Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(14):2655-2669. doi:10.7150/jca.87184 This issue Cite
Research Paper
Identification of Tumor Antigens and Immune Subtypes of High-grade Serous Ovarian Cancer for mRNA Vaccine Development
1. Department of Radiation Oncology, Cancer Hospital of Shantou University Medical College, Shantou, Guangdong, China.
2. Department of Medical Oncology, Cancer Hospital of Shantou University Medical College, Shantou, Guangdong, China.
*These 2 authors contributed equally to this project.
Received 2023-6-15; Accepted 2023-7-24; Published 2023-8-28
Abstract
High-grade serous ovarian cancer (HGSC) is the most common pathology of ovarian cancer and has aggressive characteristics and poor prognosis. mRNA vaccines are a novel tool for cancer immune treatment and may play an important role in HGSC therapy. Our study aimed to explore tumour antigens for vaccine development and identify potential populations amenable to vaccine treatment. Based on transcription data from The Cancer Genome Atlas (TCGA), we identified four tumour-specific antigens for vaccine production: ARPC1B, ELF3, VSTM2L, and IL27RA. In addition to being associated with HGSC patient prognosis, the expression of these antigens was positively correlated with the abundances of antigen-presenting cells (APCs). Furthermore, we stratified HGSC samples into three immune subtypes (IS1-IS3) with different immune characteristics. A corhort from ICGC (International Cancer Genome Consortium) was used to validate. Patients of IS3 had the best prognosis, while patients of IS1 were most likely to benefit from vaccination. There was substantial heterogeneity in immune signatures and immune-associated molecule expression in HGSC. Finally, weighted gene coexpression network analysis (WGCNA) was employed to cluster immune-related genes and explore potential biomarkers related to vaccination. In conclusion, we identified four potential tumour antigens for mRNA vaccine production for HGSC treatment, and the immune subtype could be an important indicator to select suitable HGSC patients to receive vaccination.
Introduction
Ovarian cancer (OC) is one of the most common gynaecological cancers. Statistically, 310000 patients were diagnosed with OC, and 210000 patients died of OC [1]. The mortality of OC ranks first among gynaecological cancers [2]. The most common pathology of OC is high-grade serous ovarian cancer (HGSC), which accounts for 50-60% of cases [3, 4]. The first-line treatment of early-stage HGSC is surgery followed by chemotherapy. Most HGSC cases respond to cytopathic surgery and platinum-based chemotherapy. However, more than 80% of patients experience disease recurrence, especially 18-24 months after the first treatment [5]. Moreover, 75% of OC patients are diagnosed with advanced disease because of inconspicuous symptoms and the lack of effective screening [6]. For advanced OC, the NCCN guidelines recommend platinum-based chemotherapy followed by maintenance therapy with PARP inhibitors, regardless of BRCA mutation status [7]. Recently, immune checkpoint inhibitors (ICIs), such as CTLA-4 inhibitors and PD-1/PD-L1 inhibitors, have brought promise for treating solid tumours. One important characteristic of OC is the increase in tumour-infiltrating lymphocytes (TILs), and this characteristic is a good biomarker for evaluating the effect of ICIs. Unfortunately, existing studies have not proven that single or combination ICIs benefit OC progression-free survival (PFS) or overall survival (OS) [8, 9]. OC is an aggressive malignancy with a 5-year survival rate of less than 40% [10]. As a result, more effective treatments should be studied.
The purpose of immune therapy is to activate the immunity of the host to inhibit malignancy. In addition to ICIs, other immune-related agents, such as T-cell treatments, cytokines, and vaccines, have been explored [11]. Cancer vaccines can be used to induce novel responses against tumour-specific antigens to attack and destroy malignant cells with overexpressed antigens and to achieve long-term therapeutic responses through immune memory. Therefore, cancer vaccines are specific, safe, and tolerable therapeutic agents [12]. Cancer vaccines include preventative vaccines and therapeutic vaccines. The former is mainly used in the prevention of cancer-related infections, such as hepatitis B virus vaccines and human papilloma virus (HPV) vaccines, while the latter is mainly used to treat malignancy [13]. Based on composition, tumour vaccines are divided into four groups: (1) viral vector vaccines, (2) tumour cell- and immune cell-based vaccines, (3) peptide-based vaccines, and (4) nucleic acid-based vaccines [14]. Vaccines for OC have not been fully developed. A phase I, open-label study, UPCC-11807, reported a vaccine for use as anti-angiogenic therapy and a dendritic cell (DC)-based vaccine to cure recurrent OC [15]. The included patients showed good tolerance, and 2 of them developed antitumour immunity. Sarivalasis et al. performed a phase I/II study with a MUC-1 protein-targeted DC vaccine and found that patients who obtained a complete response from second-line antitumour treatment benefited from the subsequent vaccine [16]. In addition to DC-based vaccination, several proteins or polypeptides have been utilized to produce vaccines, such as HER-2/neu, WT1, and p53 [17-19]. Essentially, the abovementioned vaccines activate CD8+ T cells to engage in cellular immunity by triggering specific targets.
Compared with other vaccines, mRNA vaccines have several significant advantages. First, DC-based and polypeptide vaccines are limited to specific antigens because of HLA restriction. In comparison, many more antigens can be encoded and modified and targeted by mRNA vaccines, unlike other traditional peptide vaccine that requires genetic analysis of cancer, which represent a feasible and precise treatment for patients. Second, mRNA is translated into protein in the cytoplasm. That mRNA does not enter the nucleus means that it avoids the molecular mistakes that can be caused by gene insertion and integration and significantly improves drug safety. Third, the production of mRNA vaccines is feasible and lacks the processes of protein and virus vector production. Finally, other types of vaccines have showed only moderate efficacy to date. Self-adjuvant propertie of mRNA vaccine increase its in vivo immunogenicity and induce a strong and persistent immune response. Due to its safety, low production cost and eminent specificity, mRNA is expected to become a new tool for precision treatment. mRNAs encoding specific biomarkers can be transported into antigen-presenting cells (APCs) by lipid nanoparticles and are presented on the surface of APCs via major histocompatibility complexes (MHCs). Subsequently, an antitumour response is evoked [20]. mRNA vaccine technology has made progress in several cancers. Two ongoing clinical trials, NCT03908671 and NCT02688686, have employed survivin-encoding mRNAs to treat non-small-cell lung cancer [21]. Another phase I/II clinical trial identified satisfactory immune activation in prostate cancer, although a subsequent study demonstrated that the CV9103 vaccine did not benefit the OS of patients [22].
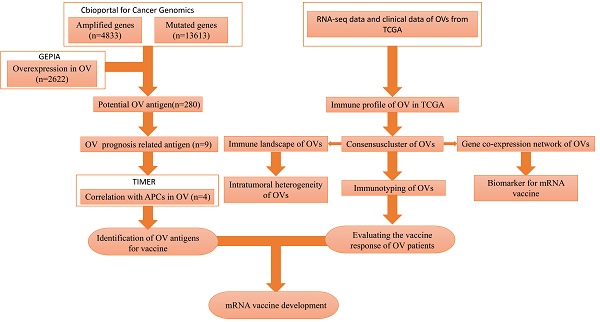
There is no published research on mRNA vaccines for HGSC. Our study attempted to identify optimal tumour antigens to produce an mRNA vaccine. In addition, we attempted to identify a population that was likely to benefit from such an mRNA vaccine. We classified patients into 3 immune subtypes to determine which patients would be likely to experience “cold” to “hot” tumour conversion with mRNA vaccination. The flowchart of this study is shown in Figure 1.
