Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(8):1362-1370. doi:10.7150/jca.83897 This issue Cite
Research Paper
MiR-363 restrain the proliferation, migration and invasion of colorectal carcinoma cell by targeting E2F3
Clinical College of Chinese Medicine, Hubei University of Chinese Medicine, Wuhan 430061, China.
#Wenyu Yang and Xiqian Yang contribute equally to this paper.
Received 2023-2-27; Accepted 2023-4-10; Published 2023-5-15
Abstract
MicroRNA (miRNA) is associated with tumor cell proliferation, migration and invasion. Studies have shown that miRNAs are closely related to the occurrence and development of colorectal cancer (CRC), but the mechanisms deserve further investigation. In this study, we aim to explore the role of miR-363 on CRC tumorigenesis. Using CRC cell lines, we tested the expression of miR-363 by using RT-PCR, and miR-363 effect on cell behavior was test by using CCK-8 assay, wound-healing assay and cell invasion assay, and western blotting. Luciferase reporter assay and western blot confirmed that E2F3 was the target gene for miR-363. We further examined the effect of E2F3 on the regulation of miR-363 on cell behavior through knockdown of E2F3. Western blot and RT-PCR assay showed that miR-363 inhibited the expression of E2F3 in HCT-116 and SW480 cell. MiR-363 overexpression or E2F3 knockdown inhibited cell proliferation, migration and invasion of CRC. This study demonstrated that miR-363 is able to suppress cell proliferation, migration and invasion by negative regulating E2F3 in CRC cells, and inhibits tumor growth in vivo.
Keywords: miR-363, E2F3, colorectal cancer, migration, invasion
Introduction
Colorectal cancer (CRC) is one of the most common gastrointestinal malignancies clinically. According to the study released by WHO's International Agency for Research on Cancer in 2020, CRC has become the third most common cancer after breast cancer and lung cancer. In China, the incidence and mortality of CRC are on the rise [1]. Up to now, China has the highest morbidity and mortality of CRC in the world, and the 5-year survival rate of CRC patients is only 31% [2]. Thus, it is of great significance to clarify the pathogenesis of CRC and find potential biomarkers for better CRC clinical treatment.
MicroRNAs (miRNAs) are small non-coding RNAs that play an important role in regulating post-transcriptional gene expression [3, 4]. MiRNAs decreased the protein translation or mRNA degradation through binding 3'UTR of their target genes [5, 6]. Aberrant expression of miRNAs is associated with the development of CRC [7-9]. MiR-362, which down-regulated in CRC, suppressed cell proliferation, migration and invasion by targeting SIX1 [10]. MiR-17 promotes cell proliferation and migration in human colorectal cancer by downregulating SIK1 [11]. MiRNA-363 is abnormally expressed in a variety of tumors, and regulates cell proliferation and apoptosis by regulating different target genes [12-15]. MiRNA-363 can affect the hepatoma cell proliferation by inhibiting the expression of HMGA2 [16]. Reduced miR-363 expression attenuates the invasion and migration of neuroblastoma cells [17]. Studies have shown that inhibiting the expression of miR-363 can promote lymph node metastasis of head and neck squamous cell carcinoma [18]. Paulína Pidíkova et al. observed that miRNAs (miR-1, miR-15, miR-16, miR-133, miR-143, miR-192, miR-195, miR-206, miR-367 and miR-497) were down-regulated in CRC, and its high expression was usually related to better clinical survival rate, suggesting that down-regulated miRNAs in CRC may be potential targets for the CRC treatment [19]. Yan et al. observed that miR-363 was underexpressed in CRC, and overexpression of miR-363 could inhibit the proliferation and migration of CRC cells [20]. Hu et al. observed that overexpression of miR-363 inhibited cell migration, invasion and EMT by negatively regulating Sox4 [21]. Overexpression of miR-363 in HT29 and HCT116 cell lines effectively inhibited cell proliferation and metastasis, and promoted apoptosis, and the mechanism is by targeting SphK2 [22]. Since the intermolecular regulation in human body is very complex, a molecule can be regulated by a variety of other molecules, or can be combined to regulate many molecules, the regulation between molecules is not one-to-one correspondence, and the occurrence and development of diseases also exist multiple targets and multiple pathways. MiR-196 can promote or suppress cancer by targeting genes such as GATA6, SOCS1, SOCS3, ANXA1, DFFA, PDCD4, ZG16 and ING5 [23]. Thus, miR-363 may bind to a variety of downstream molecules and further exert inhibitory effects. Although progress has been made in the research on the mechanism of miR-363 in the treatment of CRC, it is still of great significance to further clarify it.
