3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(7):2362-2374. doi:10.7150/jca.110981 This issue Cite
Research Paper
Identification of the role of MED6 in the development and prognosis of lung adenocarcinoma based on multi-omics profiling
1. Department of Respiratory and Critical Care Medicine, Tianjin Medical University General Hospital, Tianjin, 300052, China.
2. Department of Plastic Surgery and Medical Aesthetics, The Second Hospital of Tianjin Medical University, Tianjin, 300211, China.
3. The First Department of Breast Cancer, Key Laboratory of Cancer Prevention and Therapy, Tianjin's Clinical Research Center for Cancer, National Clinical Research Center for Cancer, Key Laboratory of Breast Cancer Prevention and Therapy, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin, 300060, China.
4. Anhui Chest Hospital, Hefei, Anhui Province, 23002, China.
5. Immunology Department, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), Tianjin Medical University, Tianjin, 300070, China.
6. Department of General Surgery, Tianjin Medical University General Hospital, Tianjin Key Laboratory of Precise Vascular Reconstruction and Organ Function Repair, Tianjin General Surgery Institute, Tianjin, 300052, China.
7. Department of Thoracic Surgery, Ningbo Medical Center Lihuili Hospital, Ningbo University, Ningbo, Zhejiang Province, 315040, China.
# These authors contributed equally to this work.
Received 2025-1-23; Accepted 2025-3-23; Published 2025-4-13
Abstract
Background: Lung adenocarcinoma (LUAD) is the most common subtype of lung cancer. Recent studies have highlighted the importance of Mediator complex subunits in cancer, but their specific roles in LUAD are still unclear.
Methods: The CRISPR-Cas9 loss-of-function data was used to assess gene dependency in cell growth. RNA-seq data were analyzed to evaluate the prognostic value of Mediator subunits and explore their downstream pathways. Single-cell sequencing data were utilized to examine the tumor microenvironment in LUAD. A drug sensitivity analysis was performed to identify potential therapeutic options.
Results: Mediator complex subunit 6 (MED6) was found to influence tumor cell growth in LUAD. Additionally, MED6 expression levels were associated with patient prognosis. MED6-positive tumor cells showed more active interactions with other cells in the LUAD microenvironment, promoting tumor progression. Based on MED6 expression, drugs such as paclitaxel, afatinib, and brivanib were identified as potential treatments.
Conclusions: This study revealed the role of MED6 in LUAD and its potential as a biomarker. Our findings suggest that MED6 has an effect on LUAD progression and provide valuable insights for patient stratification and personalized treatment strategies.
Keywords: lung adenocarcinoma, Mediator subunits, prognosis, multi-omics, tumor microenvironment, treatment
Introduction
Lung cancer is the most prevalent and lethal malignancy globally, with the highest incidence and mortality rates [1]. Among its subtypes, lung adenocarcinoma (LUAD) continues to show rising incidence, yet the overall prognosis for LUAD patients remains poor [2-4]. In recent years, multi-omics technologies have provided critical insights into gene regulation, epigenetic changes, and the tumor microenvironment in LUAD pathogenesis and progression [5-8]. While advances in treatment strategies have improved outcomes for some patients [9], tumor heterogeneity remains a significant challenge, leading to differences in disease progression and drug resistance [10-12]. As such, personalized identification, management, and treatment of LUAD patients remain urgent issues that need to be addressed.
Gene transcription is a fundamental biological process. As part of the preinitiation complex (PIC), RNA polymerase II (Pol II) catalyzes DNA transcription to synthesize mRNA, snRNA, and precursor microRNA [13, 14]. The Mediator complex, consisting of the head, middle, tail, and CDK8 kinase modules, plays a crucial role in transcription regulation [15, 16]. In collaboration with transcription factors, Mediator helps Pol II interact with enhancer-promoter loops to control gene expression [17-19]. Recent studies have highlighted the Mediator's involvement in diseases, including viral infections and cancer [20-22]. Given the separation between Pol II and the Mediator, variations in Mediator subunit expression may impact the formation of Mediator-mediated condensates in tumors [23-25]. Several studies have shown that Mediator subunits can promote tumorigenesis through various mechanisms [26-28], suggesting that they may have diverse roles in cancer and warrant further exploration.
In our study, we identified MED6 through tumor cell proliferation assays and differential expression analysis in LUAD. By integrating single-cell sequencing and RNA-seq data, we found that MED6 may interact with other cells in the tumor microenvironment. Based on MED6 expression, we also predicted potential therapeutic drugs. These results suggest that MED6 plays a key role in LUAD development and progression, highlighting its potential as both a biomarker and a therapeutic target.
Materials and Methods
Public and online data collection
The CRISPR-Cas9 loss-of-function data were obtained from the Dependency Map (DepMap) portal (https://depmap.org/portal/) [29]. A total of 49 LUAD cell lines from DepMap were selected for analysis. RNA-seq data for LUAD were retrieved from The Cancer Genome Atlas Program (TCGA) via the Xena platform [30] (https://xena.ucsc.edu/). Single-cell sequencing data were sourced from a published study [31] and downloaded from the Code Ocean capsule database (10.24433/CO.0121060.v1). Survival analysis of specific genes was performed using GEPIA2 [32] (http://gepia2.cancer-pku.cn/#index). All local data analyses were conducted using R software (version 4.4.1).
Identification of essential MED family genes
The CERES score, included in the CRISPR-Cas9 loss-of-function data, was used to estimate gene dependency for cell growth, following the method outlined in a previous study [33]. A lower CERES score indicates greater gene dependence for cell growth. The proportion of cell lines with CERES scores below -1 was used to determine gene dependency, with a threshold of >80%. Genes with a mean CERES score below -1 and a proportion >80% were classified as essential for cell growth. MED family genes were retrieved from GeneCards [34] (https://www.genecards.org/).
Differential genes and enrichment analysis
The RNA-seq data in the form of transcripts per million (TPM) were standardized using log2-transformed to obtain gene expression data for analysis. MED6 high- and low-expression groups were stratified based on the median MED6 expression level in the TCGA LUAD cohort. Differentially expressed genes (DEGs) were identified using the limma package (version 3.60.4) [35], comparing LUAD versus normal tissue and high versus low MED6 groups. DEGs were selected based on an adjusted P-value of <0.05 and a log2 fold-change (log2FC) > 0. The top 500 DEGs meeting these criteria were used for gene enrichment analysis using the clusterProfiler package (version 4.10.1) [36], based on the biological process from Gene Ontology (GO) [37, 38] and the Kyoto Encyclopedia of Genes and Genomes (KEGG) [39, 40]. Gene Set Enrichment Analysis (GSEA) was performed using the hallmark gene sets [41].
