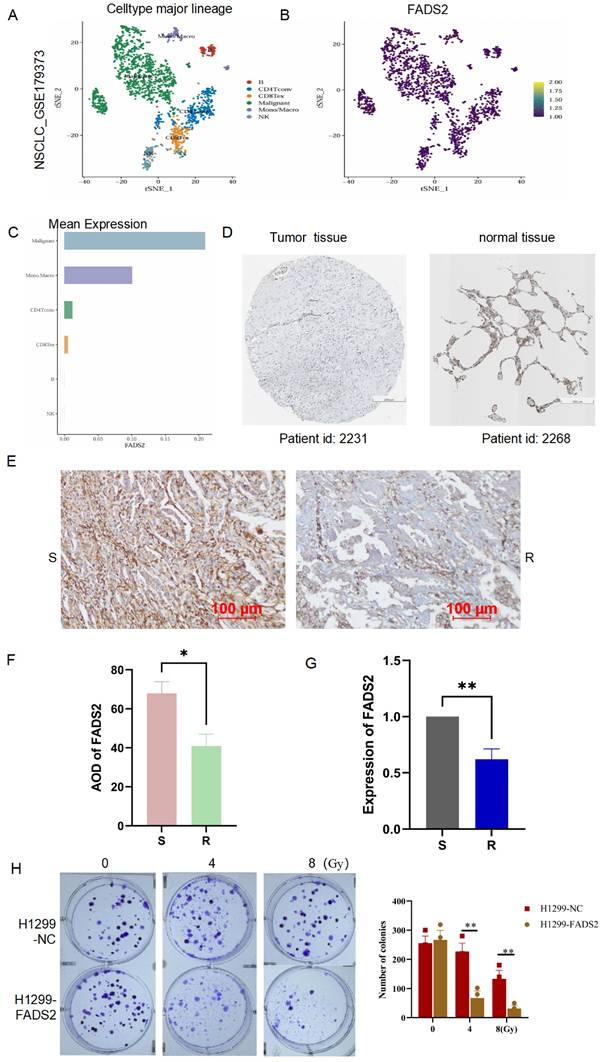
To explore the expression and localization of FADS2 in lung cancer tissues, we analyzed single-cell RNA sequencing data from lung cancer (GSE179373) were analyzed, revealing that FADS2 was predominantly expressed in malignant cells (Figure 9A-9C). In the Human Protein Atlas database, the expression of FADS2 was lower in lung cancer tissues compared to normal lung tissues (Figure 9D). To further validate these findings, 10 lung cancer tissue samples (5 radioresistant and 5 radiosensitive) were collected, and IHC analysis showed significantly lower FADS2 expression in radioresistant tissues compared to radiosensitive tissues (Figure 9E, 9F). At the cellular level, qPCR confirmed a significant decrease in FADS2 expression in radioresistant cells (Figure 9G). Furthermore, colony formation assays demonstrated that upregulation of FADS2 significantly increased the sensitivity of radioresistant cells to 4Gy and 8Gy irradiation (Figure 9H). Collectively, these results suggest that FADS2 is associated with radiotherapy resistance in lung cancer.
Expression and prognostic significance of LRDs. (A) Expression of LRDs in lung cancer patients of TCGA. (B-G) Expression of LRDs in radiotherapy patients with lung cancer of TCGA cohort. (H-J) Correlation between LRDs and OS in radiotherapy patients with lung cancer of TCGA cohort. (K-L) Correlation between LRDs and OS in radiotherapy patients with lung cancer from KM-plot database.
Validation the expression and role of FADS2 in radiotherapy resistance. (A, B, C) Single-cell sequencing analysis for the location and expression of FADS2 in lung cancer tissue from GSE179373. (D) IHC staining of FADS2 in lung cancer and para-carcinoma tissue in HPA database. (E&F) IHC analysis of FADS2 in radioresistant (n=5) and radiosensitive (n=5) tissues. (G) qPCR analysis of FADS2 in radioresistant cell (H1299) and radiosensitive cell (A549). (H) Clonogenicity assay explored the role of FADS2 in radiotherapy sensitive. H1299-FADS2: over-expression of FADS2 in H1299. *:P < 0.05, **:P < 0.01.