Method and Material
Mutation signature analysis
We explored, analysed, and visualized gene amplifications, mutation counts, and copy number variations with data from 617 HGSC patients through the cBioPortal (http://www.cbioportal.org/) platform [23]. In addition, the somatic mutation data of 436 HGSC cases from The Cancer Genome Atlas (TCGA) were downloaded via Xena Functional Genomics Explorer (https://xenabrowser.net/datapages/). The Maftools R package in R software (version 3.6.3) was used to analyse and visualize the somatic mutation signature [24].
The flowchart of our study.
Identification of potential immune biomarkers of OC
Gene Expression Profiling Interactive Analysis 2(GEPIA2) (http://gepia2.cancer-pku.cn/) is a web-based tool based on data from TCGA and Genotype-Tissue Expression (GTEx). We utilized GEPIA2 to explore differentially expressed genes between OC and normal tissue (|Log2FC| > 1, p < 0.05), as well as the relationship between the expression of potential antigen genes and overall survival (OS) in OC patients. The median expression value of each differentially expressed gene was employed as the cut-off value to identify relationships between expression and patient prognosis. Kaplan‒Meier (KM) curves and the log-rank test were used for visualization. Differentially expressed genes related to prognosis (p < 0.05) were selected as potential antigens. Furthermore, we employed Tumor Immune Estimation Resource (TIMER) (https://cistrome.shinyapps.io/timer/), a web-based tool for exploring immune cell levels in the cancer microenvironment, to investigate the relationship between antigen expression and the level of infiltrating antigen-presenting cells (APCs) in OC [25]. The correlation between infiltrating APC levels and tumour purity was determined by Spearman correlation analysis.
HGSC data download and processing
RNA-seq data and the corresponding clinicopathological information of 148 HGSC patients were obtained from the Xena database (https://xenabrowser.net/datapages/). The data cutoff for download was on March 17th, 2022. Patients with available RNA-seq data, clear survival status, and survival time were included in the study. Samples with unclear clinical information were excluded. The clinical characteristics of 148 patients were showed in Table 1. Standardized RNA-seq data were used to identify immune subtypes of OC. A total of 2422 immune-related genes (IRGs) were extracted from previous research and databases for further analysis [26, 27].
Clinic parameters of patients from the 2 cohorts
| TCGA Cohort (%) | ICGC Cohort (%) | |
|---|---|---|
| Total patients | 148 | 81 |
| alive | 59 (39.9) | 15 (18.5) |
| dead | 89 (60.1) | 66 (81.5) |
| Age | ||
| < 60y | 78 (52.7) | 43 (53.1) |
| > = 60y | 70 (47.3) | 38 (46.9) |
| Lateral | ||
| Unilateral | 57 (38.5) | - |
| Bilateral | 91 (61.5) | - |
| FIGO stage | ||
| Stage II | 7 (4.7) | 0 |
| Stage III | 113 (76.4) | 69 (85.2) |
| Stage IV | 28 (18.9) | 12 (8.1) |
Immune subtype identification
After preprocessing of the RNA-seq data, 148 HGSC samples were stratified into different immune subtypes based on the expression of 2303 IRGs. Consensus clustering was achieved by the “ConsensusClusterPlus” package in R [28]. The optimal clustering number was determined by calculating the consensus cumulative distribution function (CDF). K-means of Euclidean distance were utilized in the algorithm, with 50 subsamples and 80% of the total sample proportion used for each resampling. The cluster number varied from 2 to 9. Moreover, the KM method with the log-rank test was employed to compare the OS of immune subtypes. The relationships between immune subtype and clinicopathological factors were explored by the chi-square test. We downloaded a cohort with 81 HGSC patients from ICGC (International Cancer Genome Consortium) database (https://dcc.icgc.org/) to validate the consensus clustering algorithm. The data cutoff for download was on April 6th, 2022. The clinical characteristics of 81 patients were showed in Table 1.
Immune characteristics of the subtypes
Sixty-eight immune signatures, LM22 signatures, and 28 immune cell distributions for HGSC samples were identified to explore the differences in immune characteristics between the subtypes [29-31]. Additionally, we analysed the correlations between immune subtypes and the expression of immune checkpoint (ICP)- and immune cell death (ICD)-related genes extracted from previous research [31, 32].
Immune landscape definition
The microenvironment of HGSC changes dynamically. Therefore, we performed dimension reduction analysis to further explore the microenvironment of HGSC by employing the “monocle” package in R with a Gaussian distribution [33, 34]. This is a graph learning-based method to determine the internal structure and visualize the distribution of individual samples. Furthermore, a functional plot cell trajectory was generated to reveal and visualize the immune landscape. In the plot, different colours refer to different immune subtypes.
Gene modules associated with immune subtype
The “WGCNA” package in R was used to select the coexpression modules of the immune subtypes [35]. A value between 0 and 1 was used as the soft threshold of the adjacency matrix when constructing a biological scale-free network. Hierarchical clustering was performed based on the dissimilarity of genes in different modules (> 30 genes per module). A module that was correlated with an immune subtype was selected, and the biological functions of the genes in that module were analysed via Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional analyses.
Statistical Analysis
We used the Wilcox test to compare differences between the two groups, while the Kruskal Wallis test to compare three or more groups. The Wilcox test was us to compare data between the two groups, while the Kruskal Wallis test was used to compare three or more groups. Kaplan-Meier curves were used for OS analysis with the best cut-off value from the “survminer” package in R. P < 0.05 was considered statistically significant.
Results
Potential immune antigens for OC
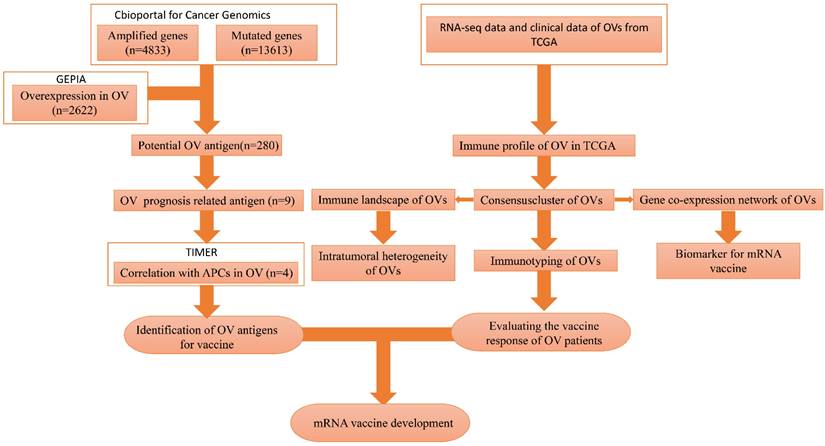
The flowchart of this study is shown in Figure 1. A total of 7638 differentially expressed genes between OC tissue and normal tissue, including 2622 overexpressed genes and 5016 underexpressed genes, were identified by GEPIA2 (Figure 2A). Genes with aberrant copy number were revealed by cBioPortal (Figure 2B), including amplified and deleted genes. The relationship between genome mutation count and altered genome fraction is shown in Figure 2C. The 10 genes with the highest alteration frequencies were SSPN, C2D5, CMAS, ETKN1, GOLT1B, KCNJ8, GYS2, SPX, LDHB, and IAPP (Figure 2D). We also explored the overall mutational landscape of TCGA-OC, and the 10 genes with the highest mutation frequency were TP53, TTN, MUC16, DST, FLG, HMCN1, SYNE1, CSMD3, MACF1, and FAT3 (Figure 2E).