Single-cell data processing and analysis
Single-cell processing followed the official Seurat R package workflow (version 4.4.0) [42]. Low-quality cells were removed based on the following criteria: total counts < 500, gene numbers < 200, and mitochondrial gene percentage > 20%. Batch effects and doublets were removed using Harmony (version 1.2.0) [43] and DoubletFinder (version 2.0.3) [44]. Gene expression normalization was performed using SCTransform command. The resolution of cell clustering was evaluated by clustree (version 0.5.1) and a proper resolution of 0.5 was set to obtain different cell clusters. Cell types were annotated based on marker gene expression, as described in previous studies [31, 45]. Copy number variation (CNV) analysis was conducted to identify malignant cells, using the infercnv package (version 1.14.2). According to previous studies [45, 46], malignant cells were defined by a CNV score > 0.001 and CNV correlation > 0.4. MED6-positive and -negative cells were separated based on a gene expression threshold of 0. Cell communication was analyzed using the CellChat package (version 1.6.1), with default parameters and the human ligand-receptor database [47].
Drug sensitivity and screening analysis
Drug sensitivity analysis was performed using the Genomics of Drug Sensitivity in Cancer (GDSC) public database [48]. The oncoPredict package (version 1.2) was used to download associated data from GDSC [49]. This package constructs a regression model for gene expression and drug sensitivity based on the known GDSC gene expression profiles of cell lines and their sensitivity to various drugs. Drug sensitivity scores were imputed based on the IC50 values for each drug in GDSC. Thus, drug sensitivity could be predicted in the MED6 high and low groups. Intergroup differences were assessed using a t-test, with P-values < 0.05 considered statistically significant.
Results
Screening the cell growth required genes in LUAD cell lines
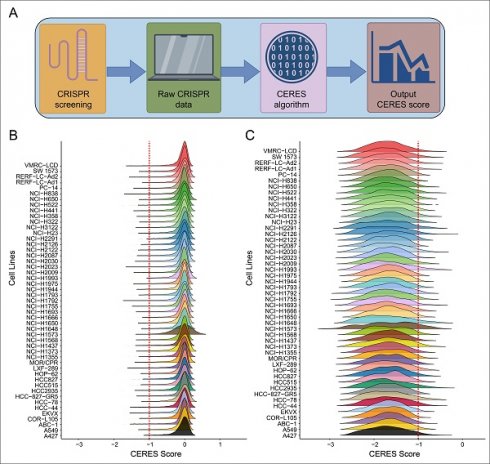
The workflow for screening cell growth-required genes is shown in Figure 1A. The CRISPR-Cas9 system was used for selective knockout of each gene to calculate CERES scores. The CERES scores of genes were predominantly distributed around zero across different LUAD cell lines (Figure 1B), indicating that most genes are not necessarily involved in tumor cell growth in LUAD. To identify genes essential for tumor cell growth, we focused on genes with CERES scores lower than -1 in over 80% of the cell lines. Genes meeting this criterion showed a concentrated distribution between -2 and -1 for their CERES scores (Figure 1C). As a result, 659 genes were identified as essential for tumor cell growth in LUAD (Table S1).
MED6 was upregulated in LUAD with prognostic value
To assess whether the MED family genes are essential for LUAD, differential expression analysis was performed between LUAD and normal tissue, focusing on the MED gene family. Among them, only three genes—MED6, MED14, and MED20—were found to be highly expressed in LUAD and essential for tumor cell growth (Figure 2A). MED6 exhibited the lowest CERES score and the highest dependency for cell growth, indicating that it is more critical for tumor proliferation than the other genes (Figure 2B). Survival analysis revealed that MED14 and MED20 did not show significant prognostic value for overall survival (OS) or disease-free survival (DFS) in LUAD patients (Figure 2C-F). However, MED6 emerged as a significant prognostic marker for both OS and DFS (Figure 2G-H). These findings suggest that high MED6 expression is associated with poor prognosis in LUAD.
Possible cellular pathways affected by MED6
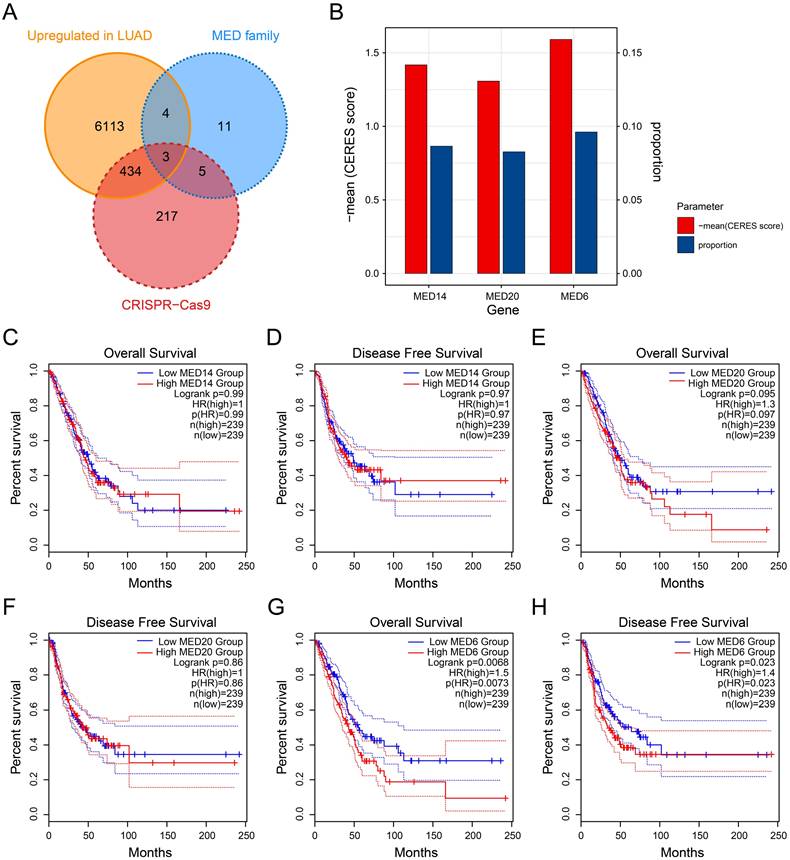
To explore the downstream functions of MED6, differential expression analysis was performed by stratifying the samples into MED6-high and MED6-low groups. The majority of DEGs were upregulated in the MED6-high group, including SNW1, EIF2S1, and ALKBH1 (Figure 3A). These results suggested that MED6 regulates numerous genes, and their expression changes correlate with MED6 expression in LUAD. The DEGs that exhibited expression patterns similar to MED6 were primarily involved in processes such as nuclear division, chromosome segregation, the cell cycle, DNA replication, and homologous recombination (Figure 3B-C). GSEA revealed significant enrichment of cell cycle-related pathways in the MED6-high group (Figure 3D-I). Additionally, the epithelial-mesenchymal transition (EMT) pathway was upregulated in the MED6-high group, suggesting that MED6 may enhance tumor cell motility and migration (Figure 3J). These findings indicate that MED6 may promote tumor growth and progression in LUAD.