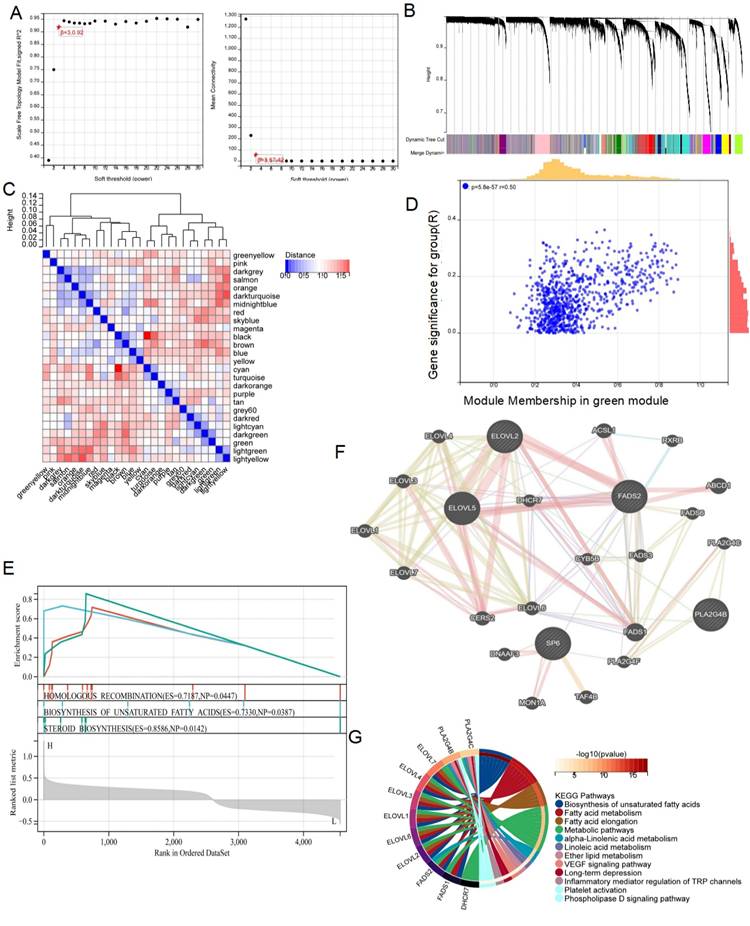
Identifying the role of FADS2 via WGCNA analysis. (A) The correlation between soft threshold and scale free topology model as well as mean connectivity. (B) Highly interconnected groups of genes were clustered and modules are represented by distinct colours in the horizontal bar. (C) The correlations between different modules were calculated. (D) The high correlation between GS and MM in the green module in radiotherapy resistance. (E) Genes in the green module were analyzed by GSEA. (F) Co-expression analysis for FADS2 with genes in the green module. (G) KEGG enrichment analysis for the key genes co-expressed with FADS2.
Exploring the potential mechanism of LRDs regulating radiotherapy resistance in lung cancer
To further elucidate the mechanism by which FADS2 regulates radiotherapy resistance in lung cancer, WGCNA was conducted based on the DEGs from the TCGA cohort, stratified by the median FADS2 expression. A soft threshold of β = 3 was applied to construct a scale-free network (Figure 10A). The DEGs were grouped into 26 co-expression modules, represented in various colors (Figure 10B, 10C). The green module exhibited the strongest correlation with radiotherapy resistance, with a module-trait relationship R value of 0.50 (P = 5.8e-17) (Figure 10D). Then, 875 DEGs from the green module underwent GSEA revealing enrichment in pathways such as homologous recombination, biosynthesis of unsaturated fatty acids, and steroid biosynthesis in the FADS2-high expression group (Figure 10E). Additionally, the GeneMANIA database identified co-expressed genes within the green module associated with FADS2 (Figure 10F), which were enriched in biosynthesis of unsaturated fatty acids fatty acid metabolism, fatty acid elongation, metabolic pathways, linoleic acid metabolism, ether lipid metabolism, VEGF signaling pathway, inflammatory mediator regulation of TRP channels, platelet activation, and phospholipase D signaling pathway (Figure 10G).