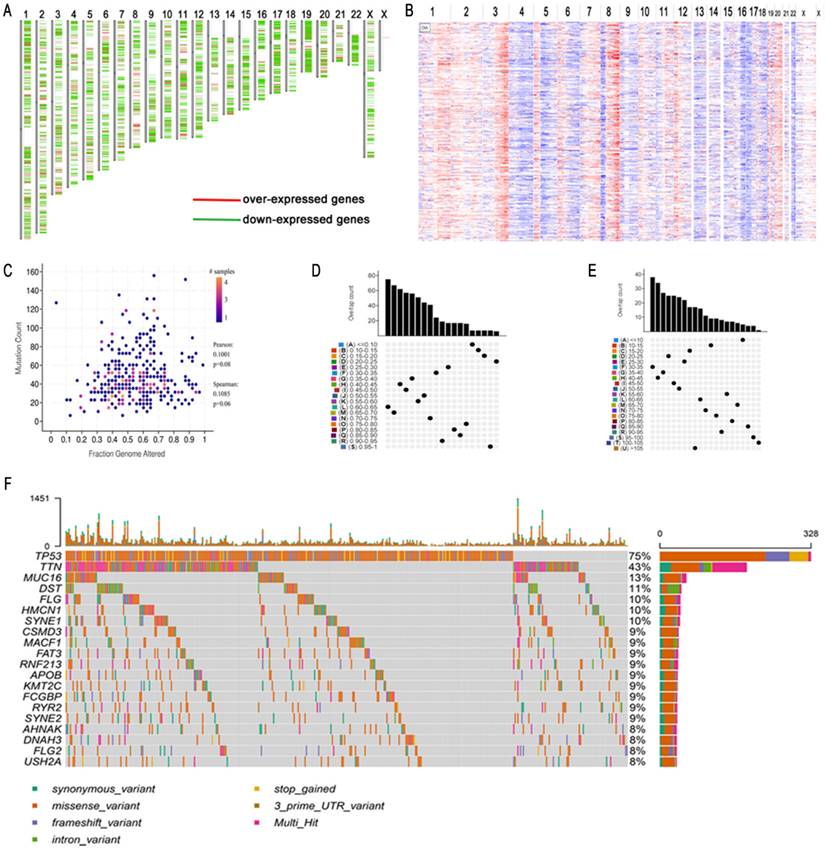
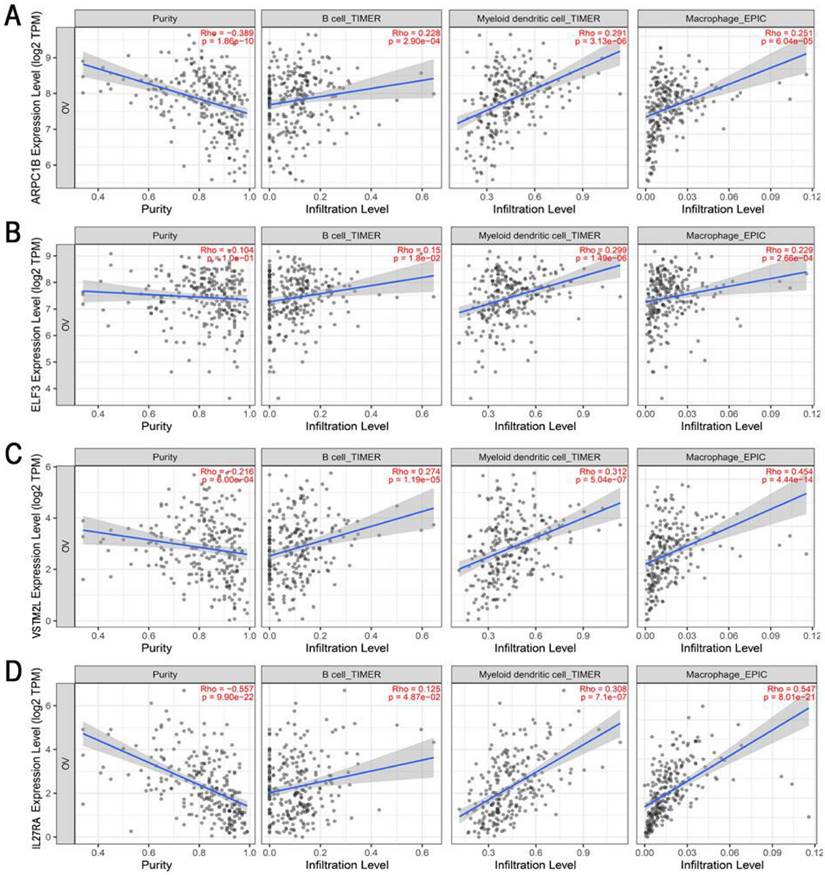
To obtain potential tumour antigens for the mRNA vaccine, we overlapped the amplified genes and the 2622 overexpressed differentially expressed genes of OC, and ultimately, we obtained 280 overexpressed, mutated genes (Figure 3A). Furthermore, we identified 9 genes related to OS (Figure 3B-3F), 4 of which (ARPC1B, ELF3, VSTM2L, and IL27RA) were positively correlated with the abundance of several immune cells (Figure 4). Specifically, overexpression of these 4 genes was related to worse OS prognosis and higher abundances of B cells, dendritic cells (DCs), and macrophages. These 4 biomarkers likely play important roles in immune activation because they are easily recognized by APCs. Thus, ARPC1B, ELF3, VSTM2L, and IL27RA were identified as potential tumour antigens for mRNA vaccine production.
Mutation landscape of high-grade serous ovarian cancer (HGSC). (A) Over- and down-expressed gens of HGSC. Red plot: over-expressed genes; Green plot: down-expressed genes. (B) The chromosomal distribution of the aberrant copy number genes in HGSCs. Red plot: amplified genes; Blue plot: deleted genes. (C) Scatter plot of samples in altered genome fraction and mutation count groups. (D) Overlapping samples in altered genome fraction group. (E) Overlapping samples in mutation count group. (F) Potential tumor biomarker-related genes with high mutation in HGSC.
Immune subtypes of HGSC and validation
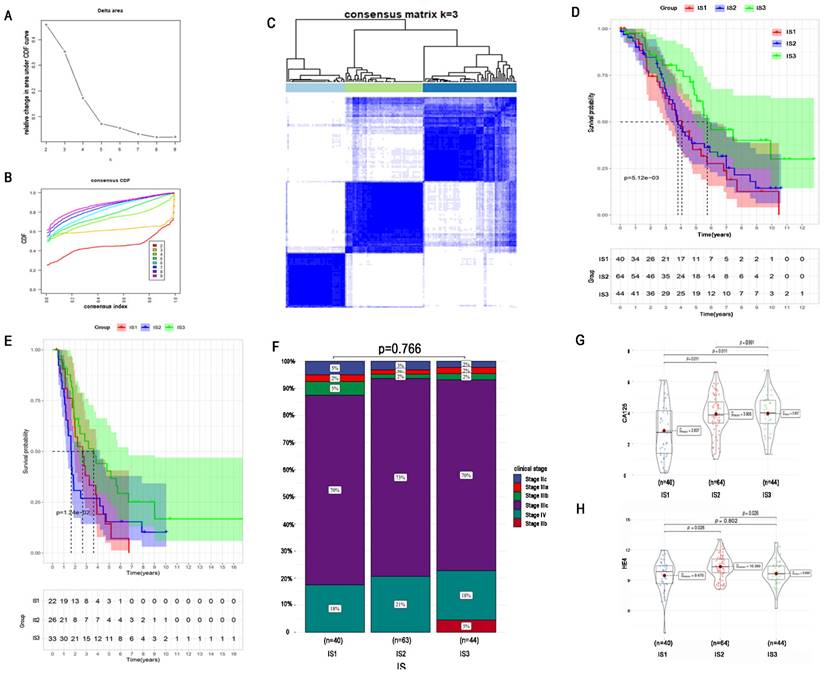
To identify an optimal population to receive the immune vaccine, we performed immunotyping with 148 TCGA HGSC patients. According to 2302 differentially expressed IRGs in HGSC, we performed consensus clustering of the 148 HGSC samples. We chose k = 3 to classify the cohort because of the limited sample number and obtained 3 immune subtypes (Figure 5A-C). There were 40, 64, and 44 patients in immune subtype 1 (IS1), IS2, and IS3, respectively. Moreover, Figure 5D shows the OS of the 3 immune subtypes. Same algorithm of consensus clustering in TCGA cohort was also performed to cluster 81 HGSC samples from ICGC, and the number of patients in IS1, IS1, and IS3 were 22, 26, and 33, respectively. OS of ICGC cohort was showed in Figure 5E. KM curves revealed that IS3 had the best survival both in TCGA and ICGC cohort. The prognosis of IS2 ranked second, and patients in IS1 had the worst prognosis.
