3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(18):3539-3549. doi:10.7150/jca.86942 This issue Cite
Research Paper
Construction of the metabolism-related models for predicting prognosis and infiltrating immune phenotype in lung squamous cell carcinoma
1. Department of Thoracic Surgery II, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Peking University Cancer Hospital and Institute, Beijing, 100142, People's Republic of China.
2. Department of Computational Oncology, Intelliphecy, Shenzhen, China; Center for Systems Biology, Intelliphecy, Shenzhen, China.
3. Department of Gastrointestinal Oncology, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Peking University Cancer Hospital and Institute, 100142, People's Republic of China.
4. Beijing International Bilingual Academy, People's Republic of China.
* Zeming Ma and Liang Wang contributed equally to this work.
Abstract
Purpose: Cancers often display disorder metabolism, which closely related to the poor outcome of patients. We aimed to establish prognostic models using metabolism-associated genes, and identify the key factor involved in metabolism in lung squamous cell carcinoma (LUSC).
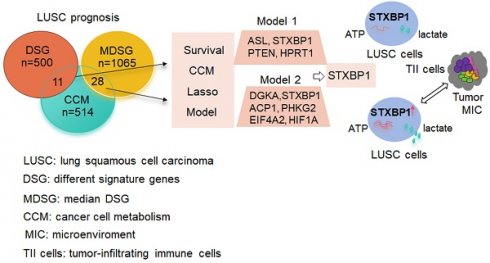
Materials and Methods: R package 'TCGA biolinks' was used to download the mRNA sequencing data of LUSC from TCGA. The clusterProfiler package was performed to analyze biological pathways. The online tool GEPIA2 and cox regression method were applied to identify the two gene lists associated with metabolism and prognosis of LUSC. The lasso modeling was conducted to establish prognostic models. The quantiseq method was used to identify the cellular abundance of expression matrix in TCGA-LUSC dataset. Immunohistochemistry and western blotting were done to evaluate the STXBP1 expression in LUSC samples. Lactate assay and ATP detection were performed to assess metabolic effect, and CCK8 assay was done to test cell proliferation in the LUSC cells with overexpression and suppression of STXBP1.
Results: Two lists of survival-metabolism-associated genes (11 and 28 genes) were identified and applied in the prognostic model 1 and model 2 construction from TCGA-LUSC dataset. High-risk LUSC patients associated with poor survival in the training cohort and the test cohort of both model 1 and model 2. Higher ROC values for 10- year survival was shown in model 2 than in model 1. In addition, macrophage M1, macrophage M2, neutrophil, and T regulatory cell were enriched in the high-risk group of model 2. STXBP1 was the only optimized gene in both model 1 and model 2, and related to the poor outcome of LUSC patients. Furthermore, STXBP1 associated with infiltrating immune cells, and increased lactate, ATP levels, and cell proliferation.
Conclusion: Our finding provides the metabolism-associated models to predict prognosis of LUSC patients. STXBP1, as the key optimized gene in the model, promotes metabolic progress to increase lactate and ATP levels in LUSC cells.
Keywords: metabolism, prognosis, immune phenotype, lung squamous cell carcinoma, STXBP1

 Global reach, higher impact
Global reach, higher impact