3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(9):2475-2485. doi:10.7150/jca.93229 This issue Cite
Research Paper
KYNU Expression Promotes Cisplatin Resistance in Esophageal Cancer
1. Department of Clinical Research Center, Xuyi People's Hospital, Affiliated Xuyi Hospital of Yangzhou University Medical College, Jiangsu, China.
2. Department of pharmacy, Xuyi People's Hospital, Affiliated Xuyi Hospital of Yangzhou University Medical College, Jiangsu, China.
3. Department of Gastroenterology, Xuyi People's Hospital, Affiliated Xuyi Hospital of Yangzhou University Medical College, Jiangsu, China.
4. Department of Endocrinology, Xuyi People's Hospital, Affiliated Xuyi Hospital of Yangzhou University Medical College, Jiangsu, China.
5. Department of Medical Research Center, The First Affiliated Hospital of Wenzhou Medical University, Zhejiang, China.
6. Department of Inspection, Xuyi People's Hospital, Affiliated Xuyi Hospital of Yangzhou University Medical College, Jiangsu, China.
7. Key Laboratory of Tropical Biological Resources of Ministry of Education, School of Pharmaceutical Sciences, Hainan University, Haikou, China.
*These authors contributed equally to this work, share first authorship.
Abstract
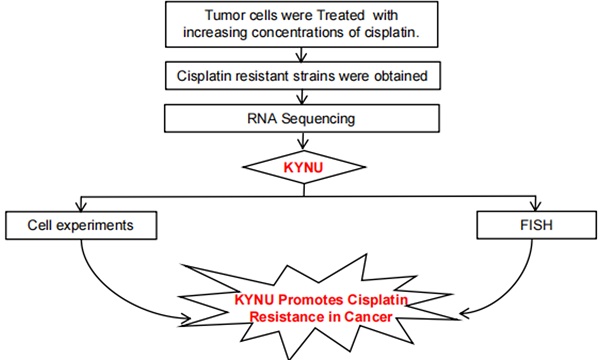
Background: Chemotherapy resistance is a barrier to effective cancer prognoses. Cisplatin (CDDP) resistance is a major challenge for esophageal cancer (EC) therapy. A deeper understanding of the fundamental mechanisms of cisplatin resistance and improved targeting strategies are required in clinical settings. This study was performed to identify and characterize a marker of cisplatin resistance in EC cells.
Method: KYSE140 and Eca-109 cells were subjected to escalating concentrations of cisplatin, resulting in the development of cisplatin-resistant KYSE140/CDDP and Eca-109/CDDP cell lines, respectively. RNA Sequencing (RNA-seq) was utilized to screen for the genes exhibiting differential expression between cisplatin-resistant and parental cells. Reverse transcription quantitative PCR was conducted to assess gene expression, and western blotting was employed to analyze protein levels. A sphere-formation assay was performed to validate tumor cell stemness. Cell counting kit-8 (CCK-8) experiments were conducted to confirm the sensitivity of cells to cisplatin. We examined the relationship between target genes and the clinicopathological features of patients with EC. Furthermore, the expression of target genes in EC tissues was evaluated via western blotting and fluorescence probe in situ hybridization (FISH).
Results: KYNU was upregulated in cisplatin-resistant EC cells (KYSE140/CDDP and Eca-109/CDDP cells) and in EC tissues compared to that in the respective parental cell lines (KYSE140 and Eca-109 cells) and non-carcinoma tissues. Downregulation of KYNU increased cell sensitivity to cisplatin and suppressed tumor stemness, whereas abnormal KYNU expression had the opposite effect. KYNU expression was correlated with the expression of tumor stemness-associated factors (SOX2, Nanog, and OCT4) and the tumor size.
Conclusions: KYNU may promote drug resistance in EC by regulating cancer stemness, and could serve as a biomarker and therapeutic target for EC.
Keywords: Cisplatin, Drug resistance, Esophageal cancer, Kynureninase, Tumor stemness

 Global reach, higher impact
Global reach, higher impact