Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(10):3094-3102. doi:10.7150/jca.116232 This issue Cite
Research Paper
Investigation of CBX4 Polymorphisms and Their Association with Clinicopathological Features in Asian Patients with Oral Squamous Cell Carcinoma
1. Institute of Medicine, Chung Shan Medical University, Taichung 40201, Taiwan.
2. Department of Radiation Oncology, Changhua Christian Hospital, Changhua 500, Taiwan.
3. Institute of Translational Medicine and New Drug Development, China Medical University, Taichung 406040, Taiwan.
4. Graduate Institute of Biomedical Sciences, China Medical University, Taichung 406040, Taiwan.
5. Center for Molecular Medicine, China Medical University Hospital, Taichung 404327, Taiwan.
6. Institute of Oral Sciences, Chung Shan Medical University, Taichung 40201, Taiwan.
7. School of Medicine, Chung Shan Medical University, Taichung 40201, Taiwan.
8. Department of Otolaryngology, Chung Shan Medical University Hospital, Taichung 40201, Taiwan.
9. Department of Otolaryngology Head and Neck Surgery China Medical University Hospital, Taichung 404327, Taiwan.
10. School of Medicine, College of Medicine, China Medical University, Taichung 40402, Taiwan.
11. Department of Surgery, China Medical University Hospital, Taichung 404327, Taiwan.
12. Department of Medical Research, Chung Shan Medical University Hospital, Taichung 40201, Taiwan.
13. Department of Medical Laboratory Science and Biotechnology, Asia University, Taichung 413305, Taiwan.
# These authors contributed equally.
Received 2025-4-22; Accepted 2025-6-21; Published 2025-7-4
Abstract
Head and neck squamous cell carcinomas (HNSCCs) are derived from the mucosal epithelium in the oral cavity, pharynx and larynx, and are predominantly linked to behavioral risk factors such as tobacco use and excessive alcohol consumption. In Taiwan, oral squamous cell carcinoma (OSCC) is one of the most prevalent subtypes of HNSCC and ranks as the fifth leading cause of cancer-related mortality in the country. The Chromobox (CBX) gene family encodes core subunits of the canonical Polycomb Repressive Complex 1 (cPRC1), a key epigenetic regulator mediating chromatin compaction and transcriptional silencing through histone modification. Dysregulation of CBX gene expression, whether at the transcriptional or post-transcriptional level, has been increasingly linked to tumorigenesis, metastasis, and cancer recurrence in a variety of malignancies. In particular, CBX4 has been implicated in the progression of gastric cancer, with growing evidence suggesting its broader role in cancer development. From the TCGA database, we found that levels of CBX4 are lower in Asian OSCC patients compared with other races. This prompted us to further investigate the genetic landscape of CBX4 in oral cancer. We conducted a case-control study involving 1184 male OSCC patients and 1188 healthy controls to evaluate whether genetic variants in CBX4, in conjunction with environmental exposures, contribute to OSCC susceptibility and progression in an Asian population. Four single-nucleotide polymorphisms (SNPs) of CBX4 (rs1285251, rs2289728, rs3764374 and rs77447679) were selected for genotyping using real-time PCR. After adjusting for confounding factors such as age, smoking, alcohol intake, and betel-nut chewing, our analysis revealed that individuals harboring the CC or CT genotype at rs3764374 had a significantly increased risk of developing OSCC compared to those with the wild-type genotype. Notably, among male patients who had no history of betel-quid chewing, those carrying the CC/CT genotype at rs3764374 or the CA/AA genotype at rs77447679 exhibited a greater likelihood of presenting with stage II or III OSCC, indicating more advanced disease. These findings support the notion that specific CBX4 polymorphisms may contribute to the pathogenesis and clinical progression of OSCC, particularly in populations exposed to distinct environmental risk factors. This study provides new insights into the gene-environment interplay in OSCC and suggests potential biomarkers for risk stratification and disease monitoring in Asian male populations.
Keywords: oral squamous cell carcinoma, CBX4, cancer progression, single-nucleotide polymorphisms
Introduction
Head and neck squamous cell carcinomas (HNSCCs) represent a heterogeneous group of epithelial malignancies that arise from the mucosal linings of the oral cavity, nasopharynx, oropharynx, hypopharynx, and larynx, with squamous cell histology being the predominant pathological subtype. Globally, HNSCC ranks as the sixth most prevalent cancer, with oral squamous cell carcinoma (OSCC) constituting a major clinical subtype [1-4]. Although advances in multimodal therapeutic strategies, including surgery, radiotherapy, and chemotherapy, have improved disease management, the overall five-year survival rate remains unsatisfactory, ranging between 30% and 50%, largely due to frequent locoregional recurrence and distant metastatic dissemination [5, 6]. The etiology of OSCC is multifactorial, involving cumulative genetic and epigenetic alterations triggered by prolonged exposure to environmental carcinogens. Key contributing risk factors include persistent mucosal inflammation, tobacco and alcohol use, areca (betel) nut chewing, and oncogenic viral infections. The Chromobox (CBX) gene family encodes core subunits of the canonical Polycomb Repressive Complex 1 (cPRC1), a key epigenetic regulator mediating chromatin compaction and transcriptional silencing through histone modification. Altered transcriptional or post-transcriptional regulation of CBX members has been increasingly associated with oncogenic processes across diverse malignancies.
