3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(12):3663-3674. doi:10.7150/jca.95979 This issue Cite
Research Paper
Multi-omic profiling of breast tumor microenvironment uncovers a role of mitochondrial calcium gatekeepers
1. Institute of Biomedical Sciences, National Sun Yat-sen University, Kaohsiung 804, Taiwan.
2. Department of Surgery, Kaohsiung Veterans General Hospital, Kaohsiung 813, Taiwan.
3. Department of Pathology, Show Chwan Memorial Hospital, Changhua 500, Taiwan.
4. Department of Allergy and Immunology, China Medical University Children's Hospital, Taichung 404, Taiwan.
5. Research Center for Allergy, Immunology, and Microbiome, China Medical University Hospital, Taichung 404, Taiwan.
6. Institute of Molecular & Cellular Biology, National Tsing Hua University, Hsinchu 300, Taiwan.
7. Comprehensive Breast Health Center, Taipei Veterans General Hospital, Taipei 112, Taiwan.
8. Division of Breast Surgery, Department of Surgery, Center for Cancer Research, Kaohsiung Medical University Chung-Ho Memorial Hospital, Kaohsiung 807, Taiwan.
9. Institute of Clinical Medicine, National Cheng Kung University, Tainan 704, Taiwan.
10. Department of Surgery, Kaohsiung Municipal Minsheng Hospital, Kaohsiung 802, Taiwan.
11. Department of Obstetrics and Gynecology, Kaohsiung Veterans General Hospital, Kaohsiung 813, Taiwan.
12. Institute of BioPharmaceutical Sciences, National Sun Yat-sen University, Kaohsiung 804, Taiwan.
Abstract
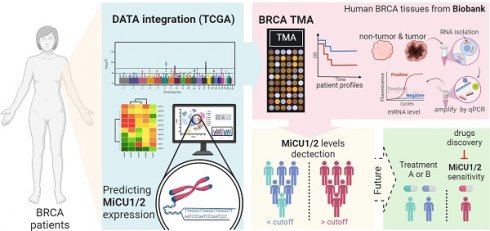
In this study, we aimed to elucidate the role of mitochondrial calcium uptake 1/2 (MiCU1/2) in breast cancer (BRCA) by employing a comprehensive multi-omics approach. Unlike previous research, we utilized a novel web application tailored for whole tumor tissue, single-cell, and spatial transcriptomics analysis to investigate the association between MiCU1/2 and the tumor immune microenvironment (TIME). Our gene set enrichment analysis (GSEA) provided insights into the primary biological effects of MiCU1/2, while our CRISPR-based drug screening repository identified potential effective drugs. Our study revealed that high MiCU1/2 expression serves as an independent diagnostic biomarker, correlating with advanced clinical status and indicating poorer recurrence-free survival (RFS) rates in BRCA patients. Additionally, spatial transcriptome analysis highlighted the heightened expression of MiCU1/2 in tumors and its relevance in surrounding immune cells. Furthermore, using the CIBERSORT algorithm, we discovered a positive correlation between MiCU1/2 levels and macrophage infiltration, underscoring their potential impact on immune infiltration. We also identified expression patterns of immune-related genes associated with responses against various immune cell types, including CXCL, MIF, GDF, SPP1, and IL16. Finally, our pharmacogenomic screening identified potential small molecule drugs capable of effectively targeting breast cancer cells with elevated MiCU1/2 expression. Overall, our study establishes MiCU1/2 as a promising novel biomarker for BRCA diagnosis and prognostic prediction, as well as a potential therapeutic target, highlighting the importance of exploring these pathways to advance patient care and outcomes in BRCA treatment.
Keywords: Breast cancer, Spatial transcriptomic, MiCU, Single-cell RNA-Sequence, Precision medicine

 Global reach, higher impact
Global reach, higher impact