Molecular docking of small molecule compounds for improving radiotherapy effect
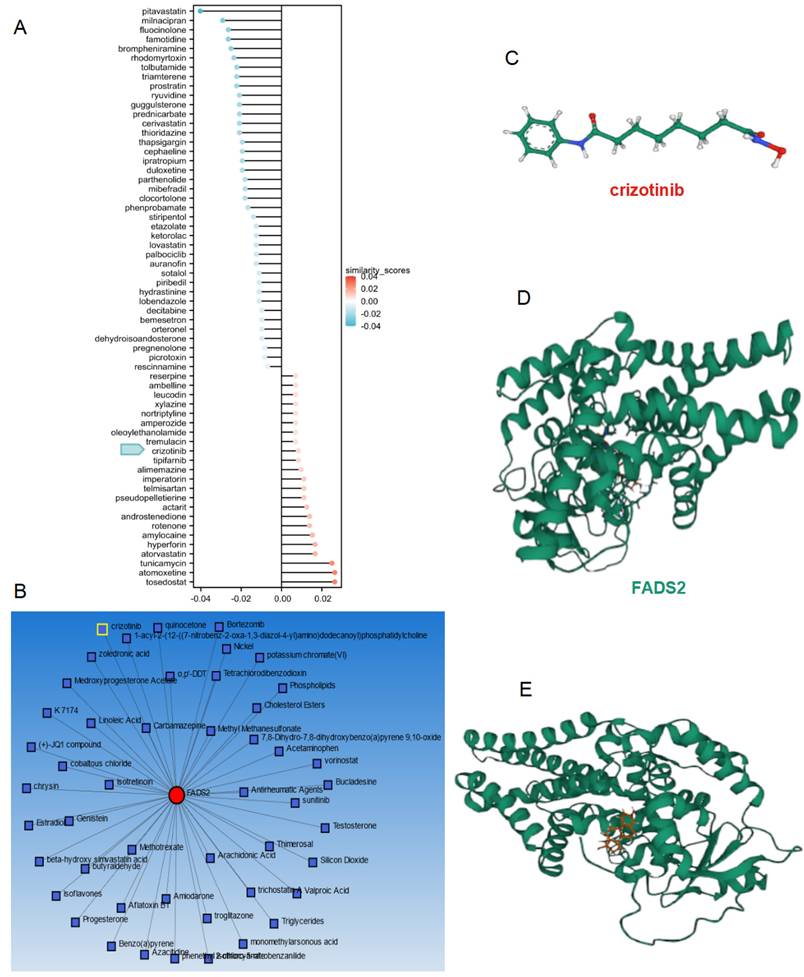
To improve the prognosis of patients in the high-risk score subgroup, the L1000FWD database was utilized to identify small-molecule compounds with potential therapeutic effects (Figure 11A). The NetworkAnalyst database was employed to explore compounds targeting FADS2 (Figure 11B). Crizotinib emerged as a common drug in both databases, exhibiting inhibition of FADS2 expression. The structure of crizotinib and its interaction with FADS2 were visualized (Figure 11C-11E). Molecular docking analysis confirmed the direct interaction between FADS2 and crizotinib. The hydrophobic pockets of both targets were successfully occupied, with a low binding energy of -7.896 kcal/mol, indicating a highly stable binding. These results suggest that crizotinib may serve as a potential radiosensitizer for lung cancer.
Discussion
Lung cancer, a leading cause of cancer-related mortality globally, presents a significant challenge in clinical management. Radiotherapy plays a critical role in managing advanced-stage lung cancer, offering symptom relief and controlling disease progression[29]. However, the development of radiotherapy resistance remains a substantial barrier, contributing to rapid disease progression. Understanding the mechanisms underlying this resistance, particularly the role of lactate metabolism, is essential for developing strategies to overcome this challenge.
This study identified DNA repair and lactate metabolism-associated biological processes and signaling pathways as key factors in radiotherapy resistance in lung cancer. On one hand, DNA repair is a well-established contributor to radiotherapy resistance[30], supporting the validity of our findings. On the other hand, this result underscores the importance of lactate-related genes in modulating the radiotherapy response.