Identification of MED6 in malignant cells
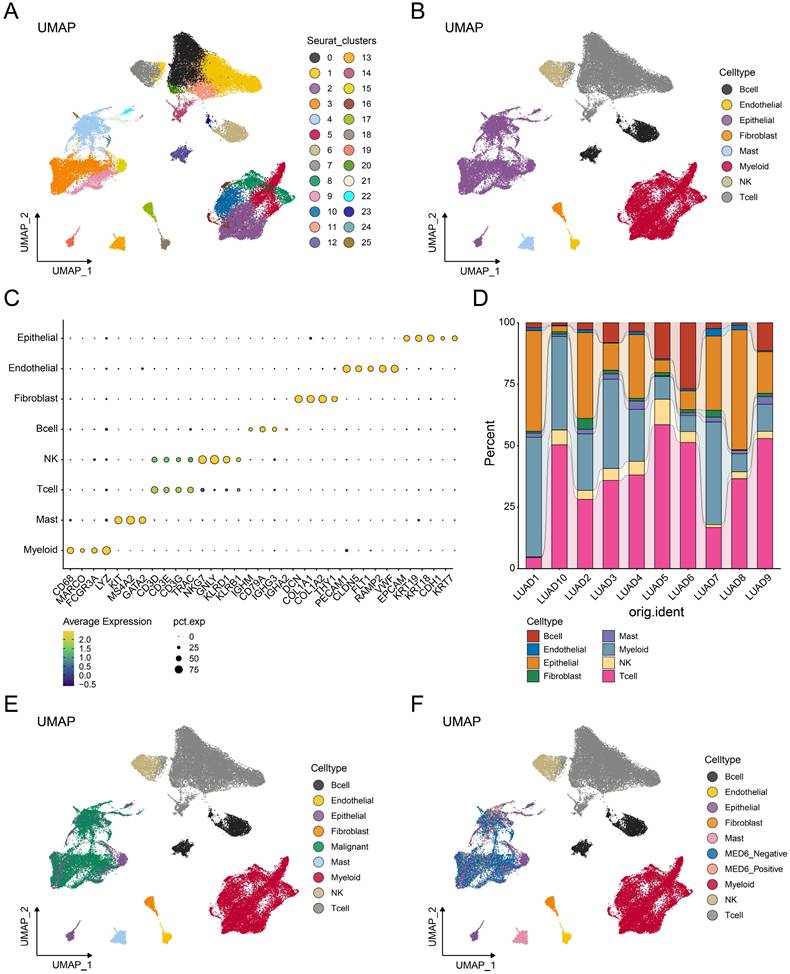
Single-cell analysis was performed to investigate the role of MED6 in tumor cells. A total of 58,191 cells and 25 clusters were obtained after processing and removing batch effects (Figure 4A). Eight cell types were identified and clearly distinguished from each other (Figure 4B), with cell type markers consistent with previous studies [31, 45] (Figure 4C). The proportion of each cell type slightly varied across different samples (Figure 4D). However, epithelial, myeloid, and T cells were the predominant cell types in each sample. Since LUAD is primarily derived from epithelial cells [50], the CNV analysis was performed in epithelial cells. Malignant tumor cells were identified based on high CNV scores and correlations (Figure S1A-J). Consistently, these malignant cells were separated from other cell types (Figure 4E). Based on MED6 expression, tumor cells were divided into MED6-positive and MED6-negative cells, resulting in 10 distinct cell types (Figure 4F). These findings suggest that MED6 expression may provide new molecular insights into LUAD.
Interaction between MED6 positive tumor cells and others
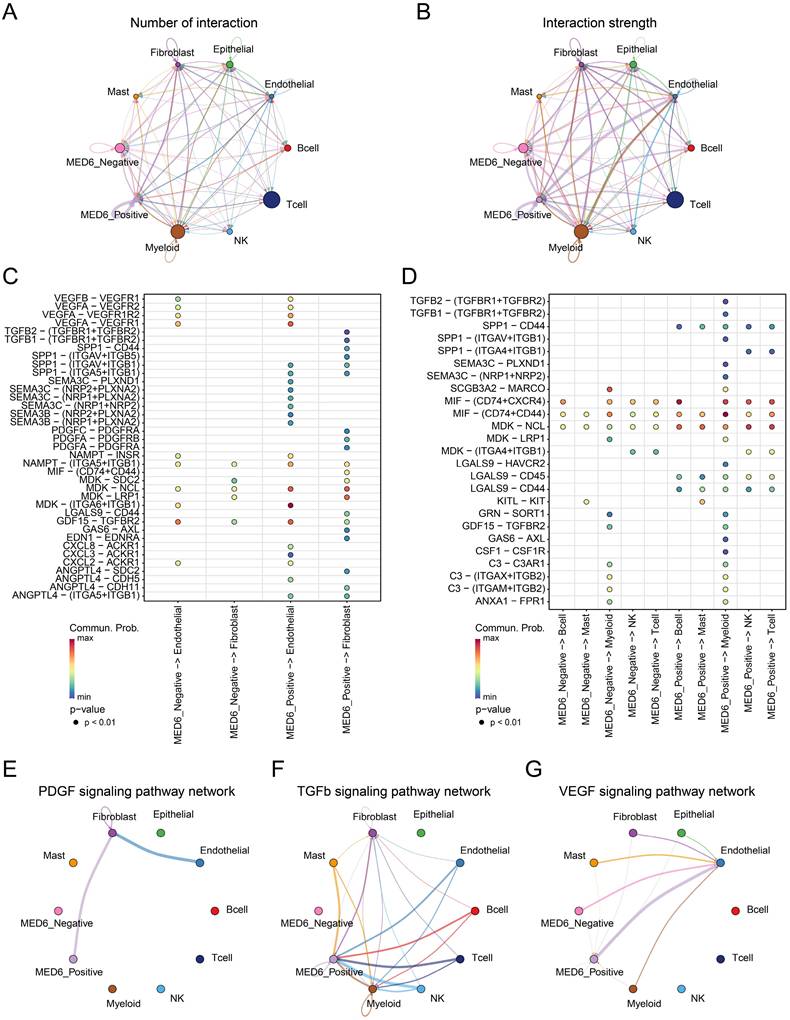
Cell-cell interactions within the tumor microenvironment (TME) are closely linked to tumor initiation and growth [51, 52]. Therefore, cell communication analysis was conducted between MED6-positive, MED6-negative, and other cells. The analysis revealed that MED6-positive cells exhibited a higher number of interactions compared to MED6-negative cells, suggesting that more ligand-receptor pairs were detected in interactions involving MED6-positive cells (Figure 5A). Furthermore, the interaction strength was stronger with MED6-positive cells, indicating that MED6-positive cells play a central role in cell communication within the TME (Figure 5B). In interactions with fibroblasts and endothelial cells, MED6-positive cells were more likely to communicate through signaling pathways such as transforming growth factor (TGF)-β, vascular endothelial growth factor (VEGF), semaphorin (SEMA), and midkine (MDK) (Figure 5C). Additionally, MED6-positive cells exhibited stronger interactions with immune cells, particularly through the SPP1 signaling pathway (Figure 5D). The platelet-derived growth factor (PDGF) and TGFβ pathways were exclusively present in MED6-positive cells (Figure 5E-F), and MED6-positive cells showed higher intensity in the VEGF pathway (Figure 5G). These results suggest that MED6-positive tumor cells have significant interactions with other cells in the TME.
CRISPR-Cas9 Screening in LUAD Cell Lines. (A) Workflow of the CRISPR-Cas9 screening process. (B) Distribution of overall CERES scores for all genes across LUAD cell lines. (C) Distribution of CERES scores for genes with scores below -1 in more than 80% of LUAD cell lines.