Reports suggest that CBX3 has been identified as a key player in lung cancer progression by interacting with pathways such as phosphoinositide 3-kinase (PI3K)/ protein kinase B (AKT), rat sarcoma virus (Ras)/ kirsten rat sarcoma virus (KRAS), Wnt/β-catenin, mitogen-activated protein kinase (MAPK), Notch, and p53, leading to increased proliferation and therapy resistance [7]. Similarly, CBX2 overexpression has been associated with poor survival outcomes in cancer patients, potentially by maintaining cancer stem cells in an undifferentiated state and repressing tumor suppressor genes [8]. In ovarian cancer, components of PRC1, including CBX proteins, contribute to disease development, recurrence, and chemoresistance by silencing tumor suppressor genes and regulating stem cell compartments [8]. The diverse roles of CBX proteins in cancer underscore their potential as therapeutic targets. Understanding the specific functions and mechanisms of each CBX member is crucial for developing targeted cancer therapies [9]. CBX4, a pivotal component of the PRC1, plays multifaceted roles in cancer development and progression. Its functions vary across cancer types, exhibiting both oncogenic and tumor-suppressive activities. In colorectal carcinoma, CBX4 suppresses metastasis by recruiting histone deacetylase 3 (HDAC3) to the runt-related transcription factor 2 (Runx2) promoter, leading to transcriptional repression of Runx2, a key factor in cancer metastasis. This mechanism underscores the tumor-suppressive role of CBX4 in this context [10]. Conversely, in breast cancer, CBX4 exhibits oncogenic properties by downregulating miR-137, resulting in the activation of the Notch1 signaling pathway, which promotes tumor progression [11]. Furthermore, CBX4 has been implicated in colon cancer development through its potential influence on circadian rhythm and immune infiltration, suggesting a complex role in tumorigenesis [12]. Although these studies collectively demonstrate that the function of CBX4 in cancer is context-dependent, acting as either an oncogene or tumor suppressor depending on the cancer type and molecular environment. Its mechanistic contribution to OSCC development and progression remains insufficiently characterized.
Recent studies have identified various genetic susceptibility factors that may contribute to the development of head and neck squamous cell carcinoma (HNSCC). Among these, single-nucleotide polymorphisms (SNPs), the most common type of genetic variation, have shown significant potential as predictive biomarkers for assessing individual cancer risk [13-15]. By modulating transcriptional activity or protein structure, SNPs can impact cellular pathways critical to tumor initiation and development. Recent evidence has identified a significant association between the CBX4 rs77447679 polymorphism and the risk of gastric cancer. CBX4 encodes a component of the PRC1 and also functions as a SUMO E3 ligase, implicating it in chromatin modification and post-translational regulation, both of which are critical in cancer development. The observed correlation between rs77447679 and gastric cancer risk underscores the potential biological relevance of CBX4-mediated sumoylation in gastrointestinal malignancies [16]. However, to date, no studies have specifically addressed the role of CBX4 SNPs in OSCC. Therefore, our present study aims to explore this previously uncharacterized association and provide novel insights into the genetic determinants of OSCC pathogenesis.
Materials and Methods
Study participants and specimen collection
This study enrolled a total of 1,184 male patients diagnosed with oral squamous cell carcinoma (OSCC) from Chung Shan Medical University Hospital in Taichung, Taiwan. All participants provided written informed consent after receiving a comprehensive explanation of the study protocol. Clinical staging for OSCC was determined at diagnosis according to the tumor-node-metastasis (TNM) classification system established by the American Joint Committee on Cancer (AJCC, 2002). A comparison group of 1,188 cancer-free individuals, aged 20 to 70, was recruited from the Taiwan Biobank (https://www.twbiobank.org.tw/new_web_en/index.php). Detailed information regarding age, sex, and lifestyle behaviors (including betel nut chewing, tobacco use, and alcohol intake) was collected through structured interviews. Participants who reported drinking more than two alcoholic beverages daily were categorized as alcohol users, while those who had smoked at least one cigarette per day within the past three months were identified as habitual smokers. Ethical approval for this research was granted by the Institutional Review Board of Chung Shan Medical University Hospital.
Comprehensive analysis of CBX4 from The Cancer Genome Atlas (TCGA)
The UALCAN platform (http://ualcan.path.uab.edu/index.html) offers an intuitive and interactive interface for exploring cancer transcriptomics and clinical datasets. It leverages level 3 RNA sequencing and associated clinical information from The Cancer Genome Atlas (TCGA), covering 31 distinct tumor types [17]. In this study, UALCAN was utilized to assess CBX4 expression differences between tumor and normal tissues, as well as to evaluate its prognostic relevance based on overall survival in patients with HNSCC. Bioinformatics analyses were conducted using UALCAN, platforms known for its validated and reproducible methodologies in cancer research [17].
Identifying gene set enrichments of CBX4 in TCGA database
Large-scale initiatives such as TCGA have generated extensive genomic profiles linked to various tumor types. While several online tools have been developed to facilitate data access, many still require a foundational understanding of bioinformatics to perform more advanced analyses. To investigate the gene set enrichment associated with CBX4 expression within TCGA datasets, we utilized the Gene ENrichment Identifier (GENI; https://www.shaullab.com/geni), a web-based resource that efficiently computes transcriptome-wide correlations for a target gene and ranks them according to curated biological gene sets [18].
Selection of cbx4 polymorphisms
In this investigation, four SNPs within the CBX4 (NM_003655.3) were selected based on data from the International HapMap Project: rs1285251, rs2289728, rs3764374, and rs77447679. These SNPs were selected to investigate their potential roles in modulating CBX4 expression and function, which may contribute to disease susceptibility and pathogenesis.