Building on these insights, LRDs were identified, and a lactate-related risk score model was constructed using LASSO regression and stepwise Cox regression. Time-dependent ROC and survival analyses demonstrated the strong predictive and diagnostic value of the risk score in patients with lung cancer undergoing radiotherapy. To explore the underlying mechanisms, GO, KEGG, GSEA, and GSVA analyses confirmed a significant enrichment of biological processes related to energy metabolism, including response to hypoxia, ATP metabolism, respiratory electron transport, and lipid catabolic regulation. Under hypoxic conditions, the hypoxia-inducible factor (HIF) induces LDHA expression, driving anaerobic metabolism by converting pyruvate to lactate to generate ATP, which enhances cell proliferation and defines radiotherapy resistance in prostate cancer[31]. The Warburg effect, which persists even under normoxic conditions, promotes DNA repair and enhances cell survival, ultimately contributing to resistance in various cancers, including head and neck cancer[32], esophageal squamous cell carcinoma[33], lung cancer[34]. These findings suggest that lactate-related genes may regulate radiotherapy resistance in lung cancer by promoting the Warburg effect, thus providing energy necessary for DNA repair.
Additionally, this study revealed differences in immune cell infiltration between lactate-related risk score subgroups. CD4 memory T cells and M1 macrophages were inhibited, while M2 macrophages were significantly activated in the high-risk score subgroup. Furthermore, differences in keratinocytes, NK cells, and osteoblasts were noted between the subgroups. These findings align with previous studies showing that CD4 T cells and M1 macrophages play a role in inhibiting tumor progression, as seen in glioblastoma[35], and prostate cancer[36]. Noonepalle, et al. demonstrated that macrophage M1 responses induced acute pro-inflammatory reactions immediately following radiotherapy. However, these macrophages transitioned into anti-inflammatory, pro-tumor M2 macrophages in the following days, contributing to cancer resistance[37]. Additionally, Yang et al. reported that lactate facilitates the formation of an immunosuppressive tumor microenvironment by lowering the pH, which accelerates tumor progression and impairs anti-tumor immunity[10]. These findings suggest that lactate-related genes promote radiotherapy resistance by enhancing the Warburg effect and suppressing immune regulation.
Drug analysis and molecular docking for inhibiting radiotherapy resistance in lung cancer. (A) The potential drugs for lung cancer treatment. (B) Network analyst database predicting the potential drugs reacting with FADS2. (C) The 2D structure of crizotinib. (D) The protein structure of FADS2. (E) Molecular docking for crizotinib and FADS2.
Fatty acid desaturase 2 (FADS2), a key gene in the risk score model, was found to be downregulated, indicating a poor prognosis for radiotherapy patients with lung cancer. FADS2 is essential for the biosynthetic pathways that produce long-chain polyunsaturated fatty acids (PUFAs)[25]. PUFAs play important roles in tumor progression, enhancing the efficacy of chemotherapy and radiotherapy while reducing the risk of recurrence by modulating inflammation and immune responses in lung and colorectal cancers[26, 38]. Besides these, PUFAs significantly impact mitochondrial oxidative phosphorylation (OXPHOS), stimulating the tricarboxylic acid cycle in the mitochondria to produce more ATP and ROS than glycolysis[39]. Elevated FADS2 expression was shown to inhibit radiotherapy resistance in lung cancer cells, similar to its role in reducing DNA damage in esophageal adenocarcinoma[40]. It is hypothesized that decreased FADS2 expression reduces PUFA production, leading to downregulation of OXPHOS and promoting the Warburg effect, thereby facilitating radiotherapy resistance in lung cancer.
The study has several limitations. Our analysis was based solely on cohorts from TCGA and GEO, and future research should include additional real-world cohorts to further validate the lactate-related risk score. Additionally, further experiments are required to fully elucidate the underlying mechanisms. In summary, a novel lactate-related risk score model for predicting radiotherapy outcomes has been developed, with FADS2 identified as a potent biomarker for predicting radiotherapy resistance in lung cancer. While previous studies have primarily focused on lactate-related genes for prognostic significance in lung cancer, this study is the first to demonstrate their potential utility in predicting radiotherapy efficacy through integrative multi-database analysis. Assessing FADS2 expression in surgical or biopsy specimens imposes no additional burden on patients, underscoring its strong clinical applicability. This study offers a promising therapeutic strategy for clinicians and is likely to stimulate interest among drug developers.
Abbreviations
DEG: differentially expressed genes; LRDs: lactate-related differentially expressed genes; TCGA: The Cancer Genome Atlas; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; GSEA: Gene set enrichment analysis; GSVA: gene set variation analysis; LASSO: least absolute shrinkage and selection operator; ROC: receiver operating characteristic; IHC: immunohistochemical; qPCR: quantitative real-time PCR; WGCNA: weighted gene co-expression network analysis; TOM: topological overlap matrix; FADS2: Fatty acid desaturases 2; PUFA: polyunsaturated fatty acids; OXPHOS: oxidative phosphorylation.
