3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(8):2431-2441. doi:10.7150/jca.93055 This issue Cite
Research Paper
Uncovering Microbial Composition of the Tissue Microenvironment in Bladder Cancer using RNA Sequencing Data
1. School of Health Science and Engineering, University of Shanghai for Science and Technology, Shanghai 200093, China.
2. Department of Medical Genetics, Naval Medical University, Xiang-Yin Road, 800, Shanghai 200433, China.
3. Department of Precision Medicine, Translational Medicine Research Center, Naval Medical University, Xiang-Yin Road, 800, Shanghai 200433, China.
4. Shanghai Key Laboratory of Cell Engineering, Shanghai, China.
5. Department of Urology, Huadong Hospital, Fudan University, Shanghai, China.
6. Department of Urology, Shanghai General Hospital Affiliated to Nanjing Medical University, Shanghai, 200080, China.
7. Department of Urology, Shanghai General Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, 200080, China.
# First author: Ruiqian Yao, Bin Ai, and Zeyi Wang contributed equally to this work.
Abstract
Purpose: Bladder cancer (BC) is one of the top 10 common tumors in the world. It has been reported that microbiota can colonize tissues and play important roles in tumorigenesis and progression. However, the current understanding of microorganisms in the BC tissue microenvironment remains unclear.
Methods: In this study, we integrated the RNA-seq data of 479 BC tissue samples from seven datasets combined with a range of bioinformatics tools to explore the landscape of microbiome in the BC tissue microenvironment.
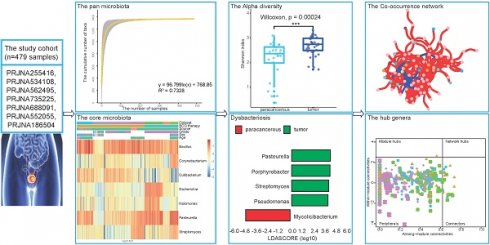
Results: The pan-microbiome was estimated to surpass 1,400 genera. A total of seven core microbiota (Bacillus, Corynebacterium, Cutibacterium, Escherichia, Halomonas, Pasteurella, and Streptomyces) were identified. Among them, Bacillus was widely distributed in all datasets with a high relative abundance (10.11% of all samples on average). Moreover, some biological factors, including tissue source and tumor grade, were found significant effects on the microbial composition of the bladder tissue. Pseudomonas, Porphyrobacter, and Acinetobacter were enriched in tumor tissues, while Mycolicibacterium and Streptomyces were enriched in patients who showed durable response to BCG therapy. In addition, we established microbial co-occurrence networks and found that the BCG therapy may attenuate the microbiological interactions.
Conclusions: This study clearly provided a microbial landscape of the BC tissue microenvironment, which was important for exploring the interactions between microorganisms and BC tissues. The identified specific taxa might be potential biomarkers for BC.
Keywords: Bladder cancer, Cancer microbiome, Biological factors, Co-occurrence network analysis

 Global reach, higher impact
Global reach, higher impact