CBX4 genotyping
Genotyping of the CBX4 variants rs1285251, rs2289728, rs3764374, and rs77447679 was performed using the TaqMan allelic discrimination method on the ABI StepOne Real-Time PCR platform (Applied Biosystems). The reactions were analyzed with SDS software version 3.0, following the manufacturer's protocol to ensure accurate detection of each SNP [13].
Statistical analysis
Differences in age and baseline demographic variables between OSCC cases and control participants were assessed using the nonparametric Mann-Whitney U test. Logistic regression analysis was conducted to calculate odds ratios (ORs) and corresponding 95% confidence intervals (CIs) to determine the strength of associations. A threshold of p < 0.05 was applied to define statistical significance. All analyses were performed using SAS software.
Results
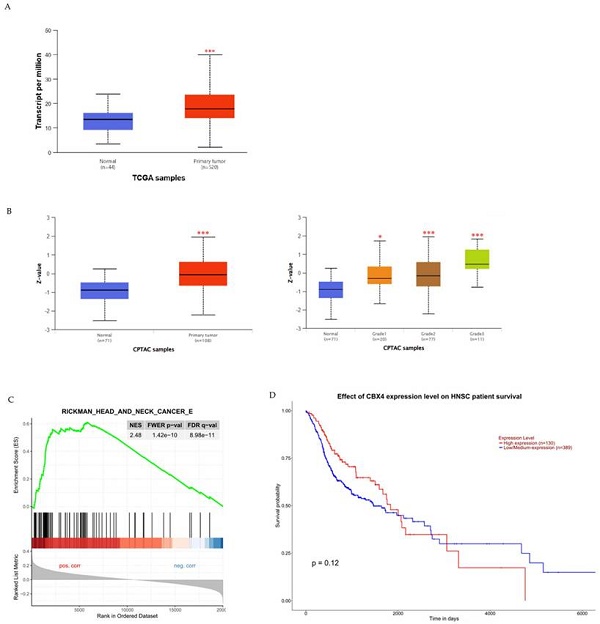
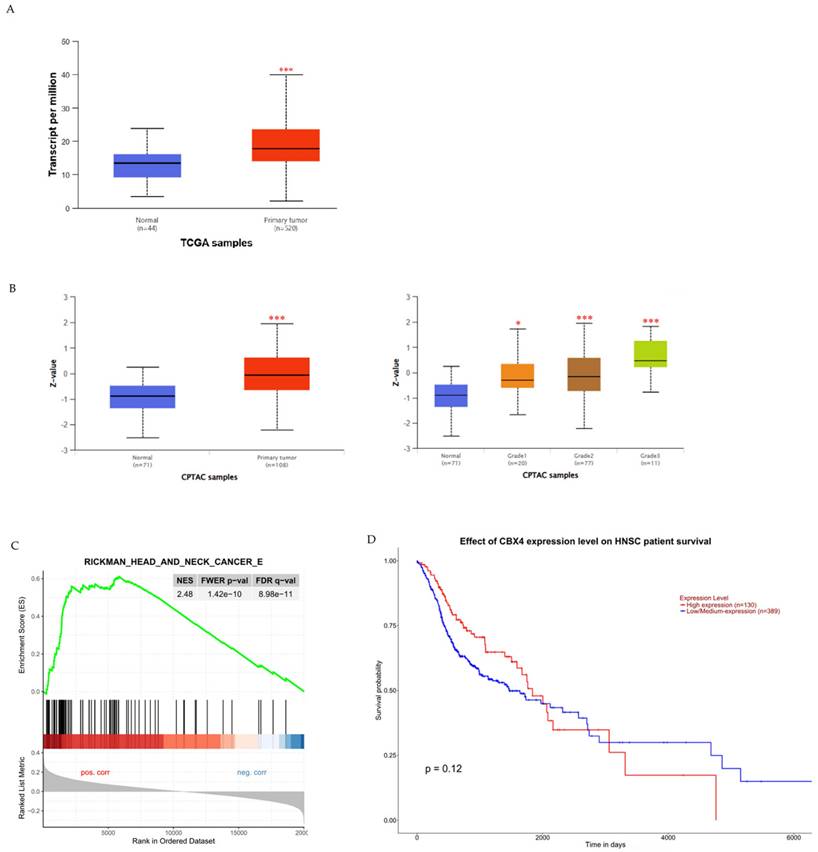
To examine the potential clinical significance of CBX4 expression in HNSCC, we utilized the UALCAN platform to compare CBX4 mRNA and protein levels between healthy controls and HNSCC patients, and to investigate any associations with patient survival. Initially, our analysis revealed a significant elevation in CBX4 transcript levels among HNSCC patients compared to control individuals (p < 0.001), indicating a possible oncogenic role for CBX4 in the progression of HNSCC (Figure 1A). Additionally, to further substantiate these findings, we assessed CBX4 protein expression utilizing data from the Clinical Proteomic Tumor Analysis Consortium (CPTAC). Protein abundance, expressed as Z-scores normalized against median values from normal samples, demonstrated significantly higher CBX4 protein levels within tumor tissues, particularly in moderately (grade 2) to poorly differentiated (grade 3) tumors (Figure 1B). Moreover, subsequent gene enrichment analysis indicated significant enrichment of head and neck cancer-related pathways in the group characterized by high CBX4 expression (Normalized Enrichment Score, NES = 2.48), suggesting the involvement of CBX4 in promoting aggressive tumor behavior (Figure 1C). Interestingly, despite the clear association between elevated CBX4 expression and aggressive HNSCC phenotypes, our survival analysis indicated no statistically significant impact of CBX4 expression levels on overall patient survival (p = 0.12, Figure 1D). Collectively, these results imply complex regulatory mechanisms of CBX4 in HNSCC progression that remain to be fully elucidated. Further research should be directed toward uncovering the molecular pathways through which CBX4 contributes to tumor aggressiveness, independently of survival outcomes.