Regarding the distribution of patient characteristics, we found no difference in disease stage between the immune subtypes (Figure 5F). Compared with patients in IS1, patients in IS2 and IS3 had higher expression of the tumour biomarker CA125 (Figure 5G). Regarding the tumour biomarker HE4, patients in IS2 had the highest HE4 expression, and there was no significant difference in HE4 expression between IS1 and IS3 (Figure 5H).
Identification of tumor antigens associated with HGSC prognosis. (A) Potential tumor antigens (total 280) with overexpression, mutation, and amplification in HGSC, and significant association with OS and immune infiltration (total 4 candidates). (B) Differential expression of 4 candidates in normal ovarian tissue and HGSCs. *, significant difference. Kaplan-Meier curves showing OS of HGSC patients stratified on the basis of (C) ARPC1B, (D). ELF3, (E). VSTM2L, and (F) IL27RA expression levels. A 50% (Median) cutoff was set up for dividing low- and high-expression groups. Log-rank test was used for hypothesis testing, and a p-value <0.05 was considered statistically significant.
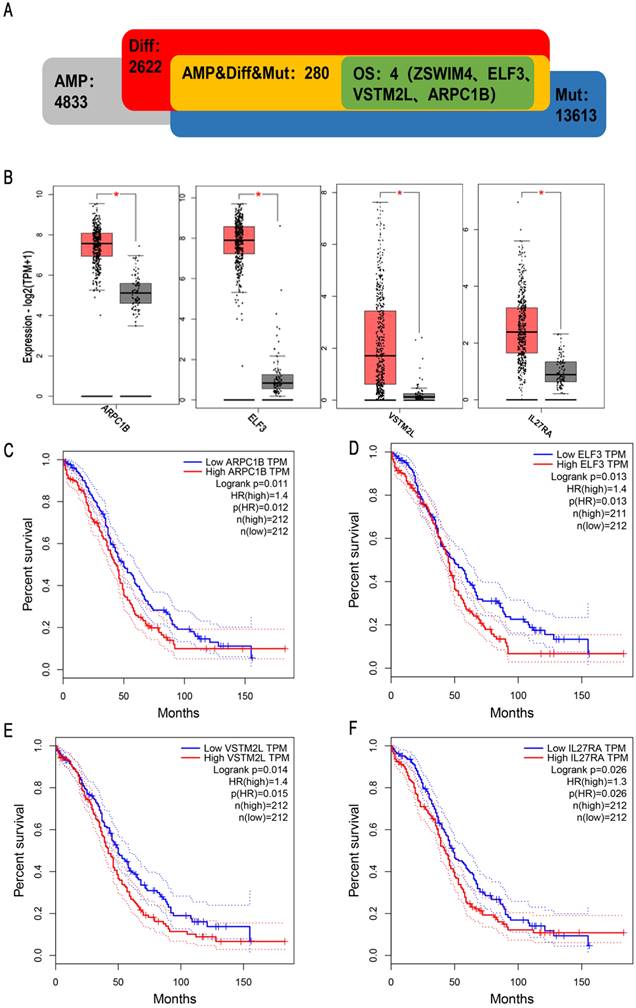
Identification of tumor antigens associated with enrichment of antigen-presenting cells (APCs). Correlation between the expression levels of (A) ARPC1B, (B) ELF3, (C) VSTM2L, and (D) IL27RA and infiltration of APCs (B cells, dendritic cells, and macrophages) in HGSC.
The immune cell levels and immune landscape of the immune subtypes
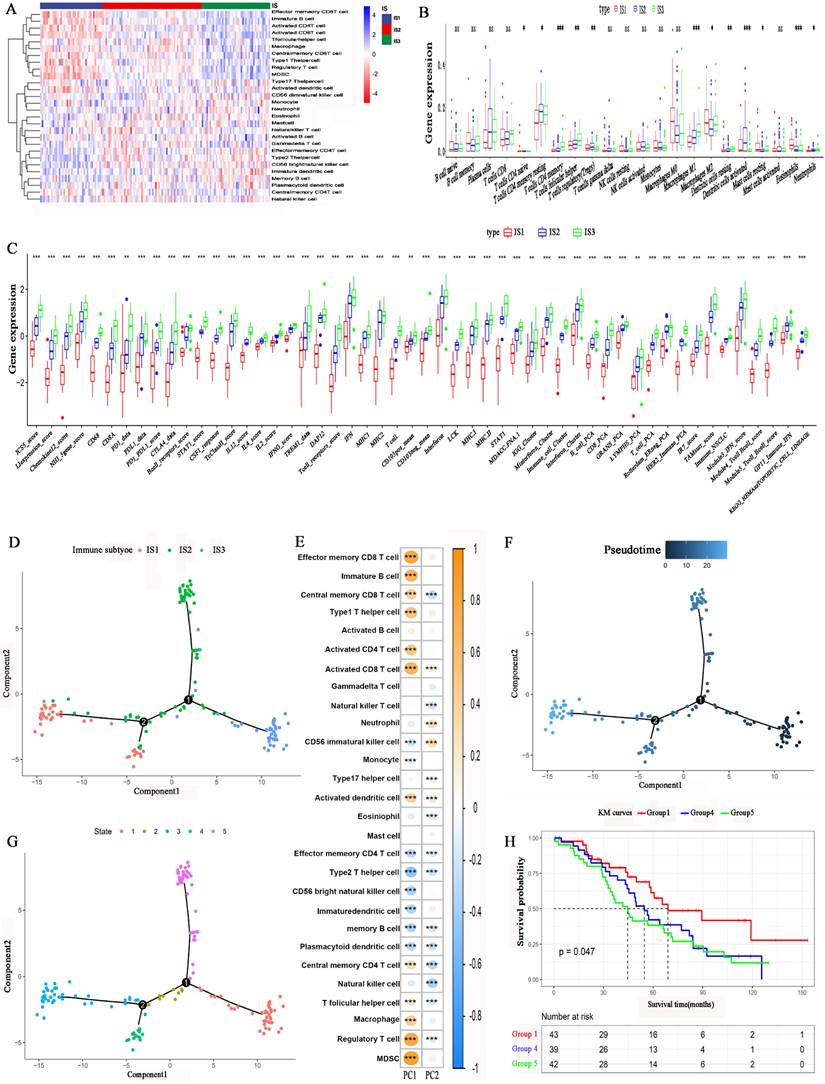
We employed the single-sample gene set enrichment analysis (ssGSEA) method to calculate the 28 immune signatures. Figure 6A, shows the distribution of 28 immune cell levels in the immune subtypes. The abundances of effector memory CD8 T cells, immature B cells, activated CD4 T cells, activated CD8 T cells, type 1 T helper cells, regulatory T cells (Tregs), and myeloid-derived suppressor cells (MDSCs) were higher in IS1. For IS3, the abundances of effector memory CD4 T cells, type 2 T helper cells, CD56 bright natural killer cells, neutrophils, immature DCs, and memory B cells were higher. Moreover, the abundances of T follicular helper cells, natural killer T cells, gamma delta T cells, and natural killer cells were higher in IS2. CIBERSORT analysis revealed that the abundances of immunosuppressive regulatory cells, such as Tregs, M2 macrophages and DCs, were higher in IS1 and IS2 (Figure 6B). In contrast, the abundances of immune-activating cells, such as M1 macrophages, were higher in IS3. Furthermore, the expression of immune-related inflammatory factors was higher in IS3 than in IS1 and IS2 (Figure 6C). In conclusion, IS3 was an immunologically “hot” subtype, while IS1 was an immunologically “cold” subtype; IS2 was an intermediate subtype.