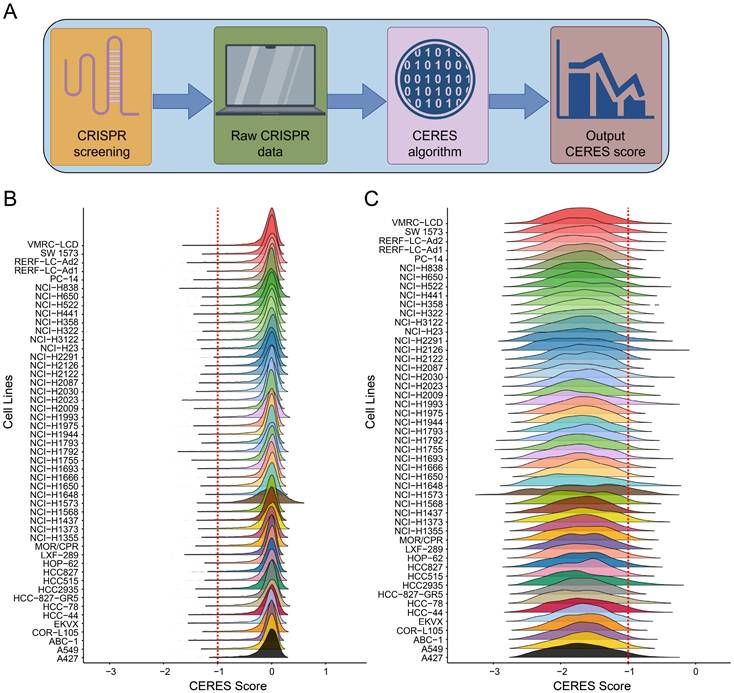
Identification of MED6 as a Prognostic Marker. (A) CRISPR-Cas9 screening of key MED family genes upregulated in LUAD. (B) Mean CERES scores and proportion parameters of MED family genes. (C-H) Kaplan-Meier survival analysis for high/low expression groups of MED14 (C-D), MED20 (E-F), and MED6 (G-H) with respect to overall survival (OS) (C, E, G) and disease-free survival (DFS) (D, F, H).
Potential drugs were screened based on MED6
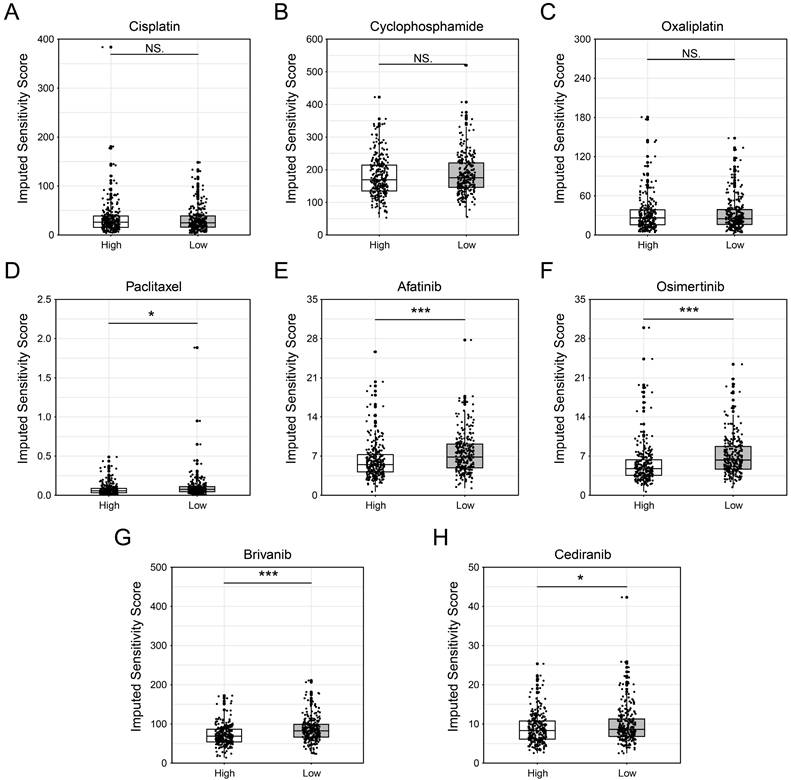
Given that MED6 is a cell growth-dependent and prognostic gene in LUAD, potential drugs were screened for patients with high MED6 expression. Drug sensitivity was assessed using compounds from GDSC database, with IC50 values used to represent drug sensitivity in the MED6 high and low groups. Chemotherapeutic drugs such as cisplatin, cyclophosphamide, and oxaliplatin showed no significant differences between the MED6 high and low groups (Figure 6A-C). However, paclitaxel demonstrated a lower IC50 in the MED6-high group, suggesting that patients with high MED6 expression may be more sensitive to paclitaxel (Figure 6D). Additionally, the IC50 values for afatinib and osimertinib, which inhibit epidermal growth factor receptor (EGFR), were significantly lower in the MED6-high group (Figure 6E-F). Furthermore, the VEGFR and PDGFR dual inhibitors brivanib and cediranib also showed lower IC50 values in the MED6-high group (Figure 6G-H). These findings identify several drugs that may be potentially effective for LUAD patients with high MED6 expression.
Potential Pathways Affected by MED6. (A) Differentially expressed genes (DEGs) between high and low MED6 groups. (B) GO enrichment analysis of DEGs. (C) KEGG pathway enrichment analysis of DEGs (D-H) GSEA revealing significant pathways in the high versus low MED6 groups, (D) G2/M checkpoint, (E) Mitotic spindle, (F) E2F targets, (G) Myc targets V1, (H) Epithelial-mesenchymal transition. NES corresponds to normalized enrichment score.
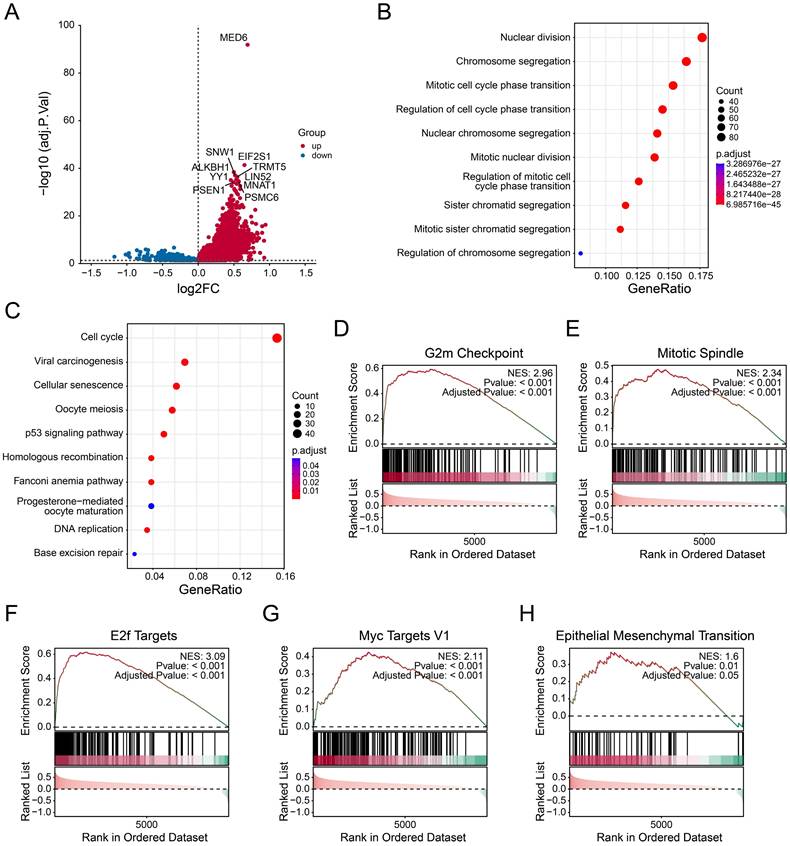
Single-Cell Transcriptome Analysis of LUAD. (A) Uniform Manifold Approximation and Projection (UMAP) showing different cell clusters. (B) UMAP depicting different cell types. (C) Annotation markers for each identified cell type. (D) Proportions of each cell type across different samples. (E) UMAP of various cell types, including malignant cells. (F) UMAP of cell types, highlighting MED6-positive and MED6-negative cells.