The E2F family was first discovered in the E2 gene of adenovirus, and a total of eight E2F members have been discovered. Due to the differences in structure, the functions of different members are not the same [24]. E2F3 is involved in the regulation of G1/S cell cycle and affects the rate of DNA synthesis, which is closely related to the occurrence of liver cancer, breast cancer, colon cancer, etc. [25], and is well known as oncogenes in cancer initiation and progression [26]. Clinical studies have found that the higher the clinical stage and pathological grade of the tumor, the higher the positive rate of E2F3 expression [27]. Olsson et al. also conducted a study on bladder cancer and prostate cancer cells, and the results showed that overexpression of E2F3 in bladder cancer cells significantly improved the proliferation ability of tumor cells [26]. MiR-128 inhibited CRC proliferation and metastasis through E2F3 [28]. MiR-503 inhibits cell proliferation and induces apoptosis in colorectal cancer cells by targeting E2F3 [29]. Jiang et al. showed that miR-363 can target the expression of E2F3 and inhibit the proliferation and invasion of HepG2 cells [30], but whether miR-363 can play its role by inhibiting the expression of E2F3 in CRC cells has not been reported, and further studies are needed.
In this study, we demonstrated that miR-363 was down-regulated in CRC, and miR-363 overexpression significantly inhibited CRC cell proliferation, migration and invasion, and this effect may be achieved by inhibiting E2F3 expression.
Materials and methods
Cell culture and transfection
The HCT-116, SW480 human CRC cell lines and the human normal colonic epithelial cell line NCM460 were purchased from Shanghai Cell Bank, Chinese Academy of Sciences. HCT-116 and NCM460 cells were maintained in McCoy's 5A medium (Gibco, Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS, Gibco, Thermo Fisher Scientific, Inc.), SW480 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS, and all cells were incubated at 37℃ in a 5% CO2 incubator. Cells were seeded in six-well plates and transfected with miR-363 mimics, E2F3-siRNA and negative control using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. The sequences of miR-363 mimics were: 5'-AAUUGCACGGUAUCCAUCUGUA-3', and the sequences of negative control were: 5'-UUCUCCGAACGUGUCACGU-3'.
RT-PCR analysis
Cells were lysed in Trizol (Invitrogen, Carlsbad, CA, USA) for total RNA extraction using reverse transcriptase kit (TAKARA, USA). qRT-PCR was performed using a real-time system (Bio-Rad) using the SYBR Green PCR Kit (KM4101, KAPA Biosystems). The results analysis by 2-△△Ct method, and the miR-363 expression was normalized to the endogenous expression of U6 small nuclear RNA. The mRNA expression level of E2F3 normalized to the endogenous expression of GAPDH. All primers were designed and produced by Nanjing Kingsy Biotechnology Co., Ltd. and are summarized in Table 1.
Primer sequences
| Primer | Sequence (5'-3') |
|---|---|
| miR-363-F | ACACTCCAGCTGGGAATTGCACGGTATCCA |
| miR-363-R | TGGTGTCGTGGAGTCG |
| E2F3-F | GTATGATACGTCTCTTGGTCTGC |
| E2F3-R | CAAATCCAATACCCCATCGGG |
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCG |
| GAPDH-F | ATTCCATGGCACCGTCAAGGCTGA |
| GAPDH-R | TTCTCCATGGTGGTGAAGACGCCA |
Luciferase reporter assay
Targetscan (www.targetscan.org) bioinformatics software was utilized to predict the potential target genes of miR-363. The result showed that there were seven base binding sites between miR-363 and E2F3 3'-UTR. The putative target sites of the human E2F3 3ʹ-UTR segments for miR-363 were amplified by PCR. 5 × 105 cells were seeded in 24-well plates and co-transfected with E2F3 3ʹ-UTR (0.25 µg) or mutant E2F3 3ʹ-UTR and miR-363 mimics (50 nM) or NC using lipofectamine 2000. The activities of firefly and Renilla luciferase were detected using the dual-luciferase reporter assay system (Genecopoeia, USA) and evaluated by SynergyH multiscan spectrum (Bio-Tek, USA).
Cell proliferation
Cell proliferation was identified using CCK8 assay performed as described [31]. 24 hours later with transfection, cells were seeded into 96-well plates (5×103 cells/well). CCK-8 solution (10 μl) was added to each well, and after 4 h, the absorbance was measured at 450 nm using Multiskan FC (Thermo Fisher, USA).