Supplementary Material
Supplementary tables.
Acknowledgements
This research was supported by the Clinical Medicine Special Program from Nantong University (Grant no.2023JY012), the Health Committee of Nantong (grant no. QN2022031) and Suzhou Science and Technology Plan (Grant no. SZM2023031), Science and Technology Bureau of Nantong (Grant no.MS2023041).
Author contributions
Canyu Liu, Qin Ge and Qiong Yi: financial support and final approval of the manuscript; Qianqian Li, Lili Shen and Yanguang Yang: manuscript writing, data analysis; Guomei Tai, Qiwei Zhu, Canyu Liu: IHC staining, cell experiment, and statistical analysis.
Ethics committee approval and/or patient consent
The study was approved by the Research Ethics Board of the Tumor Hospital Affiliated with Nantong University (No.2023-053). All patients signed informed consent.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-63
2. Zhou L, Dong C, Xu Z, Wang X, Zhang L, Chen S. et al. NEDD8-conjugating enzyme E2 UBE2F confers radiation resistance by protecting lung cancer cells from apoptosis. J Zhejiang Univ Sci B. 2021;22:959-65
3. Liang L, Wang Z, Duan H, He Z, Lu J, Jiang X. et al. Survival Benefits of Radiotherapy and Surgery in Lung Cancer Brain Metastases with Poor Prognosis Factors. Curr Oncol. 2023;30:2227-36
4. Zeng Y, Zhang L, Liang Y, Zhang X, Li L, Wang M. et al. Evaluation safety and efficacy of immune checkpoint blockers (ICB) and radiotherapy combination versus ICB in non-small cell lung cancer patients with recurrence or metastasis: A systematic review and meta-analysis. Cancer Med. 2023;12:13928-41
5. Xiong J, Barayan R, Louie AV, Lok BH. Novel therapeutic combinations with PARP inhibitors for small cell lung cancer: A bench-to-bedside review. Semin Cancer Biol. 2022;86:521-42
6. Busato F, Khouzai BE, Mognato M. Biological Mechanisms to Reduce Radioresistance and Increase the Efficacy of Radiotherapy: State of the Art. Int J Mol Sci. 2022;23:10211
7. Zhou T, Zhang LY, He JZ, Miao ZM, Li YY, Zhang YM. et al. Review: Mechanisms and perspective treatment of radioresistance in non-small cell lung cancer. Front Immunol. 2023;14:1133899
8. Tan X, Huang X, Niu B, Guo X, Lei X, Qu B. Targeting GSTP1-dependent ferroptosis in lung cancer radiotherapy: Existing evidence and future directions. Front Mol Biosci. 2022;9:1102158
9. Liao M, Yao D, Wu L, Luo C, Wang Z, Zhang J. et al. Targeting the Warburg effect: A revisited perspective from molecular mechanisms to traditional and innovative therapeutic strategies in cancer. Acta Pharm Sin B. 2024;14:953-1008
10. Yang Y, Gao Y, Xiong Y, Gong Y, Lu J, Zhang Y. et al. Research Progress of Warburg Effect in Hepatocellular Carcinoma. Front Biosci (Landmark Ed). 2024;29:178
11. Chelakkot C, Chelakkot VS, Shin Y, Song K. Modulating Glycolysis to Improve Cancer Therapy. Int J Mol Sci. 2023;24:2606
12. Li L, Liu H, Du L, Xi P, Wang Q, Li Y. et al. miR-449a Suppresses LDHA-Mediated Glycolysis to Enhance the Sensitivity of Non-Small Cell Lung Cancer Cells to Ionizing Radiation. Oncol Res. 2018;26:547-56
13. Skaripa-Koukelli I, Hauton D, Walsby-Tickle J, Thomas E, Owen J, Lakshminarayanan A. et al. 3-Bromopyruvate-mediated MCT1-dependent metabolic perturbation sensitizes triple negative breast cancer cells to ionizing radiation. Cancer Metab. 2021;9:37
14. Shang S, Wang MZ, Xing Z, He N, Li S. Lactate regulators contribute to tumor microenvironment and predict prognosis in lung adenocarcinoma. Front Immunol. 2022;13:1024925