Cell-Cell Communication Between Different Cell Types. (A) Number of interactions between various cell types. (B) Interaction strength between different cell types. (C) Interaction network between MED6-related tumor cells and stromal cells. (D) Interaction network between MED6-related tumor cells and immune cells. (E-G) Specific signaling pathway networks in distinct cell types, (E) PDGF signaling pathway, (F) TGF-β signaling pathway, (G) VEGF signaling pathway. "Commun. Prob." refers to communication probability.
Drug Screening Between High and Low MED6 Expression Groups. (A-H) Predicted sensitivity scores for various drugs across high and low MED6 groups, (A) Cisplatin, (B) Cyclophosphamide, (C) Oxaliplatin, (D) Paclitaxel, (E) Afatinib, (F) Osimertinib, (G) Brivanib, (H) Cediranib. * P < 0.05, *** P < 0.001, NS indicates no significance.
Discussion
The MED gene family encodes several Mediator subunits, which are essential for Pol II transcription [16, 53]. These subunits come together to form the Mediator complex, a crucial transcriptional regulator that bridges transcription factors with other components of the pre-initiation complex (PIC), including Pol II [54]. While some studies suggest that MED subunits might have oncogenic potential, the specific role of MED6 in tumors remains underexplored [55, 56]. To the best of our knowledge, this is the first study to investigate the role of MED6 in lung cancer and propose its potential as a prognostic marker in LUAD.
To identify genes in the MED family, we used the CRISPR-Cas9 system to knockout genes in 49 LUAD cell lines from public databases. In over 80% of these lines, MED6 knockout significantly inhibited tumor cell growth, indicating that MED6 is closely linked to tumor cell proliferation and growth. Moreover, MED6 expression correlated with both OS and DFS in LUAD patients, suggesting its potential as a prognostic marker. This effect likely stems from the role of MED6 within the Mediator complex, which is involved in transcription. MED6, located at the head of Mediator [57, 58], coordinates PIC assembly [59], and interactions between Mediator and transcription initiation factors stabilize the complex [60]. Previous studies have shown that MED6 interacts with the C-terminal domain of Pol II, a critical step in transcription initiation [61]. Thus, Mediator serves as a key link, transmitting transcription factor signals to Pol II to start transcription [62]. Interestingly, Mediator may also be involved in transcription-coupled export (TREX2), affecting the nuclear pore complex [63], which could influence mRNA export and further broaden Mediator's regulatory role in transcription [64]. Additionally, other Mediator subunits have been linked to tumor pathology and prognosis [65, 66], reinforcing the potential of MED6 as a key player in tumor biology. These findings suggest that MED6, as part of the Mediator complex, plays a crucial role in LUAD by regulating the assembly, positioning, and transcription of the pre-transcriptional complex, making it a promising prognostic biomarker.
Since MED6 is essential for transcription, it may also affect LUAD through the expression of downstream genes. Our results showed that genes like SNW1, ALKBH1, and EIF2S1 exhibited expression patterns similar to MED6. In neuroblastoma, SNW1, a molecular chaperone, regulates the NOTCH pathway and correlates with poor prognosis [67]. ALKBH1 promotes lung cancer cell migration and invasion by demethylating m6A RNA [68]. EIF2S1 controls GPX4 and SLC7A11 expression to protect tumor cells from ferroptosis, thus promoting tumor growth [69]. These genes may synergistically enhance LUAD progression. Additionally, we found that in MED6-high LUAD cells, the expression of cell cycle-related genes was elevated, and related pathways were significantly enriched. Consistent with previous studies, the upregulation of E2f target genes and pathways suggests independent pro-tumor activities [70, 71]. Moreover, Myc-target V1 plays a critical role in tumor proliferation, migration, and angiogenesis, further promoting tumor progression [72]. Abnormalities in the G2M checkpoint and mitotic spindle have been well-documented and are associated with poor prognosis in cancer patients [73, 74]. Notably, the EMT pathway, which is critical for tumorigenesis and metastasis, was also enriched in the MED6-high-expression group [75], suggesting that high MED6 levels may facilitate tumor invasion and metastasis. In summary, MED6 likely promotes LUAD progression by regulating downstream gene expression and tumor cell growth.
Building on the advantages of bioinformatics technologies in cancer research [46, 76, 77], our study explored how MED6 affects interactions between tumor cells and other cells in TME at the single-cell level. Compared to MED6-negative tumor cells, MED6-positive tumor cells showed more frequent interactions with other TME cell types. Tumor-host cell interactions are crucial for tumor initiation and progression [78]. Similar to previous studies, our single-cell analysis revealed diverse and complex interactions in the LUAD TME, particularly for MED6-positive tumor cells [79, 80]. Secreted signals, such as ligand-receptor binding, play a major role in intercellular communication in the TME [51]. For example, melanoma can induce a pro-angiogenic phenotype in bone marrow progenitor cells through exosomes, increasing metastasis [81]. Additionally, CC and CXC chemokines and their receptors are expressed across various TME cell types, promoting tumor growth and immune regulation [82]. Our results showed that MED6-positive tumor cells exhibited more significant activity in interactions with stromal and immune cells via secreted signals like SEMA, MDK, and SPP1. Even in pathways related to angiogenesis, invasion, metastasis, and immune suppression—such as VEGF, PDGF, and TGF-β[83-85]—MED6-positive tumor cells showed stronger intercellular communication. Previous research indicates that MDK signaling promotes tumor growth, metastasis, and angiogenesis [86], while SPP1 influences tumor-associated macrophages (TAMs) and immune suppression in the TME [87, 88]. The SPP1-CD44 interaction impairs immune cell antigen presentation and immune responses, facilitating tumor progression [89]. Although different types of SEMA have distinct roles in angiogenesis [90, 91], this highlights the complex role of MED6 in tumor biology. In conclusion, MED6 may regulate tumor cell interactions in the TME, contributing to tumor progression. Furthermore, changes in MED6 expression provide new insights into TME heterogeneity in LUAD.