Association of CBX4 expression with HNSCC progression and patient survival outcomes. (A) Comparative analysis of CBX4 transcript levels in normal tissues versus HNSCC patient samples, demonstrating a significant elevation in tumor tissues. (B) Quantification of CBX4 protein expression across different histological grades of HNSCC, revealing progressively increased levels in grade 2 and grade 3 tumors compared to normal controls. (C) Gene Set Enrichment Analysis (GSEA) plot illustrating a strong enrichment of head and neck cancer-related gene signatures in samples with high CBX4 expression, suggesting a potential role in promoting malignant transformation. (D) Kaplan-Meier survival curves comparing overall survival among HNSCC patients stratified by CBX4 expression levels, based on UALCAN database analysis. * p < 0.05, ** p < 0.01, *** p < 0.001.
The distributions of demographical characteristics in 1188 controls and 1184 male patients with oral cancer.
| Variable | Controls (N=1188) | Patients (N=1184) | p value |
|---|---|---|---|
| Age (yrs) | p = 0.028* | ||
| < 55 | 561 (47.2%) | 506 (42.7%) | |
| ≥ 55 | 627 (52.8%) | 678 (57.3%) | |
| Betel quid chewing | p < 0.001* | ||
| No | 990 (83.3%) | 387 (32.7%) | |
| Yes | 198 (16.7%) | 797 (67.3%) | |
| Cigarette smoking | p < 0.001* | ||
| No | 556 (46.8%) | 249 (21.0%) | |
| Yes | 632 (53.2%) | 935 (79.0%) | |
| Alcohol drinking | p < 0.001* | ||
| No | 952 (80.1%) | 734 (62.0%) | |
| Yes | 236 (19.9%) | 450 (38.0%) | |
| Stage | |||
| I+II | 514 (43.4%) | ||
| III+IV | 670 (56.6%) | ||
| Tumor T status | |||
| T1+T2 | 580 (49.0%) | ||
| T3+T4 | 604 (51.0%) | ||
| Lymph node status | |||
| N0 | 788 (66.6%) | ||
| N1+N2+N3 | 396 (33.4%) | ||
| Metastasis | |||
| M0 | 1177 (99.4%) | ||
| M1 | 7 (0.6%) | ||
| Cell differentiation | |||
| Well differentiated | 186 (15.7%) | ||
| Moderately or poorly differentiated | 998 (84.3%) |
Mann-Whitney U test or Chi-square test was used between healthy controls and patients with oral cancer. * p value < 0.05 as statistically significant.
To explore potential risk factors contributing to the onset of OSCC in a clinical context, we conducted a case-control study involving 1184 male patients diagnosed with OSCC and 1188 matched healthy male controls. Comparative analysis of demographic and lifestyle factors between these two groups revealed several significant differences. Specifically, OSCC patients were notably older than healthy controls (p < 0.001). Moreover, lifestyle habits traditionally linked to OSCC, such as betel-nut chewing, tobacco smoking, and alcohol intake, were significantly more prevalent among OSCC patients compared to controls (all p-values < 0.001, Table 1). These findings reinforce the critical roles of age and lifestyle behaviors as significant contributors to OSCC development, highlighting the necessity for targeted preventive interventions and health education to mitigate risk among susceptible populations. Future investigations should further elucidate interactions between genetic susceptibility and environmental exposures to enhance risk prediction and inform personalized treatment strategies.
To mitigate potential confounding effects arising from various environmental exposures, we performed multivariate logistic regression analyses adjusting for critical risk factors, including age, betel-quid chewing, tobacco smoking, and alcohol consumption. Adjusted odds ratios (AORs) along with their 95% confidence intervals (CIs) were calculated to accurately reflect the associations between CBX4 polymorphisms and OSCC. The detailed genotype distributions and their relationships to OSCC susceptibility are presented comprehensively in Table 2. Specifically, the most prevalent allelic genotypes identified in CBX4 polymorphisms rs1285251, rs2289728, rs3764374, and rs77447679 were homozygous C/C, homozygous G/G, homozygous C/C, and homozygous C/C, respectively, observed consistently among both OSCC cases and control participants. Nevertheless, statistical analysis indicated that carriers of these polymorphisms (rs1285251, rs2289728, rs3764374, and rs77447679) did not demonstrate a statistically significant altered risk of developing OSCC compared to individuals possessing the wild-type genotypes.