The immune landscape of HGSC was explored according to the IRG expression profile. A graph learning-based dimensionality reduction method was utilized. The immune component distribution of individual patients is shown in Figure 6D. Patients with different immune subtypes were distributed in the respective branches of the tree. The horizontal ordinate (principal component 1) was positively associated with the levels of memory CD8 T cells, activated CD8 T cells, type 1 T helper cells, and macrophages, while the vertical ordinate (principal component 2) was negatively associated with the levels of natural killer T cells, memory B cells and memory T cells (Figure 6E). The IS3 cases were scattered throughout the area with a high horizontal ordinate score and a low horizontal ordinate score, suggesting a higher abundance of immune cells in IS3. Figure 6F shows the distribution of HGSC patients on the DDRTree. Furthermore, the distribution of outlying HGSC patients is also shown in Figure 6G. We can see that the prognosis of patients in IS3 was significantly better than that of patients in IS1 or IS2 (Figure 6H). The abovementioned results imply that the level of immune cell infiltration in IS3 was high. The immune landscape can reflect immune infiltration and prognosis, providing an important reference for the selection of patient populations that are likely to experience a “cold” tumour transitioning into a “hot” tumour with mRNA vaccination.
Identification of potential immune subtypes of HGSC. A. Cumulative distribution function curve and (B) delta area of immune-related genes in TCGA cohort. (C) Sample clustering heat map. (D) Kaplan-Meier curves showing OS of HGSC immune subtypes in TCGA cohort. (E) Kaplan-Meier curves showing OS of HGSC immune subtypes in ICGC cohort. (F)Distribution of four different clinical stages across IS1-IS3 in the cohort. (G) CA-125 expression of IS1-IS3. (H) HE4 expression of IS1-IS3.
Cellular characteristics and immune landscape of immune subtypes. (A) Differential enrichment scores of 28 immune cell signatures among HGSC immune subtypes. (B) Differential enrichment scores of CIBERSORT 22 immune cell signatures. (C) Differential enrichment scores of 56 immune signatures among HGSC immune subtypes. (D) Immune landscape of HGSCs where each point represents a patient, and the immune subtypes are color-coded. The horizontal axis represents the first principal component, and the vertical axis represents the second principal component. (E) The relationship of two principal components and 28 immune cell signatures. (F) Immune landscape of samples from two extreme locations. (G) Immune landscape of the subsets of HGSC immune subtypes. (H) Different subsets in three extreme locations associated with different prognoses. *p < 0.05, **p < 0.01, ***p < 0.001.
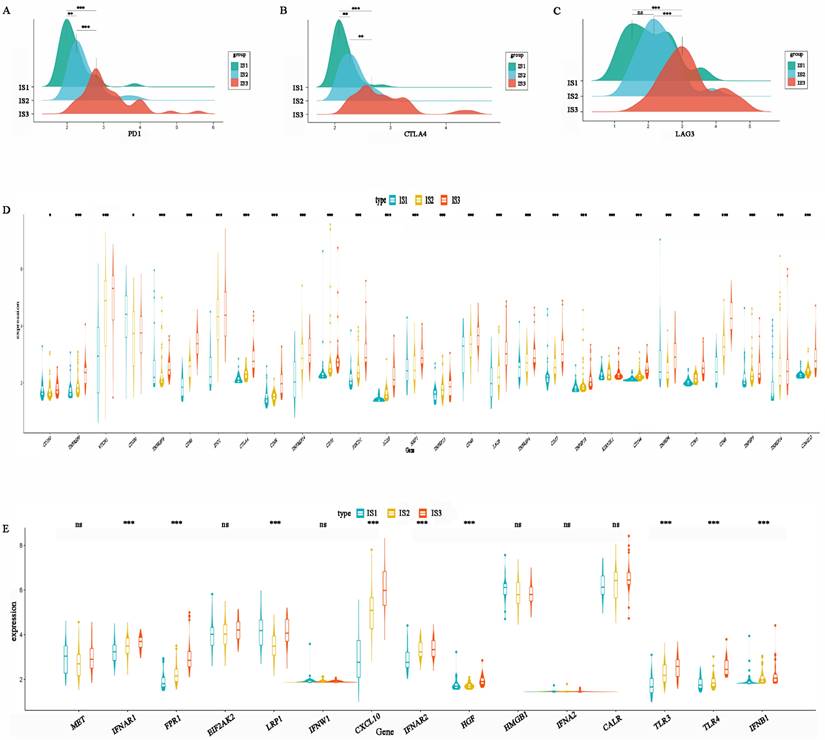
Association between immune subtypes and immune molecular, ICPs and ICD modulators. (A) Differential expression of PD-1 among the HGSC immune subtypes. (B) Differential expression of CTLA-4 among the HGSC immune subtypes. (C) Differential expression of LAG3 among the HGSC immune subtypes. (D) Differential expression of ICP genes among the HGSC immune subtypes. (E) Differential expression of ICD genes among the HGSC immune subtypes. *p < 0.05, **p < 0.01, ***p < 0.001.
Immune characteristics of the immune subtypes
ICP and ICD-related genes are important indicators for evaluating the immune response. The levels of PD1, CTLA-4, and LAG3 in IS3 were higher than those in IS2, while IS1 had the lowest expression levels (Figure 7A-C). This result suggests that patients with the IS3 subtype might be more likely to benefit from PD1, CTLA4, and LAG3 inhibitors. In addition, the expression of other ICPs, including TNFRSF9, CD86, IDO1, CD28, TNFRSF14, CD70, PDCD1, ICOS, TNFRSF15, CD40, LAG3, TNFRSF4, CD27, KIR3DL1, CD244, CD80, CD48, and CD4OLG, was significantly higher in IS3 than IS2 or IS1 (Figure 7D). In addition, ICD-related genes, including FPR1, CXCL10, IFNAR2, TLR3, TLR4, and IFNB1, were upregulated in IS3 (Figure 7E). In summary, IS3 was found to be a subtype with immune “hot” characteristics but an immunosuppressive status that might benefit from ICIs.