15. Madji Hounoum B, Mavel S, Coque E, Patin F, Vourc'h P, Marouillat S. et al. Wildtype motoneurons, ALS-Linked SOD1 mutation and glutamate profoundly modify astrocyte metabolism and lactate shuttling. Glia. 2017;65:592-605
16. Jiang W, Zhang F, Tang Z, Xu S, Zhang Y, Liu L. et al. Prediction of prognosis and immune response in lung adenocarcinoma based on mitophagy and lactate-related gene signatures. Int J Clin Oncol. 2025;30:277-97
17. Chang X, Lu T, Xu R, Wang C, Zhao J, Zhang L. Identification of lactate metabolism-related subtypes and development of a lactate-related prognostic indicator of lung adenocarcinoma. Front Genet. 2022;13:949310
18. Guo Z, Hu L, Wang Q, Wang Y, Liu XP, Chen C. et al. Molecular Characterization and Prognosis of Lactate-Related Genes in Lung Adenocarcinoma. Curr Oncol. 2023;30:2845-61
19. Qian H, Ji R, Shen C, Wei Y, Sheng C, Ni Q. et al. ATRX is a predictive marker for endocrinotherapy and chemotherapy resistance in HER2-/HR+ breast cancer through the regulation of the AR, GLI3 and GATA2 transcriptional network. Aging (Albany NY). 2023;15:14996-5024
20. Kaur S, Nag A, Gangenahalli G, Sharma K. Peroxisome Proliferator Activated Receptor Gamma Sensitizes Non-small Cell Lung Carcinoma to Gamma Irradiation Induced Apoptosis. Front Genet. 2019;10:554
21. Ryu SH, Lee SW, Yang YJ, Song SY, Kim JH, Choi EK. et al. Intracytoplasmic epidermal growth factor receptor shows poor response to the cetuximab antitumor effect in irradiated non-small cell lung cancer cell lines. Lung Cancer. 2012;77:482-7
22. Xu MD, Liu SL, Zheng BB, Wu J, Wu MY, Zhang Y. et al. The radiotherapy-sensitization effect of cantharidin: Mechanisms involving cell cycle regulation, enhanced DNA damage, and inhibited DNA damage repair. Pancreatology. 2018;18:822-32
23. Deng S, Vlatkovic T, Li M, Zhan T, Veldwijk MR, Herskind C. Targeting the DNA Damage Response and DNA Repair Pathways to Enhance Radiosensitivity in Colorectal Cancer. Cancers (Basel). 2022;14:4874
24. Li X, Yang Y, Zhang B, Lin X, Fu X, An Y. et al. Lactate metabolism in human health and disease. Signal Transduct Target Ther. 2022;7:305
25. Xuan Y, Wang H, Yung MM, Chen F, Chan WS, Chan YS. et al. SCD1/FADS2 fatty acid desaturases equipoise lipid metabolic activity and redox-driven ferroptosis in ascites-derived ovarian cancer cells. Theranostics. 2022;12:3534-52
26. Finocchiaro C, Segre O, Fadda M, Monge T, Scigliano M, Schena M. et al. Effect of n-3 fatty acids on patients with advanced lung cancer: a double-blind, placebo-controlled study. Br J Nutr. 2012;108:327-33
27. Feng P, Huang C. Phospholipase D-mTOR signaling is compromised in a rat model of depression. J Psychiatr Res. 2013;47:579-85
28. Hopperton KE, Duncan RE, Bazinet RP, Archer MC. Fatty acid synthase plays a role in cancer metabolism beyond providing fatty acids for phospholipid synthesis or sustaining elevations in glycolytic activity. Exp Cell Res. 2014;320:302-10
29. Yao Y, Fareed R, Zafar A, Saleem K, Huang T, Duan Y. et al. State-of-the-art combination treatment strategies for advanced stage non-small cell lung cancer. Front Oncol. 2022;12:958505
30. Xu Q, Zhang H, Liu H, Han Y, Qiu W, Li Z. Inhibiting autophagy flux and DNA repair of tumor cells to boost radiotherapy of orthotopic glioblastoma. Biomaterials. 2022;280:121287
31. Koukourakis MI, Giatromanolaki A, Panteliadou M, Pouliliou SE, Chondrou PS, Mavropoulou S. et al. Lactate dehydrogenase 5 isoenzyme overexpression defines resistance of prostate cancer to radiotherapy. Br J Cancer. 2014;110:2217-23