Given the potential role of MED6 in LUAD tumor progression, we performed drug screening for MED6-high-expressing populations. We found no significant difference in sensitivity to cisplatin, oxaliplatin, and cyclophosphamide. As platinum-based drugs, cisplatin and oxaliplatin likely target defective DNA repair processes in tumor cells [92], while cyclophosphamide likely works by inactivating aldehyde dehydrogenase [93]. These mechanisms may differ from the role of MED6 in transcription, possibly explaining the lack of significant effects in the MED6-high group. Paclitaxel, which induces mitotic arrest and apoptosis [94], aligns with the role of MED6 in promoting cell cycle activity and mitotic spindle formation. Interestingly, both brivanib and cediranib, dual inhibitors of VEGFR and PDGFR [95-98], correspond with stronger interactions between MED6-high tumor cells and stromal cells via VEGFR and PDGFR. Furthermore, crosstalk between the EGFR and VEGFR pathways promotes angiogenesis and tumor proliferation [99, 100]. This suggests that EGFR inhibitors like afatinib and osimertinib may be effective for MED6-high-expression patients. In conclusion, the drugs identified in this study may offer therapeutic potential for MED6-high-expressing LUAD patients.
Although our bioinformatics analysis of MED6 provides valuable insights, there are several limitations. Further in vivo and in vitro studies are needed to validate the role of MED6 in LUAD tumor cells and its downstream effects. Additionally, the mechanisms behind MED6-positive tumor cell interactions in the TME require further investigation. Since our study samples were derived solely from TCGA, this may potentially lead to bias in the ability of MED6 to serve as a prognostic marker for LUAD patients. Future study should involve large-scale, multi-center trials to overcome the issues of small sample size and sample selection bias, in order to validate the prognostic role of MED6 and assess its robustness as a predictive biomarker. Additionally, the dataset used in this study lacks corresponding clinical information for the patients, which limits our ability to accurately evaluate the correlation and relationship between MED6 and clinical variables. Finally, the drugs identified for MED6-high-expressing patients should undergo additional testing to confirm their therapeutic efficacy and safety in clinical trials.
In conclusion, our study provides a comprehensive investigation into the role of MED6 in LUAD. Our results suggest that MED6 promotes tumor progression by influencing tumor cell proliferation and intercellular interactions within the TME, thereby impacting LUAD prognosis. Based on MED6 expression levels, more effective, personalized treatment strategies may be developed. These findings underscore MED6 as a key molecule in LUAD development, offering new insights into identifying patients with varying prognoses and exploring personalized treatment options.
Abbreviations
LUAD: lung adenocarcinoma; MED6: Mediator complex subunit 6; PIC: preinitiation complex; Pol II: RNA polymerase II; DepMap: Dependency Map; TCGA: The Cancer Genome Atlas Program; DEGs: differentially expressed genes; log2FC: log2 fold-change; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; GSEA: Gene Set Enrichment Analysis; CNV: copy number variation; GDSC: Genomics of Drug Sensitivity in Cancer; OS: overall survival; DFS: disease-free survival; EMT: epithelial-mesenchymal transition; TME: tumor microenvironment; TGF-β: transforming growth factor-β; VEGF: vascular endothelial growth factor; SEMA: semaphorin; MDK: midkine; PDGF: platelet-derived growth factor; EGFR: epidermal growth factor receptor; TREX2: transcription-coupled export; TAMs: tumor-associated macrophages.
Supplementary Material
Supplementary figure and table.
Acknowledgements
We thank The Cancer Genome Atlas Program (TCGA), the DepMap portal, the Code Ocean capsule and the GEPIA2 databases for providing platforms and contributors for sharing their data.
Funding
This work was supported by The Medical Science and Technology Project of Zhejiang Province (2024KY284).
Availability of data and materials
The public datasets were obtained from TCGA via Xena platform (https://xena.ucsc.edu/), DepMap portal (https://depmap.org/portal/), Code Ocean capsule database (10.24433/CO.0121060.v1) and GEPIA2 (http://gepia2.cancer-pku.cn/#index).
Author contributions
CQY, DC, and SW contributed to conception and design of the study. BCW and YXL performed the statistical analysis. GXW and XKW wrote the first draft of the manuscript. CCS and YT wrote sections of the manuscript. KYZ and JF provided the overall guidance. All authors contributed to manuscript revision, read, and approved the submitted version.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Liu HI, Chiang CJ, Su SY, Jhuang JR, Tsai DR, Yang YW. et al. Incidence trends and spatial distributions of lung adenocarcinoma and squamous cell carcinoma in Taiwan. Sci Rep. 2023;13:1655
3. Chien LH, Jiang HF, Tsai FY, Chang HY, Freedman ND, Rothman N. et al. Incidence of Lung Adenocarcinoma by Age, Sex, and Smoking Status in Taiwan. JAMA Netw Open. 2023;6:e2340704
4. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48
5. Lee YF, Phua CZJ, Yuan J, Zhang B, Lee MY, Kannan S. et al. PARP4 interacts with hnRNPM to regulate splicing during lung cancer progression. Genome Med. 2024;16:91
6. La'ah AS, Tsai PH, Yarmishyn AA, Ching LJ, Chen CY, Chien Y. et al. Neutrophils Recruited by NKX2-1 Suppression via Activation of CXCLs/CXCR2 Axis Promote Lung Adenocarcinoma Progression. Adv Sci (Weinh). 2024;11:e2400370
7. Wang S, Zeng Y, Zhu L, Zhang M, Zhou L, Yang W. et al. The N6-methyladenosine Epitranscriptomic Landscape of Lung Adenocarcinoma. Cancer Discov. 2024;14:2279-99
8. Tavernari D, Battistello E, Dheilly E, Petruzzella AS, Mina M, Sordet-Dessimoz J. et al. Nongenetic Evolution Drives Lung Adenocarcinoma Spatial Heterogeneity and Progression. Cancer Discov. 2021;11:1490-507
9. Meyer ML, Fitzgerald BG, Paz-Ares L, Cappuzzo F, Janne PA, Peters S. et al. New promises and challenges in the treatment of advanced non-small-cell lung cancer. Lancet. 2024;404:803-22
10. Zhang J, Huang Y, Han Y, Dong D, Cao Y, Chen X. et al. Immune microenvironment heterogeneity of concurrent adenocarcinoma and squamous cell carcinoma in multiple primary lung cancers. NPJ Precis Oncol. 2024;8:55
11. Goldstraw P, Chansky K, Crowley J, Rami-Porta R, Asamura H, Eberhardt WE. et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol. 2016;11:39-51
12. Xing P, Wang S, Cao Y, Liu B, Zheng F, Guo W. et al. Treatment strategies and drug resistance mechanisms in adenocarcinoma of different organs. Drug Resist Updat. 2023;71:101002
13. Sims RJ 3rd, Mandal SS, Reinberg D. Recent highlights of RNA-polymerase-II-mediated transcription. Curr Opin Cell Biol. 2004;16:263-71