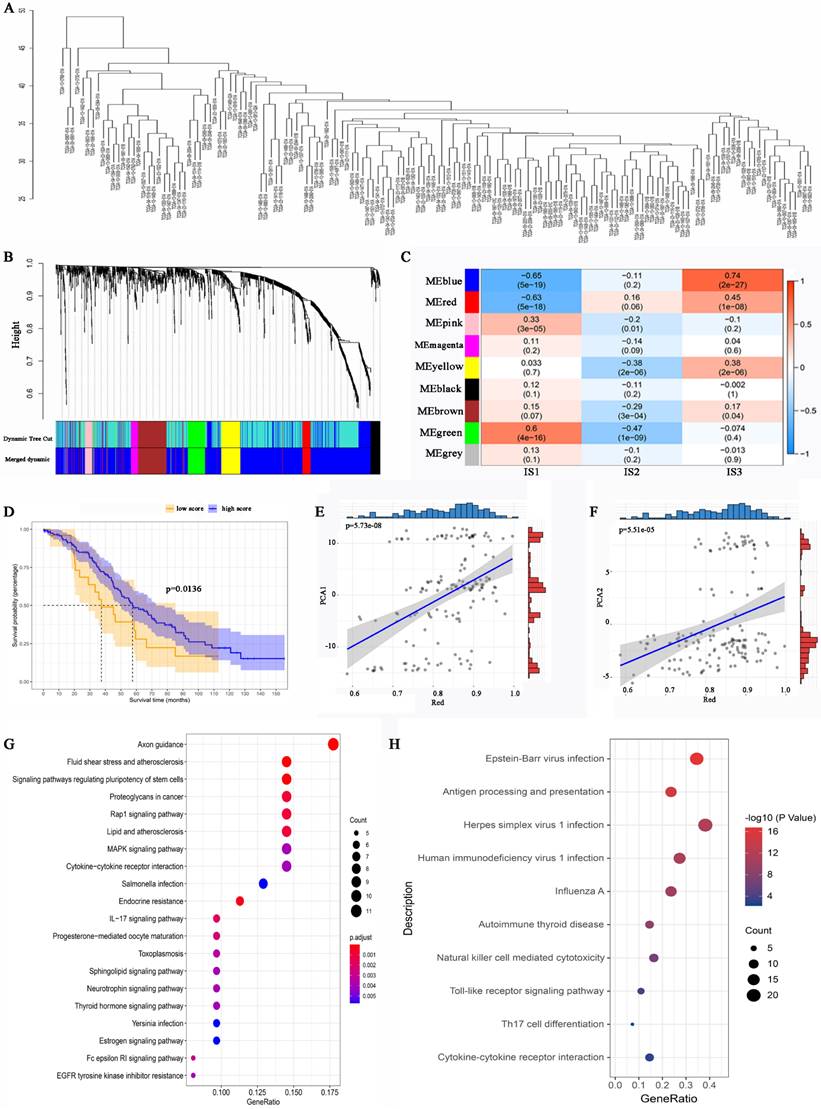
WGCNA identification of functional immune gene modules and hub genes in HGSC
To identify IRGs in HGSC, WGCNA was performed, and a dendrogram was employed to visualize the results (Figure 8A). The soft threshold power was defined as “3”. Different dendrogram branch colours represent different gene clusters (Figure 8B). Ultimately, 8 functional immune gene modules (all modules except for the grey module) were clustered and visualized (Figure 8C). The genes of the pink and green modules were positively associated with IS1, while the genes of the blue and red modules were negatively associated with IS1. Moreover, the genes of the red, blue, yellow, and brown modules were positively associated with IS3. The KM analysis identified that the expression of genes in the red module was related to the OS of HGSC patients (Figure 8D). Notably, the genes of the red module were positively associated with component 1 and component 2 (Figure 8E, 8F). GO analysis revealed that the hub genes in the red module were associated with proteoglycans in cancer, cytokine‒cytokine receptor interactions, and the IL-17 signalling pathway (Figure 8G). KEGG analysis suggested that the genes of the red module were related to antigen processing and presentation, natural killer cell-mediated cytotoxicity, Th17-cell differentiation, and the IL-17 signalling pathway (Figure 8H). The expression level of the hub genes in the red module may be a biomarker that can be used to identify HGSC patients who are likely to benefit from mRNA vaccines.
Discussion
OC is an aggressive gynaecologic malignancy and has the highest mortality among such malignancies. Currently, early-stage OC is treated with a combination of surgery and chemotherapy. HGSC is the most common pathology of OC. Platinum-based chemotherapy regimens and PARP inhibitors are important treatments for HGSC. However, the invasiveness and mortality of HGSC are still high. As a research focus, immune therapy has been identified to be effective in a variety of solid tumours, but not in HGSC. The identification of novel and effective treatments for HGSC is necessary. An important molecular feature of HGSC is the enrichment of tumour-infiltrating lymphocytes (TILs), implying that immune therapy may be useful for treating HGSC. Tumour vaccines, such as mRNA vaccines, as a new type antitumour therapy, have been found to be useful in several cancers. Specific antigens expressed on the surface of the tumour can be targets of such vaccines. Recently, several studies have found mRNA vaccine targets to cure pancreatic cancer, cholangiocellular carcinoma, and prostate cancer [27, 32]. However, related HGSC studies are lacking. Our study is the first to explore potential mRNA vaccine targets for the immune treatment of HGSC, offering important references for identifying novel treatments for HGSC in the future.
Our research demonstrated that ARPC1B, ELF3, VSTM2L, and IL27RA are potential targets for mRNA vaccine production. High expression of these genes in HGSC is related to worse prognosis and higher abundances of APCs, including B cells, macrophages, and DCs. Previous studies on tumor antigens have shown that mutated genes are more easily recognized by immune cells [36]. High expression of the protein in the tumor is associated with poor prognosis for the patient, suggesting that it may be related to tumor development and growth. Meanwhile, the expression level of these genes in ovarian cancer is positively correlated with the degree of antigen presentation and immune cell infiltration, indicating that it is more easily recognized by the immune system and can induce a strong immune response to kill tumor cells [27, 37]. Based on the above views, we believe that an appropriate mRNA should have these characteristics: mutation, immune infiltration correlation, and prognostic correlation. ARPC1B is an actin-related protein that binds and activates Aurora A to regulate centrosome integrity [38]. ARPC1B is necessary for actin reorganization and lamellipodia formation, and its mutation induces cytotoxic T-cell dysfunction [39]. Importantly, a previous study showed that ARPC1B expression was positively correlated with tumour burden in OC [40]. ELF3, another identified potential antigen, is a transcription factor for IGF1. ELF3 has been identified as carcinogenic in several solid tumours, including lung adenocarcinoma, thyroid cancer, and colorectal cancer [41-43]. ELF3 is highly expressed in epithelial OC during hypoxia, leading to the increased secretion of IGF1 and VEGF and ultimately promoting endothelial cell proliferation, migration, and angiogenesis in epithelial OC [44]. Moreover, ELF3 is an immune gene. Hao Xu et al. found that ELF3 expression was associated with the infiltration of various immune cells, including cytotoxic cells, CD8 T cells, and macrophages [45]. ELF3 may be an optimal antigen for vaccine production. IL27RA is a heterodimeric receptor complex of IL27. It plays an important role in inflammation induction and has anti-inflammatory and immunomodulatory effects [46]. IL-27 activates Janus kinase (JAK), signal transducer and activator of transcription 1 (STAT1), and STAT3, regulating the differentiation of T helper (Th) cells through the activity of related downstream transcription factors [47-49]. IL27RA is positively correlated with the infiltration of CD8 T cells, DCs and neutrophils in rectal cancer [50]. VSTM2L is a secreted protein that antagonizes human neuroprotective peptides [51].
Identification of immune gene co-expression modules of HGSC. (A) Sample clustering. (B) Dendrogram of all differentially expressed genes clustered based on a dissimilarity measure (1-TOM). (C) Relationship between gene modules and immune subtypes. (D) Differential prognosis in res module with high and low mean. Correlation between red module feature vector and first (E) or second (F) principal component in immune landscape. (G) GO analysis in the red module. (H) KEGG analysis in the red module.
It has been found to be associated with digestive carcinomas, such as gastric cancer and rectal carcinoma. VSTM2L induces resistance to radiotherapy and chemotherapy through the IL-4 pathway [52]. Shuyi Zhang et al. found that VSTM2L plays a role in the tumour immune microenvironment (TIME) and may be a potential immune target [53]. VSTM2L is related to poor prognosis in OC because it is related to tumour initiation and migration [54]. In summary, high expression of the abovementioned genes has been identified to contribute to a poor prognosis, and all the genes have been found to affect the TIME of cancer. Our study demonstrated that these genes induce aggregation of APCs, suggesting that they can be used as specific antigens to produce mRNA vaccines. The underlying molecular mechanisms should be explored in the future.
mRNA vaccines can be effective in select populations. We divided 148 HGSC patients into 3 immune subtypes based on the expression of immune genes. IS3 had the best survival, followed by IS2, and the prognosis of IS1 was the worst. This immune subtype system can be utilized to evaluate the prognosis of patients. We also analysed the immune landscape, immune cell abundances, and immune molecule expression in the different immune subtypes to distinguish “cold” and “hot” tumours. IS1 was found to be a “cold” subtype that will likely benefit from mRNA vaccine treatment because it will improve immune cell infiltration. IS3 tumours are “hot” tumours in a state of immunosuppression. Patients in IS3 are likely to obtain survival benefits from ICP blockade. Previous studies have demonstrated that only a small group of patients obtain survival advantages from immune treatment. Therefore, it is challenging but necessary to find a way to identify this population [54]. Moreover, understanding the unique, complex immune microenvironment of each subtype is necessary for guiding individualized treatment. Finally, we performed WGCNA to identify the gene modules of the 3 subtypes. Low expression of the genes in the red module was related to low abundance of immune cells and poor prognosis. Patients with this molecular signature are likely to benefit from mRNA vaccination.