Genotyping and allele frequency of CBX4 single nucleotide polymorphism (SNP) in oral cancer and normal controls.
| Variable | Controls (N=1188) n (%) | Patients (N=1184) n (%) | AOR (95% CI)a |
|---|---|---|---|
| rs1285251 | |||
| CC | 525 (44.2%) | 532 (44.9%) | 1.000 (reference) |
| CT | 532 (44.8%) | 510 (43.1%) | 0.990 (0.811-1.210) |
| TT | 131 (11.0%) | 142 (12.0%) | 1.106 (0.811-1.510) |
| CT+TT | 663 (55.8%) | 652 (55.1%) | 1.007 (0.916-1.107) |
| rs2289728 | |||
| GG | 370 (31.1%) | 394 (33.3%) | 1.000 (reference) |
| GA | 594 (50.3%) | 556 (47.0%) | 0.812 (0.655-1.005) |
| AA | 221 (18.6%) | 234 (19.7%) | 0.956 (0.729-1.253) |
| GA+AA | 818 (68.9%) | 790 (66.7%) | 0.922 (0.834-1.020) |
| rs3764374 | |||
| CC | 742 (62.5%) | 744 (62.8%) | 1.000 (reference) |
| CT | 396 (33.3%) | 395 (33.4%) | 1.044 (0.853-1.278) |
| TT | 50 (4.2%) | 45 (3.8%) | 0.927 (0.568-1.511) |
| CT+TT | 446 (37.5%) | 440 (37.2%) | 1.015 (0.921-1.119) |
| rs77447679 | |||
| CC | 747 (62.9%) | 746 (63.0%) | 1.000 (reference) |
| CA | 393 (33.1%) | 398 (33.6%) | 1.112 (0.909-1.361) |
| AA | 48 (4.0%) | 40 (3.4%) | 0.770 (0.460-1.288) |
| CA+AA | 441 (37.1%) | 438 (37.0%) | 1.036 (0.940-1.142) |
The ORs with analyzed by their 95% CIs were estimated by logistic regression models.
a Adjusted for the effects of age, betel quid chewing, cigarette smoking and alcohol drinking.
To further investigate potential clinical implications of CBX4 polymorphisms in OSCC, we analyzed associations between various CBX4 SNP genotypes and clinical characteristics of the patients. Our analysis highlighted one particular SNP, rs3764374, that appears to influence OSCC disease progression. Specifically, patients harboring the variant genotypes (C/T or T/T) of the rs3764374 polymorphism exhibited a significantly elevated risk of developing OSCC compared to those with the wild-type C/C genotype (AOR = 1.434; 95% CI: 1.02-2.012; p = 0.037, Table 3). This finding suggests that the rs3764374 variant of CBX4 could serve as a predictive genetic marker for OSCC susceptibility and might contribute to identifying individuals at greater risk of aggressive disease. Future research exploring the functional implications and underlying molecular mechanisms of this SNP would be valuable for enhancing early risk assessment and developing personalized therapeutic strategies for OSCC patients.
Betel nut chewing has consistently been identified as a crucial environmental factor influencing the onset and advancement of OSCC. To more precisely evaluate its impact in our study, we categorized OSCC patients into two distinct groups based on their history of betel nut chewing. Subsequently, we investigated the distribution of CBX4 genotypes and explored potential correlations with clinicopathological features within these groups. Our results notably revealed that CBX4 SNPs, specifically rs3764374 and rs77447679, exhibited significant associations with increased rates of OSCC progression exclusively among individuals who had never engaged in betel nut chewing (Tables 4 and 5). These findings suggest a complex interplay between genetic susceptibility and environmental influences, highlighting that genetic predisposition through CBX4 variants may independently drive disease progression in the absence of traditional risk factors. Further studies are necessary to delineate the biological mechanisms underlying these genetic interactions, providing opportunities for tailored prevention strategies and targeted therapies in genetically susceptible populations.
Adjusted odds ratio (AOR) and 95% confidence intervals (CI) of clinical statuses associated with genotypic frequencies of CBX4 rs3764374 in male oral cancer patients.
| Variable | CC (N=744) | CT+TT (N=440) | AOR (95% CI) | p value |
|---|---|---|---|---|
| Clinical stage | ||||
| Stage I+II | 319 (42.9%) | 195 (44.3%) | 1.000 (reference) | 0.665 |
| Stage III+IV | 425 (57.1%) | 245 (55.7%) | 0.947 (0.746-1.202) | |
| Tumor size | ||||
| ≤ T2 | 364 (48.9%) | 216 (49.1%) | 1.000 (reference) | 0.985 |
| > T2 | 380 (51.1%) | 224 (50.9%) | 1.002 (0.791-1.270) | |
| Lymph node metastasis | ||||
| No | 486 (65.3%) | 302 (68.6%) | 1.000 (reference) | 0.296 |
| Yes | 258 (34.7%) | 138 (31.4%) | 0.874 (0.678-1.125) | |
| Cell differentiated grade | ||||
| Grade I | 129 (17.3%) | 57 (10.0%) | 1.000 (reference) | 0.037* |
| Grade II or III | 615 (82.7%) | 383 (87.0%) | 1.434 (1.021-2.012) |
Cell differentiated grade: grade I: well differentiated; grade II: moderately differentiated; grade III: poorly differentiated.
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for age, betel quid chewing, cigarette smoking, and alcohol drinking. * p value < 0.05 as statistically significant.