Colony formation assay
To assess colony formation, 500 cells were seeded in each well of a 6-well plate and maintained in medium containing 10% FBS at 37°C. The medium was replaced every three days. After 14 days, colonies were fixed with methanol and stained with 0.1% crystal violet (Sigma-Aldrich, Milwaukee, USA). Visible colonies were manually counted and triplicate measurements were acquired for each treatment group.
Wound-healing assay
Cells were seeded in 6-well plates at 5 × 105 cells per well. Before cell seeding, a line was drawn at the bottom of the well plate every 0.5 cm using a marker, and four lines were drawn in each well. On the next day, a scratch was made perpendicular to the well using a pipette tip against a ruler. The cells were washed with PBS and culture medium was added to the wells. Photographs were taken at 24 h to observe scratch healing. Image J software was used to measure the migration distances and calculate the cell migration ability.
Cell invasion assay
For cell invasion assay, the upper Transwell chambers were pre-coated with 80 µl of Matrigel (354230; BD Biosciences, USA). The chambers were incubated at 37 °C for 30 minutes for gel formation and hydrated in FBS for 4 h before use. In the lower chambers, 600 µl of DMEM or McCoy's 5A medium containing 10% FBS was added. Then, cells were added to the upper chambers at a density of 1 × 105 cells/well and incubated for 24 h at 4 °C. Subsequently, count the invaded cells under a microscope.
Flow cytometry
Cells in each group were cultured for 24 h and then harvested, added in 1ml pre-cooled PBS, and centrifuge at 1000g, cell cycle and apoptosis of HCT-116 and SW480 cells were analyzed using flow cytometry according to the manufacturer's instructions and the data were analyzed by flow cytometry (Beckman Coulter, USA).
Western blotting
Cells were washed with PBS and subjected to a lysis buffer. Protein lysates were separated using 15% SDS-PAGE and transferred to PVDF membranes. The membranes were blocked with a buffer containing 10% non-fat milk in PBS with 0.05% Tween-20 for 2 h and incubated with E2F3 antibody (1:1000, ab152126, Abcam), anti MMP-2 antibody (1:1000, ab97779, Abcam) and β-actin (1:1000, PAB36265, Bioswamp). Then the membranes were incubated with specific secondary antibodies attached to horseradish peroxidase at 4°C. Evaluation of the expression of proteins was performed using Image J version 1.38.
Tumor formation assay in a nude mouse model
To validate the results in vitro, the tumorigenic ability of miR-363 was investigated in vivo. Immunodeficient BALB/C nu/nu male mice (n = 10; 5 weeks) are obtained from the Hubei Laboratory Animal Centre (Wuhan, China). All animals used in this study were strictly conformed to the international health and medical research guidelines for animal welfare and approved by the animal ethics committee of Wuhan Myhalic Biotechnological Co., Ltd (HLK-20210501-01). To evaluate the role of miR-363 in vivo, the stable overexpressed miR-363 CRC cells line was constructed by recombinant retrovirus at first. CRC cells of overexpressed miR-363 (miR-363-mimics), NC (negative control) and C (control) were respectively injected into the left and right forelimb of nude mice by subcutaneous. The tumor volume (V) was calculated according to the formula: length × (width2) / 2. The tumor formation in mice was monitored by the calipers every 3 days. Nude mice were euthanized with sodium pentobarbital (150 mg/kg, IP) at the experimental end-point, and then the tumors were harvested. When the mice rapidly lost 15-20% of their original body weight, could not feed themselves, or the tumor grew more than 10% of the animal's original weight, and other conditions caused extreme pain to the mice, the mice were euthanized after rigorous "Cost/Benefit Analysis".
Statistical analysis
All statistical analysis was performed using SPSS 19.0 software (IBM Corp., Armonk). One-way analysis of variance followed by the Tukey's post hoc test was performed to compare differences between multiple groups. P < 0.05 was considered to indicate a statistically significant difference.
Results
MiR-363 was significantly down-regulated in CRC cell lines
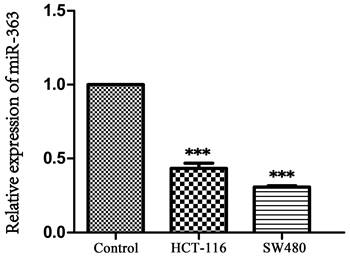
As the result showed in Figure 1, expression of miR-363 in HCT-116 and SW480 was significantly decreased compared with NCM460 (P < 0.001), indicating that miR-363 was significantly down-regulated in CRC cells.
Expression of miR-363 in human CRC cell lines. The expression of miR-363 were detected in human CRC cell lines (HCT-116 and SW480) and the normal cell line (NCM460) by RT-PCR assay. n = 3. ***P<0.001. Take the average of normal group or C group as 1.