Given tumour heterogeneity, it is important to identify the immune landscape of individual tumours before administering immunotherapies, including mRNA vaccines. The TIME is an important element related to the immune response. We identified 3 immune subtypes of HGSC. Lymphocytes and APCs, such as type M1 macrophages, were enriched in IS3, and there were higher levels of specific cellular factors and HLA molecules, implying an active immune environment. Furthermore, ICP and ICD-related gene expression was high in IS3, implying that this subtype will respond well to ICP blockade. In contrast, IS1 and IS2 tumours were “cold” tumours, especially IS1 tumours. IS1 tumours showed characteristic low abundances of TILs, cytotoxic T cells, natural killer cells, and macrophages. Moreover, IS1 showed low expression levels of immune factors, including IL-2, IL-6, IFN, MHCI, and MHCII. Generally, APCs infiltrate tumours under the guidance of chemokines, engulf tumour cells, and present antigens to T cells. Subsequently, the T cells are induced to differentiate to kill tumour cells [55]. As “cold” tumours, IS1 tumours lacks mature APCs and thus cannot induce a further inflammatory response. An effective vaccine could stimulate IS1 tumours to produce more APCs, thereby inducing an immune environment conducive to tumour killing. Patients of IS1 are likely to benefit from receiving mRNA vaccines.
Although our study found potential targets to produce mRNA vaccines and identified populations that are likely to benefit from such vaccines, there were several limitations. First, our study focused on patients from the HGSC cohort of TCGA, and validation of the findings with data from patients from other countries is needed. Second, assessment of the mRNA sequences to determine mRNA stability, immunogenicity, and ability to elicit an immune response is needed [56], but our study did not provide the mRNA sequence of the potential vaccine targets or the most effective gene fragments, which should be explored in the future.
Conclusion
Our study found that ARPC1B, ELF3, VSTM2L, and IL27RA are potential targets for HGSC mRNA vaccines. We also identified 3 immune subtypes with different prognoses by analysing the immune environment of HGSC patients. Patients of IS3 could benefit from ICP blockade treatment, while patients of IS1 or IS2 are likely to benefit from receiving mRNA vaccines because of their low levels of infiltrating immune cells and immune factors.
Abbreviations
HGSC: High-grade serous ovarian cancer; TCGA: The Cancer Genome Atlas; APCs: antigen-presenting cells; ICGC: International Cancer Genome Consortium; WGCNA: weighted gene coexpression network analysis; OC: Ovarian cancer; ICIs: immune checkpoint inhibitors; TILs: tumour-infiltrating lymphocytes; PFS: progression-free survival; OS: overall survival; HPV: human papilloma viru; DC: dendritic cell; MHCs: major histocompatibility complexes; GEPIA2: Gene Expression Profiling Interactive Analysis 2; GTEx: Genotype-Tissue Expression; KM: Kaplan‒Meier; TIMER: Tumor Immune Estimation Resource; IRGs: immune-related genes; CDF: cumulative distribution function; ICP: immune checkpoint; ICD: immune cell death; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; IS: immune subtype; ssGSEA: single-sample gene set enrichment analysis; Tregs: regulatory T cells; MDSCs: myeloid-derived suppressor cells; TILs: tumour-infiltrating lymphocytes; JAK: Janus kinase; STAT1: signal transducer and activator of transcription 1.
Acknowledgements
The authors would like to thank TCGA and ICGC for open access to the database.
Disclosure
Personal identifying information was not covered in the TCGA and ICGC database. It was not necessary to require Institutional Review Board approval or patient informed consent.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-49
2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33
3. Cree IA, White VA, Indave BI, Lokuhetty D. Revising the WHO classification: female genital tract tumours. Histopathology. 2020;76:151-6
4. Kim J, Park EY, Kim O, Schilder JM, Coffey DM, Cho CH. et al. Cell Origins of High-Grade Serous Ovarian Cancer. Cancers (Basel). 2018 10
5. Lisio MA, Fu L, Goyeneche A, Gao ZH, Telleria C. High-Grade Serous Ovarian Cancer: Basic Sciences, Clinical and Therapeutic Standpoints. Int J Mol Sci. 2019 20
6. Roett MA, Evans P. Ovarian cancer: an overview. Am Fam Physician. 2009;80:609-16
7. Tew WP, Lacchetti C, Ellis A, Maxian K, Banerjee S, Bookman M. et al. PARP Inhibitors in the Management of Ovarian Cancer: ASCO Guideline. J Clin Oncol. 2020;38:3468-93
8. Matulonis UA, Shapira-Frommer R, Santin AD, Lisyanskaya AS, Pignata S, Vergote I. et al. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: results from the phase II KEYNOTE-100 study. Ann Oncol. 2019;30:1080-7
9. Zamarin D, Burger RA, Sill MW, Powell DJ Jr, Lankes HA, Feldman MD. et al. Randomized Phase II Trial of Nivolumab Versus Nivolumab and Ipilimumab for Recurrent or Persistent Ovarian Cancer: An NRG Oncology Study. J Clin Oncol. 2020;38:1814-23
10. Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD. et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284-96
11. Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480-9
12. Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat Rev Genet. 2008;9:776-88
13. Bayó C, Jung G, Español-Rego M, Balaguer F, Benitez-Ribas D. Vaccines for Non-Viral Cancer Prevention. Int J Mol Sci. 2021 22
14. Faghfuri E, Pourfarzi F, Faghfouri AH, Abdoli Shadbad M, Hajiasgharzadeh K, Baradaran B. Recent developments of RNA-based vaccines in cancer immunotherapy. Expert Opin Biol Ther. 2021;21:201-18
15. Kandalaft LE, Powell DJ Jr, Chiang CL, Tanyi J, Kim S, Bosch M. et al. Autologous lysate-pulsed dendritic cell vaccination followed by adoptive transfer of vaccine-primed ex vivo co-stimulated T cells in recurrent ovarian cancer. Oncoimmunology. 2013;2:e22664
16. Sarivalasis A, Boudousquié C, Balint K, Stevenson BJ, Gannon PO, Iancu EM. et al. A Phase I/II trial comparing autologous dendritic cell vaccine pulsed either with personalized peptides (PEP-DC) or with tumor lysate (OC-DC) in patients with advanced high-grade ovarian serous carcinoma. J Transl Med. 2019;17:391
17. Ghiringhelli F, Menard C, Puig PE, Ladoire S, Roux S, Martin F. et al. Metronomic cyclophosphamide regimen selectively depletes CD4+CD25+ regulatory T cells and restores T and NK effector functions in end stage cancer patients. Cancer Immunol Immunother. 2007;56:641-8
18. Knutson KL, Schiffman K, Cheever MA, Disis ML. Immunization of cancer patients with a HER-2/neu, HLA-A2 peptide, p369-377, results in short-lived peptide-specific immunity. Clin Cancer Res. 2002;8:1014-8