Adjusted odds ratio (AOR) and 95% confidence intervals (CI) of clinical statuses associated with genotypic frequencies of CBX4 rs3764374 in male oral cancer patients among with betel quid chewers or without betel quid chewers.
| Non-Betel Quid Chewers (N=387) | Betel Quid Chewers (N=797) | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | CC (N=240) | CT+TT (N=147) | AOR (95% CI) | p value | CC (N=504) | CT+TT (N=293) | AOR (95% CI) | p value |
| Clinical Stage | ||||||||
| Stage I+II | 99 (41.2%) | 60 (40.8%) | 1.000 (reference) | 0.945 | 220 (43.7%) | 135 (46.1%) | 1.000 (reference) | 0.519 |
| Stage III+IV | 141 (58.8%) | 87 (59.2%) | 1.015 (0.666-1.546) | 284 (56.3%) | 158 (53.9%) | 0.909 (0.680-1.215) | ||
| Tumor size | ||||||||
| ≤ T2 | 115 (47.9%) | 64 (43.5%) | 1.000 (reference) | 0.349 | 249 (49.4%) | 152 (51.9%) | 1.000 (reference) | 0.494 |
| > T2 | 125 (52.1%) | 83 (56.5%) | 1.220 (0.805-1.850) | 255 (50.6%) | 141 (48.1%) | 0.904 (0.677-1.208) | ||
| Lymph node metastasis | ||||||||
| No | 158 (65.8%) | 96 (65.3%) | 1.000 (reference) | 0.909 | 328 (65.1%) | 206 (70.3%) | 1.000 (reference) | 0.169 |
| Yes | 82 (34.2%) | 51 (34.7%) | 1.026 (0.664-1.586) | 176 (34.9%) | 87 (29.7%) | 0.803 (0.588-1.097) | ||
| Cell differentiated grade | ||||||||
| Grade I | 35 (14.6%) | 11 (7.5%) | 1.000 (reference) | 0.036* | 94 (18.7%) | 46 (15.7%) | 1.000 (reference) | 0.252 |
| Grade II or III | 205 (85.4%) | 136 (92.5%) | 2.152 (1.051-4.406) | 410 (81.3%) | 247 (84.3%) | 1.255 (0.851-1.850) | ||
Cell differentiated grade: grade I: well differentiated; grade II: moderately differentiated; grade III: poorly differentiated.
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for age, cigarette smoking, and alcohol drinking. * p value < 0.05 as statistically significant.
Adjusted odds ratio (AOR) and 95% confidence intervals (CI) of clinical statuses associated with genotypic frequencies of CBX4 rs77447679 in male oral cancer patients among with betel quid chewers or without betel quid chewers.
| Non-Betel Quid Chewers (N=387) | Betel Quid Chewers (N=797) | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | CC (N=233) | CA+AA (N=154) | AOR (95% CI) | p value | CC (N=513) | CA+AA (N=284) | AOR (95% CI) | p value |
| Clinical Stage | ||||||||
| Stage I+II | 100 (42.9%) | 59 (38.3%) | 1.000 (reference) | 0.394 | 224 (43.7%) | 131 (46.1%) | 1.000 (reference) | 0.506 |
| Stage III+IV | 133 (57.1%) | 95 (61.7%) | 1.201 (0.788-1.830) | 289 (56.3%) | 153 (53.9%) | 0.905 (0.676-1.213) | ||
| Tumor size | ||||||||
| ≤ T2 | 114 (48.9%) | 65 (42.2%) | 1.000 (reference) | 0.145 | 259 (50.5%) | 142 (50.0%) | 1.000 (reference) | 0.939 |
| > T2 | 119 (51.1%) | 89 (57.8%) | 1.363 (0.899-2.067) | 254 (49.5%) | 142 (50.0%) | 1.011 (0.755-1.354) | ||
| Lymph node metastasis | ||||||||
| No | 157 (67.4%) | 97 (63.0%) | 1.000 (reference) | 0.390 | 334 (65.1%) | 200 (70.4%) | 1.000 (reference) | 0.169 |
| Yes | 76 (32.6%) | 57 (37.0%) | 1.209 (0.784-1.862) | 179 (34.9%) | 84 (29.6%) | 0.802 (0.586-1.099) | ||
| Cell differentiated grade | ||||||||
| Grade I | 34 (14.6%) | 12 (7.8%) | 1.000 (reference) | 0.039* | 94 (18.3%) | 46 (16.2%) | 1.000 (reference) | 0.402 |
| Grade II or III | 199 (85.4%) | 142 (92.2%) | 2.094 (1.040-4.219) | 419 (81.7%) | 238 (83.8%) | 1.181 (0.800-1.743) | ||
Cell differentiated grade: grade I: well differentiated; grade II: moderately differentiated; grade III: poorly differentiated.
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for age, cigarette smoking, and alcohol drinking. * p value < 0.05 as statistically significant.
Discussion
Single-nucleotide polymorphisms (SNPs) represent common genetic variations occurring at a single nucleotide position in the human genome. Unlike rare mutations, SNPs are defined by their relatively high frequency within the population and are often shaped by environmental influences. These genetic variants contribute to phenotypic diversity and have been linked to an individual's predisposition to various diseases, including malignancies [19, 20]. Alterations in gene function resulting from SNPs or other genetic changes can modify disease susceptibility and are associated with an elevated risk of cancer and distinct pathological features [21, 22].