Overexpression of miR-363 inhibited cell proliferation, migration and invasion in CRC cells
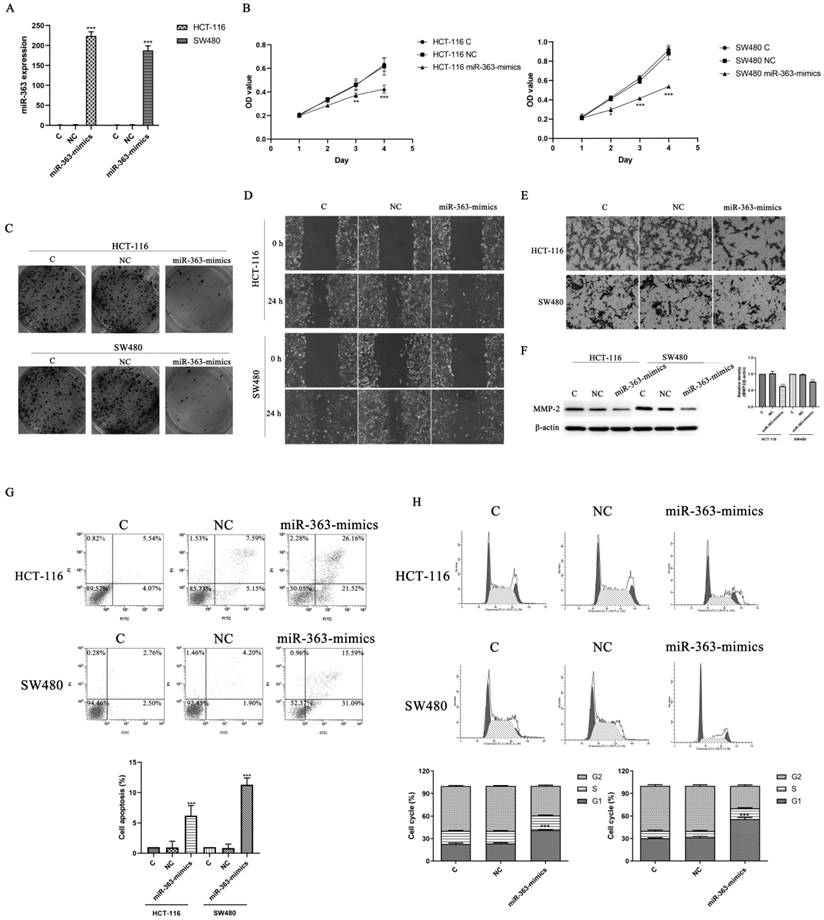
To explore the biological role of miR-363, the HCT-116 and SW480 cell were transfected with miR-363 mimics and negative control, and detected by using RT-PCR analysis (Figure 2A). The transfected HCT-116 and SW480 cells were tested for cell proliferation using CCK-8 assay. Compared with the negative control, the cell proliferation in miR-363 mimics group was significantly decreased (P<0.05, P<0.01, P<0.001) (Figure 2B), and colony formation assay exhibited that cell colony number was decreased in miR-363 mimics group compared with NC group (Figure 2C). Thus, miR-363 overexpression inhibited HCT-116 and SW480 cell proliferation.
Next, we studied the effects of miR-363 on cell migration and invasion in the HCT-116 and SW480 cell by using wound-healing assay and cell invasion assay. The results indicated that miR-363 overexpression inhibited the cell migration and invasion in the HCT-116 and SW480 cell compared to the negative controls (Figure 2D and 2E). Moreover, overexpression of miR-363 significantly reduced the expression level of invasion-related protein MMP-2 (P < 0.001) (Figure 2F). We further detected the effect of overexpression of miR-363 on apoptosis and cell cycle. As shown in Figure 2G and 2H, compared with the C and NC group, the apoptosis rate of miR-363-mimics group was significantly increased (P < 0.001), and G1 phase was significantly blocked (P < 0.001). Thus, miR-363 overexpression inhibited the CRC cell proliferation, migration and invasion, and promoted cell apoptosis.