32. Mirestean CC, Iancu RI, Iancu DPT. New horizons in modulating the radio-sensitivity of head and neck cancer - 100 years after Warburg' effect discovery. Front Oncol. 2022;12:908695
33. Chen Y, Fan P, Chen Z, Zheng Z, He M, Zhao X. et al. Long non-coding RNA SRA1 suppresses radiotherapy resistance in esophageal squamous cell carcinoma by modulating glycolytic reprogramming. Open Med (Wars). 2024;19:20240946
34. Allen KT, Chin-Sinex H, DeLuca T, Pomerening JR, Sherer J, Watkins JB 3rd. et al. Dichloroacetate alters Warburg metabolism, inhibits cell growth, and increases the X-ray sensitivity of human A549 and H1299 NSC lung cancer cells. Free Radic Biol Med. 2015;89:263-73
35. Chen D, Varanasi SK, Hara T, Traina K, Sun M, McDonald B. et al. CTLA-4 blockade induces a microglia-Th1 cell partnership that stimulates microglia phagocytosis and anti-tumor function in glioblastoma. Immunity. 2023;56:2086-104 e8
36. Feng D, Shi X, You J, Xiong Q, Zhu W, Wei Q. et al. A cellular senescence-related gene prognostic index for biochemical recurrence and drug resistance in patients with prostate cancer. Am J Cancer Res. 2022;12:3811-28
37. Noonepalle SKR, Grindrod S, Aghdam N, Li X, Gracia-Hernandez M, Zevallos-Delgado C. et al. Radiotherapy-induced Immune Response Enhanced by Selective HDAC6 Inhibition. Mol Cancer Ther. 2023;22:1376-89
38. Miccadei S, Masella R, Mileo AM, Gessani S. omega3 Polyunsaturated Fatty Acids as Immunomodulators in Colorectal Cancer: New Potential Role in Adjuvant Therapies. Front Immunol. 2016;7:486
39. Varela-Lopez A, Vera-Ramirez L, Giampieri F, Navarro-Hortal MD, Forbes-Hernandez TY, Battino M. et al. The central role of mitochondria in the relationship between dietary lipids and cancer progression. Semin Cancer Biol. 2021;73:86-100
40. Molendijk J, Kolka CM, Cairns H, Brosda S, Mohamed A, Shah AK. et al. Elevation of fatty acid desaturase 2 in esophageal adenocarcinoma increases polyunsaturated lipids and may exacerbate bile acid-induced DNA damage. Clin Transl Med. 2022;12:e810
Author contact
![]() Corresponding authors: Canyu Liu: Department of Radiation Oncology, The Fourth Affiliated Hospital of Soochow University, Suzhou, Dushu Lake Hospital, Medical Center of Soochow University, 9th.Chongwen Road, Suzhou, 215000, China. Tel.: +86-15251347010. Email: canyuliucom. Qin Ge: Department of Radiation Oncology, Nantong Tumor Hospital, Affiliated Tumor Hospital of Nantong University, 30 Tong'yang Road, Nantong, Jiangsu 226321, China. Tel.: +86-13813606819. Email: geqin.kelleycom. Qiong Yi: Department of Radiation Oncology, Nantong Tumor Hospital, Affiliated Tumor Hospital of Nantong University, 30 Tong'yang Road, Nantong, Jiangsu 226321, China. Tel.: +86-(0513)86729186. Email: yiqiong86edu.cn.
Corresponding authors: Canyu Liu: Department of Radiation Oncology, The Fourth Affiliated Hospital of Soochow University, Suzhou, Dushu Lake Hospital, Medical Center of Soochow University, 9th.Chongwen Road, Suzhou, 215000, China. Tel.: +86-15251347010. Email: canyuliucom. Qin Ge: Department of Radiation Oncology, Nantong Tumor Hospital, Affiliated Tumor Hospital of Nantong University, 30 Tong'yang Road, Nantong, Jiangsu 226321, China. Tel.: +86-13813606819. Email: geqin.kelleycom. Qiong Yi: Department of Radiation Oncology, Nantong Tumor Hospital, Affiliated Tumor Hospital of Nantong University, 30 Tong'yang Road, Nantong, Jiangsu 226321, China. Tel.: +86-(0513)86729186. Email: yiqiong86edu.cn.

 Global reach, higher impact
Global reach, higher impact