19. Ohno S, Kyo S, Myojo S, Dohi S, Ishizaki J, Miyamoto K. et al. Wilms' tumor 1 (WT1) peptide immunotherapy for gynecological malignancy. Anticancer Res. 2009;29:4779-84
20. Miao L, Zhang Y, Huang L. mRNA vaccine for cancer immunotherapy. Mol Cancer. 2021;20:41
21. Wang Y, Tan H, Yu T, Chen X, Jing F, Shi H. Potential Immune Biomarker Candidates and Immune Subtypes of Lung Adenocarcinoma for Developing mRNA Vaccines. Front Immunol. 2021;12:755401
22. Kübler H, Scheel B, Gnad-Vogt U, Miller K, Schultze-Seemann W, Vom Dorp F. et al. Self-adjuvanted mRNA vaccination in advanced prostate cancer patients: a first-in-man phase I/IIa study. J Immunother Cancer. 2015;3:26
23. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-4
24. Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747-56
25. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS. et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:e108-e10
26. Bhattacharya S, Dunn P, Thomas CG, Smith B, Schaefer H, Chen J. et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data. 2018;5:180015
27. Huang X, Zhang G, Tang T, Liang T. Identification of tumor antigens and immune subtypes of pancreatic adenocarcinoma for mRNA vaccine development. Mol Cancer. 2021;20:44
28. Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572-3
29. Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D. et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017;18:248-62
30. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol. 2018;1711:243-59
31. Wolf DM, Lenburg ME, Yau C, Boudreau A, van 't Veer LJ. Gene co-expression modules as clinically relevant hallmarks of breast cancer diversity. PLoS One. 2014;9:e88309
32. Huang X, Tang T, Zhang G, Liang T. Identification of tumor antigens and immune subtypes of cholangiocarcinoma for mRNA vaccine development. Mol Cancer. 2021;20:50
33. Elking D, Darden T, Woods RJ. Gaussian induced dipole polarization model. J Comput Chem. 2007;28:1261-74
34. Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381-6
35. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559
36. Wagner S, Mullins CS, Linnebacher M. Colorectal cancer vaccines: Tumor-associated antigens vs neoantigens. World J Gastroenterol. 2018;24:5418-32
37. Lin H, Wang K, Xiong Y, Zhou L, Yang Y, Chen S. et al. Identification of Tumor Antigens and Immune Subtypes of Glioblastoma for mRNA Vaccine Development. Front Immunol. 2022;13:773264
38. Skinner M. Cell cycle: ARPC1B - a regulator of regulators. Nat Rev Mol Cell Biol. 2010;11:542
39. Randzavola LO, Strege K, Juzans M, Asano Y, Stinchcombe JC, Gawden-Bone CM. et al. Loss of ARPC1B impairs cytotoxic T lymphocyte maintenance and cytolytic activity. J Clin Invest. 2019;129:5600-14
40. Kaneda A, Kaminishi M, Nakanishi Y, Sugimura T, Ushijima T. Reduced expression of the insulin-induced protein 1 and p41 Arp2/3 complex genes in human gastric cancers. Int J Cancer. 2002;100:57-62
41. Chen H, Chen W, Zhang X, Hu L, Tang G, Kong J. et al. E26 transformation (ETS)-specific related transcription factor-3 (ELF3) orchestrates a positive feedback loop that constitutively activates the MAPK/Erk pathway to drive thyroid cancer. Oncol Rep. 2019;41:570-8
42. Enfield KSS, Marshall EA, Anderson C, Ng KW, Rahmati S, Xu Z. et al. Epithelial tumor suppressor ELF3 is a lineage-specific amplified oncogene in lung adenocarcinoma. Nat Commun. 2019;10:5438
43. Wang JL, Chen ZF, Chen HM, Wang MY, Kong X, Wang YC. et al. Elf3 drives β-catenin transactivation and associates with poor prognosis in colorectal cancer. Cell Death Dis. 2014;5:e1263
44. Seo SH, Hwang SY, Hwang S, Han S, Park H, Lee YS. et al. Hypoxia-induced ELF3 promotes tumor angiogenesis through IGF1/IGF1R. EMBO Rep. 2022;23:e52977
45. Xu H, Wang H, Li G, Jin X, Chen B. The Immune-Related Gene ELF3 is a Novel Biomarker for the Prognosis of Ovarian Cancer. Int J Gen Med. 2021;14:5537-48
46. Aparicio-Siegmund S, Garbers C. The biology of interleukin-27 reveals unique pro- and anti-inflammatory functions in immunity. Cytokine Growth Factor Rev. 2015;26:579-86
47. Diveu C, McGeachy MJ, Boniface K, Stumhofer JS, Sathe M, Joyce-Shaikh B. et al. IL-27 blocks RORc expression to inhibit lineage commitment of Th17 cells. J Immunol. 2009;182:5748-56
48. Owaki T, Asakawa M, Morishima N, Hata K, Fukai F, Matsui M. et al. A role for IL-27 in early regulation of Th1 differentiation. J Immunol. 2005;175:2191-200
49. Yoshimoto T, Yoshimoto T, Yasuda K, Mizuguchi J, Nakanishi K. IL-27 suppresses Th2 cell development and Th2 cytokines production from polarized Th2 cells: a novel therapeutic way for Th2-mediated allergic inflammation. J Immunol. 2007;179:4415-23
50. Cheng X, Wang Y, Liu L, Lv C, Liu C, Xu J. SLC7A11, a Potential Therapeutic Target Through Induced Ferroptosis in Colon Adenocarcinoma. Front Mol Biosci. 2022;9:889688
51. Rossini L, Hashimoto Y, Suzuki H, Kurita M, Gianfriddo M, Scali C. et al. VSTM2L is a novel secreted antagonist of the neuroprotective peptide Humanin. Faseb j. 2011;25:1983-2000
52. Liu H, Zhang Z, Zhen P, Zhou M. High Expression of VSTM2L Induced Resistance to Chemoradiotherapy in Rectal Cancer through Downstream IL-4 Signaling. J Immunol Res. 2021;2021:6657012
53. Zhang S, Xiong H, Yang J, Yuan X. Pan-Cancer Analysis Reveals the Multidimensional Expression and Prognostic and Immunologic Roles of VSTM2L in Cancer. Front Mol Biosci. 2021;8:792154
54. Li B, Cui Y, Nambiar DK, Sunwoo JB, Li R. The Immune Subtypes and Landscape of Squamous Cell Carcinoma. Clin Cancer Res. 2019;25:3528-37
55. Chen Z, Hambardzumyan D. Immune Microenvironment in Glioblastoma Subtypes. Front Immunol. 2018;9:1004
56. Wang Y, Zhang Z, Luo J, Han X, Wei Y, Wei X. mRNA vaccine: a potential therapeutic strategy. Mol Cancer. 2021;20:33
Author contact
![]() Corresponding author: Hong Lin, Department of Medical Oncology, Cancer Hospital of Shantou University Medical College, Shantou, Guangdong, China. No.7, Raoping Road, Shantou, Guangdong (China); E-Mail hlin13com. Hongbiao Wang; Department of Medical Oncology, Cancer Hospital of Shantou University Medical College, Shantou, Guangdong, China; No.7, Raoping Road, Shantou, Guangdong (China); E-Mail wanghongbiao123com.
Corresponding author: Hong Lin, Department of Medical Oncology, Cancer Hospital of Shantou University Medical College, Shantou, Guangdong, China. No.7, Raoping Road, Shantou, Guangdong (China); E-Mail hlin13com. Hongbiao Wang; Department of Medical Oncology, Cancer Hospital of Shantou University Medical College, Shantou, Guangdong, China; No.7, Raoping Road, Shantou, Guangdong (China); E-Mail wanghongbiao123com.

 Global reach, higher impact
Global reach, higher impact