CBX proteins, particularly CBX4, play pivotal roles in cancer biology through their involvement in epigenetic regulation. CBX4, a component of the PRC1, modulates gene expression by recognizing histone modifications and recruiting other epigenetic modifiers. In lung adenocarcinoma, CBX4 exhibits a dual function. It promotes tumor proliferation by upregulating phosphoglycerate dehydrogenase (PHGDH) expression via interaction with the histone acetyltransferase general control non-depressible 5 (GCN5), enhancing serine biosynthesis; concurrently, it suppresses metastasis by repressing zinc finger E-box binding homeobox 2 (ZEB2) transcription through PRC1-mediated H2AK119 ubiquitination [23]. Genetic variations in CBX4, such as single nucleotide polymorphisms (SNPs), have been associated with cancer susceptibility and prognosis. For instance, the CBX4 rs2289728 (G>A) SNP has been linked to a decreased risk of hepatocellular carcinoma (HCC), potentially by reducing CBX4 expression and alleviating repression of tumor suppressor genes like cyclin-dependent kinase inhibitor 2A (CDKN2A). Conversely, the rs77447679 (C>A) SNP in CBX4 has been associated with increased gastric cancer risk and poorer survival in HCC patients, correlating with higher CBX4 expression and more aggressive tumor characteristics [16, 23, 24]. CBX4 polymorphisms may modulate the regulatory functions of genes, influencing immune responses to environmental carcinogens. The role of CBX4 in suppressing programmed death-1 (PD-1) expression in T cells suggests a mechanism by which genetic and environmental factors could synergistically increase OSCC risk [25]. Taken together, current evidence supports a critical role for CBX4 in cancer development and progression through both epigenetic regulation and genetic polymorphism. Despite ongoing research, the relationship between genetic variants of CBX4 and OSCC susceptibility remains poorly understood. However, CBX4 may contribute to OSCC progression by regulating genes involved in cell cycle and stemness, such as CDC20, as observed in gastric cancer studies [26]. Further research is needed to elucidate these mechanisms in OSCC. Our investigation aims to elucidate how specific SNPs within CBX4 influence the risk and clinical manifestations associated with OSCC.
In Taiwan, head and neck cancer rates remain elevated primarily because of prevalent risk factors such as dietary patterns and the chewing of betel nuts. Our study revealed increased CBX4 expression at both the transcript and protein levels, indicating its potential involvement in promoting HNSCC progression (Figure 1). Although CBX4 expression is associated with aggressive tumor characteristics, its impact on overall survival may be influenced by treatment efficacy and other molecular factors, as observed in studies on clear cell renal cell carcinoma [17]. The four CBX4 SNPs were selected based on their minor allele frequency (>0.05) in the Chinese population and potential functional significance, as identified through the NCBI dbSNP database and subsequent analyses [16]. Interestingly, we discovered associations between specific CBX4 SNPs, rs3764374 (CT+TT genotype) and rs77447679, and more aggressive oral cancer progression, particularly among individuals without betel nut chewing habits (Tables 4 and 5). The absence of significant associations for rs1285251 and rs2289728 may be attributed to their limited impact on CBX4 function or expression, as opposed to other variants like rs77447679 which have demonstrated functional significance in cancer susceptibility [23]. Our findings suggest that CBX4 polymorphisms, such as rs77447679, may contribute to OSCC progression particularly in individuals without betel quid exposure, highlighting a potential gene-environment interaction where genetic factors play a more pronounced role in the absence of environmental carcinogens [16]. Nonetheless, further investigations are necessary to clarify the precise molecular pathways through which CBX4 SNPs influence OSCC development.
Conclusion
In summary, our findings highlight the potential clinical relevance of CBX4 genetic variants as predictive markers for susceptibility to OSCC. Our results offer novel insights into the relationship between CBX4 polymorphisms and the clinicpathological characteristics of OSCC in the Taiwanese population. These findings underscore the importance of genetic factors in OSCC pathogenesis and progression. Moreover, the identified SNPs in CBX4 could contribute to improved screening and personalized therapeutic approaches for patients with OSCC. However, the homogeneity of the Taiwanese cohort reduces the likelihood of population stratification, we recognize that unaccounted stratifications may exist and could affect the findings. Future research focusing on the underlying molecular mechanisms and functional implications of these polymorphisms is warranted to validate their clinical utility and further clarify their role in cancer biology.
Acknowledgements
Funding
This research was funded by grants from the National Science and Technology Council, Taiwan (NSTC 109-2320-B-039-013-MY3), the National Health Research Institute, Taiwan (NHRI-112A1-CACO-13222202), the China Medical University, Taiwan (CMU108-MF-01; CMU109-MF-03), the China Medical University Hospital, Taiwan (DMR-112-198; DMR-113-031; DMR-114-161).
Author contributions
L.-C.H., Y.-C.C., and S.-F.Y. were responsible for project conceptualization and data management. Formal data analysis was conducted by Y.-C.C., L.-C.H., C.-W.L., and S.-F.Y. Y.-C.C., H.-S.H., and Y.-L.Y. contributed essential resources and handled software implementation. C.-Y.C., L.-C.L., C.-W.L., and Y.-L.Y. were involved in drafting the manuscript and developing the methodology. L.-C.H., S.-F.Y., and Y.-L.Y. provided supervision and took part in critical review and manuscript revision. All authors reviewed and approved the final version of the manuscript prior to submission.