14. Schier AC, Taatjes DJ. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 2020;34:465-88
15. Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235-9
16. Soutourina J. Transcription regulation by the Mediator complex. Nat Rev Mol Cell Biol. 2018;19:262-74
17. Yin JW, Wang G. The Mediator complex: a master coordinator of transcription and cell lineage development. Development. 2014;141:977-87
18. Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol. 2015;16:155-66
19. Hua P, Badat M, Hanssen LLP, Hentges LD, Crump N, Downes DJ. et al. Defining genome architecture at base-pair resolution. Nature. 2021;595:125-9
20. Schneider WM, Luna JM, Hoffmann HH, Sanchez-Rivera FJ, Leal AA, Ashbrook AW. et al. Genome-Scale Identification of SARS-CoV-2 and Pan-coronavirus Host Factor Networks. Cell. 2021;184:120-32 e14
21. Compe E, Egly JM. The Long Road to Understanding RNAPII Transcription Initiation and Related Syndromes. Annu Rev Biochem. 2021;90:193-219
22. Li Y, Wu J, Tian Y, Zhu Q, Ge Y, Yu H. et al. MED1 Downregulation Contributes to TGFbeta-Induced Metastasis by Inhibiting SMAD2 Ubiquitination Degradation in Cutaneous Melanoma. J Invest Dermatol. 2022;142:2228-37 e4
23. Syring I, Klumper N, Offermann A, Braun M, Deng M, Boehm D. et al. Comprehensive analysis of the transcriptional profile of the Mediator complex across human cancer types. Oncotarget. 2016;7:23043-55
24. Bragelmann J, Klumper N, Offermann A, von Massenhausen A, Bohm D, Deng M. et al. Pan-Cancer Analysis of the Mediator Complex Transcriptome Identifies CDK19 and CDK8 as Therapeutic Targets in Advanced Prostate Cancer. Clin Cancer Res. 2017;23:1829-40
25. Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V. et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412-5
26. Liu Y, Tao X, Fan L, Jia L, Gu C, Feng Y. Knockdown of mediator complex subunit 19 inhibits the growth of ovarian cancer. Mol Med Rep. 2012;6:1050-6
27. Yu W, Zhang Z, Min D, Yang Q, Du X, Tang L. et al. Mediator of RNA polymerase II transcription subunit 19 promotes osteosarcoma growth and metastasis and associates with prognosis. Eur J Cancer. 2014;50:1125-36
28. Huang CY, Hsieh NT, Li CI, Weng YT, Liu HS, Lee MF. MED28 Regulates Epithelial-Mesenchymal Transition Through NFkappaB in Human Breast Cancer Cells. J Cell Physiol. 2017;232:1337-45
29. Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS. et al. Defining a Cancer Dependency Map. Cell. 2017;170:564-76 e16
30. Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675-8
31. Bischoff P, Trinks A, Obermayer B, Pett JP, Wiederspahn J, Uhlitz F. et al. Single-cell RNA sequencing reveals distinct tumor microenvironmental patterns in lung adenocarcinoma. Oncogene. 2021;40:6748-58
32. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556-W60
33. Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H. et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779-84
34. Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S. et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 2016;54:1 30 1-1 3
35. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47
36. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2:100141
37. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25-9
38. Gene Ontology C, Aleksander SA, Balhoff J, Carbon S, Cherry JM, Drabkin HJ. et al. The Gene Ontology knowledgebase in 2023. Genetics. 2023;224:iyad031
39. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27-30
40. Kanehisa M, Furumichi M, Sato Y, Matsuura Y, Ishiguro-Watanabe M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 2025;53:D672-D7
41. Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417-25
42. Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A. et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573-87 e29
43. Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289-96
44. McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019;8:329-37 e4
45. Kim N, Kim HK, Lee K, Hong Y, Cho JH, Choi JW. et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun. 2020;11:2285
46. Wang G, Shi C, He L, Li Y, Song W, Chen Z. et al. Identification of the tumor metastasis-related tumor subgroups overexpressed NENF in triple-negative breast cancer by single-cell transcriptomics. Cancer Cell Int. 2024;24:319
47. Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH. et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088
48. Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S. et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955-61
49. Maeser D, Gruener RF, Huang RS. oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform. 2021;22:bbab260
50. Han G, Sinjab A, Rahal Z, Lynch AM, Treekitkarnmongkol W, Liu Y. et al. An atlas of epithelial cell states and plasticity in lung adenocarcinoma. Nature. 2024;627:656-63
51. de Visser KE, Joyce JA. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell. 2023;41:374-403
52. Jin MZ, Jin WL. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct Target Ther. 2020;5:166
53. Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993-4008
54. Soutourina J, Wydau S, Ambroise Y, Boschiero C, Werner M. Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science. 2011;331:1451-4
55. Kuuselo R, Savinainen K, Sandstrom S, Autio R, Kallioniemi A. MED29, a component of the mediator complex, possesses both oncogenic and tumor suppressive characteristics in pancreatic cancer. Int J Cancer. 2011;129:2553-65
56. Vijayvargia R, May MS, Fondell JD. A coregulatory role for the mediator complex in prostate cancer cell proliferation and gene expression. Cancer Res. 2007;67:4034-41
57. Plaschka C, Lariviere L, Wenzeck L, Seizl M, Hemann M, Tegunov D. et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature. 2015;518:376-80
58. Lariviere L, Plaschka C, Seizl M, Wenzeck L, Kurth F, Cramer P. Structure of the Mediator head module. Nature. 2012;492:448-51
59. Eyboulet F, Wydau-Dematteis S, Eychenne T, Alibert O, Neil H, Boschiero C. et al. Mediator independently orchestrates multiple steps of preinitiation complex assembly in vivo. Nucleic Acids Res. 2015;43:9214-31
60. Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei PJ, Burlingame AL. et al. Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex. Cell. 2016;166:1411-22 e16
61. Robinson PJ, Bushnell DA, Trnka MJ, Burlingame AL, Kornberg RD. Structure of the mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Proc Natl Acad Sci U S A. 2012;109:17931-5
62. Poss ZC, Ebmeier CC, Taatjes DJ. The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol. 2013;48:575-608
63. Schneider M, Hellerschmied D, Schubert T, Amlacher S, Vinayachandran V, Reja R. et al. The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression. Cell. 2015;162:1016-28
64. Jani D, Valkov E, Stewart M. Structural basis for binding the TREX2 complex to nuclear pores, GAL1 localisation and mRNA export. Nucleic Acids Res. 2014;42:6686-97
65. Jin X, Zhang Y, Hu W, Liu C, Cai D, Sun J. et al. Developing a prognostic model for hepatocellular carcinoma based on MED19 and clinical stage and determining MED19 as a therapeutic target. J Cancer Res Clin Oncol. 2024;150:446
66. Wu S, Li Q, Cao Y, Luo S, Wang Z, Zhang T. Mediator complex subunit 8 is a prognostic biomarker in hepatocellular carcinoma. Am J Transl Res. 2022;14:1765-77
67. Hong M, He J, Li S. SNW1 regulates Notch signaling in neuroblastoma through interacting with RBPJ. Biochem Biophys Res Commun. 2019;509:869-76
68. Li H, Zhang Y, Guo Y, Liu R, Yu Q, Gong L. et al. ALKBH1 promotes lung cancer by regulating m6A RNA demethylation. Biochem Pharmacol. 2021;189:114284
69. Li Z, Wang Y, Liang S, Yuan T, Liu J. EIF2S1 Silencing Impedes Neuroblastoma Development Through GPX4 Inactivation and Ferroptosis Induction. Int J Genomics. 2024;2024:6594426
70. Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785-97
71. Kent LN, Rakijas JB, Pandit SK, Westendorp B, Chen HZ, Huntington JT. et al. E2f8 mediates tumor suppression in postnatal liver development. J Clin Invest. 2016;126:2955-69
72. Xu J, Sun Y, Fu W, Fu S. MYCT1 in cancer development: Gene structure, regulation, and biological implications for diagnosis and treatment. Biomed Pharmacother. 2023;165:115208
73. Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I. et al. High G2M Pathway Score Pancreatic Cancer is Associated with Worse Survival, Particularly after Margin-Positive (R1 or R2) Resection. Cancers (Basel). 2020;12:2871
74. Chen Y, Nam S, Chaudhuri O, Huang HC. The evolution of spindles and their mechanical implications for cancer metastasis. Cell Cycle. 2019;18:1671-5
75. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69-84
76. Wang Y, Song W, Li Y, Liu Z, Zhao K, Jia L. et al. Integrated analysis of tumor microenvironment features to establish a diagnostic model for papillary thyroid cancer using bulk and single-cell RNA sequencing technology. J Cancer Res Clin Oncol. 2023;149:16837-50
77. Tian Y, Liu X, Hu J, Zhang H, Wang B, Li Y. et al. Integrated Bioinformatic Analysis of the Expression and Prognosis of Caveolae-Related Genes in Human Breast Cancer. Front Oncol. 2021;11:703501
78. Sudhakar M, Vignesh H, Natarajan KN. Crosstalk between tumor and microenvironment: Insights from spatial transcriptomics. Adv Cancer Res. 2024;163:187-222
79. Almet AA, Cang Z, Jin S, Nie Q. The landscape of cell-cell communication through single-cell transcriptomics. Curr Opin Syst Biol. 2021;26:12-23
80. Armingol E, Officer A, Harismendy O, Lewis NE. Deciphering cell-cell interactions and communication from gene expression. Nat Rev Genet. 2021;22:71-88
81. Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G. et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883-91
82. Bikfalvi A, Billottet C. The CC and CXC chemokines: major regulators of tumor progression and the tumor microenvironment. Am J Physiol Cell Physiol. 2020;318:C542-C54
83. Yang Y, Cao Y. The impact of VEGF on cancer metastasis and systemic disease. Semin Cancer Biol. 2022;86:251-61
84. Zou X, Tang XY, Qu ZY, Sun ZW, Ji CF, Li YJ. et al. Targeting the PDGF/PDGFR signaling pathway for cancer therapy: A review. Int J Biol Macromol. 2022;202:539-57
85. Derynck R, Turley SJ, Akhurst RJ. TGFbeta biology in cancer progression and immunotherapy. Nat Rev Clin Oncol. 2021;18:9-34
86. Filippou PS, Karagiannis GS, Constantinidou A. Midkine (MDK) growth factor: a key player in cancer progression and a promising therapeutic target. Oncogene. 2020;39:2040-54
87. Lyu A, Fan Z, Clark M, Lea A, Luong D, Setayesh A. et al. Evolution of myeloid-mediated immunotherapy resistance in prostate cancer. Nature. 2025;637:1207-17
88. Wu J, Shen Y, Zeng G, Liang Y, Liao G. SPP1(+) TAM subpopulations in tumor microenvironment promote intravasation and metastasis of head and neck squamous cell carcinoma. Cancer Gene Ther. 2024;31:311-21
89. Zhang J, Peng Q, Fan J, Liu F, Chen H, Bi X. et al. Single-cell and spatial transcriptomics reveal SPP1-CD44 signaling drives primary resistance to immune checkpoint inhibitors in RCC. J Transl Med. 2024;22:1157
90. Sakurai A, Doci CL, Gutkind JS. Semaphorin signaling in angiogenesis, lymphangiogenesis and cancer. Cell Res. 2012;22:23-32
91. Bica C, Tirpe A, Nutu A, Ciocan C, Chira S, Gurzau ES. et al. Emerging roles and mechanisms of semaphorins activity in cancer. Life Sci. 2023;318:121499
92. Rottenberg S, Disler C, Perego P. The rediscovery of platinum-based cancer therapy. Nat Rev Cancer. 2021;21:37-50
93. Emadi A, Jones RJ, Brodsky RA. Cyclophosphamide and cancer: golden anniversary. Nat Rev Clin Oncol. 2009;6:638-47
94. Khing TM, Choi WS, Kim DM, Po WW, Thein W, Shin CY. et al. The effect of paclitaxel on apoptosis, autophagy and mitotic catastrophe in AGS cells. Sci Rep. 2021;11:23490
95. Allen E, Walters IB, Hanahan D. Brivanib, a dual FGF/VEGF inhibitor, is active both first and second line against mouse pancreatic neuroendocrine tumors developing adaptive/evasive resistance to VEGF inhibition. Clin Cancer Res. 2011;17:5299-310
96. Nakamura I, Zakharia K, Banini BA, Mikhail DS, Kim TH, Yang JD. et al. Brivanib attenuates hepatic fibrosis in vivo and stellate cell activation in vitro by inhibition of FGF, VEGF and PDGF signaling. PLoS One. 2014;9:e92273
97. Najy AJ, Jung YS, Won JJ, Conley-LaComb MK, Saliganan A, Kim CJ. et al. Cediranib inhibits both the intraosseous growth of PDGF D-positive prostate cancer cells and the associated bone reaction. Prostate. 2012;72:1328-38
98. Momeny M, Sankanian G, Hamzehlou S, Yousefi H, Esmaeili F, Alishahi Z. et al. Cediranib, an inhibitor of vascular endothelial growth factor receptor kinases, inhibits proliferation and invasion of prostate adenocarcinoma cells. Eur J Pharmacol. 2020;882:173298
99. Larsen AK, Ouaret D, El Ouadrani K, Petitprez A. Targeting EGFR and VEGF(R) pathway cross-talk in tumor survival and angiogenesis. Pharmacol Ther. 2011;131:80-90
100. Nagano H, Tomida C, Yamagishi N, Teshima-Kondo S. VEGFR-1 Regulates EGF-R to Promote Proliferation in Colon Cancer Cells. Int J Mol Sci. 2019;20:5608
Author contact
![]() Corresponding authors: Prof. Keyun Zhu, Department of Thoracic Surgery, Ningbo Medical Center Lihuili Hospital, Ningbo University, Ningbo, Zhejiang Province, 315040, China, E-mail: lhlzhukeyunedu.cn; Prof. Jing Feng, Department of Respiratory and Critical Care Medicine, Tianjin Medical University General Hospital, Tianjin, 300052, China, E-mail: tmugh_fjedu.cn.
Corresponding authors: Prof. Keyun Zhu, Department of Thoracic Surgery, Ningbo Medical Center Lihuili Hospital, Ningbo University, Ningbo, Zhejiang Province, 315040, China, E-mail: lhlzhukeyunedu.cn; Prof. Jing Feng, Department of Respiratory and Critical Care Medicine, Tianjin Medical University General Hospital, Tianjin, 300052, China, E-mail: tmugh_fjedu.cn.

 Global reach, higher impact
Global reach, higher impact