Informed consent statement
Informed consent was obtained from all subjects involved in the study.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-86
2. Hashibe M, Brennan P, Benhamou S, Castellsague X, Chen C, Curado MP. et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J Natl Cancer Inst. 2007;99:777-89
3. Shinga-Ishihara C, Nakai Y, Milgrom P, Murakami K, Matsumoto-Nakano M. Cross-cultural validity of a dietary questionnaire for studies of dental caries risk in Japanese. BMC Oral Health. 2014;14:1
4. Toporcov TN, Znaor A, Zhang ZF, Yu GP, Winn DM, Wei Q. et al. Risk factors for head and neck cancer in young adults: a pooled analysis in the INHANCE consortium. Int J Epidemiol. 2015;44:169-85
5. Jayaprakash V, Rigual NR, Moysich KB, Loree TR, Nasca MA, Menezes RJ. et al. Chemoprevention of head and neck cancer with aspirin: a case-control study. Arch Otolaryngol Head Neck Surg. 2006;132:1231-6
6. Chang WS, Tsai CW, Yang JS, Hsu YM, Shih LC, Chiu HY. et al. Resveratrol inhibited the metastatic behaviors of cisplatin-resistant human oral cancer cells via phosphorylation of ERK/p-38 and suppression of MMP-2/9. J Food Biochem. 2021;45:e13666
7. Wahab MA, Del Gaudio N, Gargiulo B, Quagliariello V, Maurea N, Nebbioso A. et al. Exploring the Role of CBX3 as a Potential Therapeutic Target in Lung Cancer. Cancers (Basel). 2024;16:3026
8. Jangal M, Lebeau B, Witcher M. Beyond EZH2: is the polycomb protein CBX2 an emerging target for anti-cancer therapy? Expert Opin Ther Targets. 2019;23:565-78
9. Ma RG, Zhang Y, Sun TT, Cheng B. Epigenetic regulation by polycomb group complexes: focus on roles of CBX proteins. J Zhejiang Univ Sci B. 2014;15:412-28
10. Wang X, Li L, Wu Y, Zhang R, Zhang M, Liao D. et al. CBX4 Suppresses Metastasis via Recruitment of HDAC3 to the Runx2 Promoter in Colorectal Carcinoma. Cancer Res. 2016;76:7277-89
11. Zeng JS, Zhang ZD, Pei L, Bai ZZ, Yang Y, Yang H. et al. Corrigendum to "CBX4 exhibits oncogenic activities in breast cancer via Notch1 signaling" [Int. J. Biochem. Cell Biol. (2018) 1-8]. Int J Biochem Cell Biol. 2018;105:145
12. Wei W, Zhao W, Zhang Y. CBX4 Provides an Alternate Mode of Colon Cancer Development via Potential Influences on Circadian Rhythm and Immune Infiltration. Front Cell Dev Biol. 2021;9:669254
13. Shastry BS. SNP alleles in human disease and evolution. J Hum Genet. 2002;47:561-6
14. Yu YL, Su KJ, Hsieh YH, Lee HL, Chen TY, Hsiao PC. et al. Effects of EZH2 polymorphisms on susceptibility to and pathological development of hepatocellular carcinoma. PLoS One. 2013;8:e74870
15. Lu S, Catalano C, Huhn S, Pardini B, Partu L, Vymetalkova V. et al. Single nucleotide polymorphisms within MUC4 are associated with colorectal cancer survival. PLoS One. 2019;14:e0216666
16. Luo Y, You S, Wang J, Fan S, Shi J, Peng A. et al. Association between Sumoylation-Related Gene rs77447679 Polymorphism and Risk of Gastric Cancer (GC) in a Chinese Population. J Cancer. 2017;8:3226-31
17. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B. et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649-58
18. Hayashi A, Ruppo S, Heilbrun EE, Mazzoni C, Adar S, Yassour M. et al. GENI: A web server to identify gene set enrichments in tumor samples. Comput Struct Biotechnol J. 2023;21:5531-7
19. Dong LM, Potter JD, White E, Ulrich CM, Cardon LR, Peters U. Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. JAMA. 2008;299:2423-36
20. Ali LA, Jemon K, Latif NA, Bakar SA, Alwi SSS. LEP G2548A polymorphism is associated with increased serum leptin and insulin resistance among T2DM Malaysian patients. Biomedicine (Taipei). 2022;12:1-11
21. Kadi FA, Yuniati T, Sribudiani Y, Rachmadi D. The association of rs187238, rs19465518 and rs1946519 IL-8 polymorphisms with acute kidney injury in preterm infants. Biomedicine (Taipei). 2021;11:43-50
22. Chupeerach C, Tapanee P, On-Nom N, Temviriyanukul P, Chantong B, Reeder N. et al. The influence of TAS2R38 bitter taste gene polymorphisms on obesity risk in three racially diverse groups. Biomedicine (Taipei). 2021;11:43-9
23. Tan C, Bei C, Zhu X, Zhang Y, Qin L, Tan S. Single Nucleotide Polymorphisms of CBX4 and CBX7 Decrease the Risk of Hepatocellular Carcinoma. Biomed Res Int. 2019;2019:6436825
24. Deng N, Zhou H, Fan H, Yuan Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget. 2017;8:110635-49
25. Chen RY, Zhu Y, Shen YY, Xu QY, Tang HY, Cui NX. et al. The role of PD-1 signaling in health and immune-related diseases. Front Immunol. 2023;14:1163633
26. Li W, Chen H, Wang Z, Liu J, Lei X, Chen W. Chromobox 4 (CBX4) promotes tumor progression and stemness via activating CDC20 in gastric cancer. J Gastrointest Oncol. 2022;13:1058-72
Author contact
![]() Corresponding authors: ylyuedu.tw (Y.-L.Y.); ysfedu.tw (S.-F.Y.).
Corresponding authors: ylyuedu.tw (Y.-L.Y.); ysfedu.tw (S.-F.Y.).

 Global reach, higher impact
Global reach, higher impact