E2F3 was a target of miR-363 in CRC cell lines
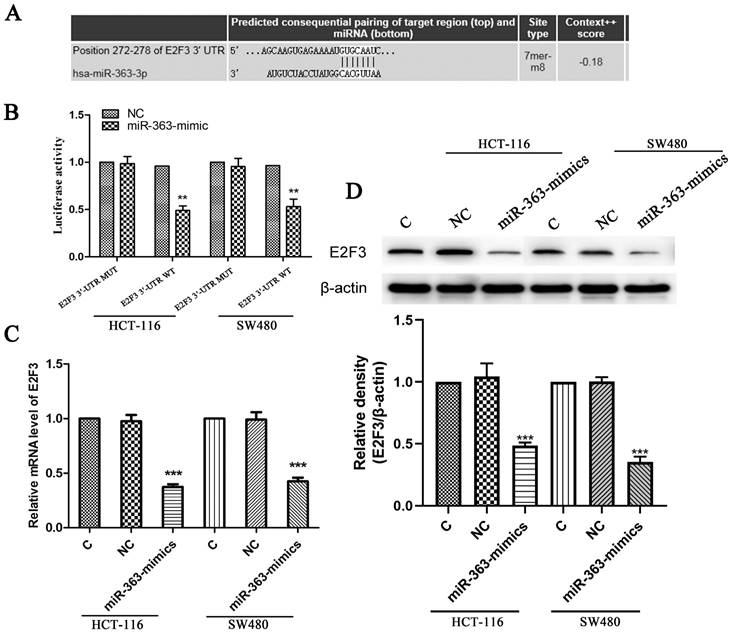
The target genes of miR-363 were searched using the TargetScan (Figure 3A). We use luciferase reporter assay to pinpoint whether E2F3 is a direct target of miR-363. The results showed that luciferase activity in the group co-transfected with miR-363 mimics and E2F3 3ʹ-UTR wild type was significantly decreased (P<0.01) (Figure 3B). This result suggested that E2F3 is miR-363's target. Next, the Figure 3C shows that the levels of E2F3 in the miR-363-mimics group was markedly decreased (P < 0.001) (Figure 3C). WB detection results showed the same trend as before (Figure 3D).
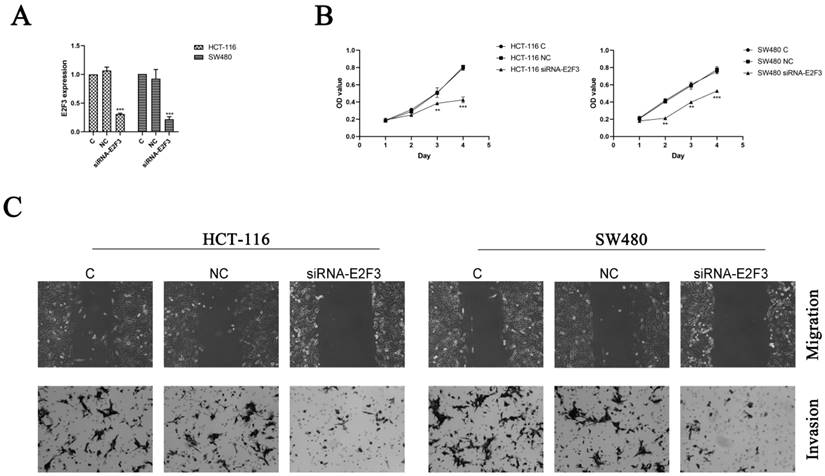
Knockdown of E2F3 significantly inhibited HCT-116 and SW480 cell proliferation, migration and invasion
To further confirm the hypothesis that overexpression of miR-363 suppresses cell behavior by targeting E2F3 in CRC, HCT-116 and SW480 cells were transfected with siRNA-E2F3 and negative control and observed by using RT-PCR analysis (Figure 4A). The transfected HCT-116 and SW480 cells were detected for cell proliferation by CCK8. Compared with the negative control, the cell proliferation in E2F3 knockdown group was significantly decreased (P < 0.01, P < 0.001) (Figure 4B).
E2F3 effects on cell migration and invasion in the HCT-116 and SW480 cells were observed by using wound-healing assay and cell invasion assay, and the results indicated knockdown of E2F3 restrained the cell migration and invasion in the HCT-116 and SW480 cells compared to the NC group (Figure 4C). Thus, miR-363 suppresses cell behavior by targeting E2F3 in CRC cells.
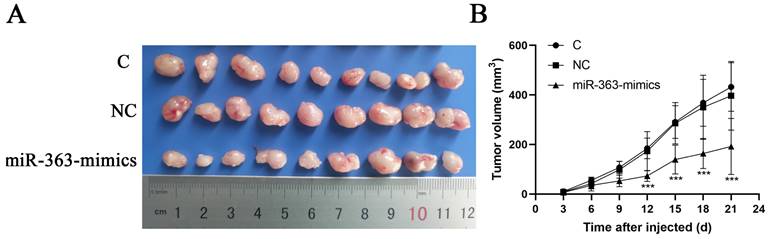
MiR-363 suppressed tumor growth in vivo
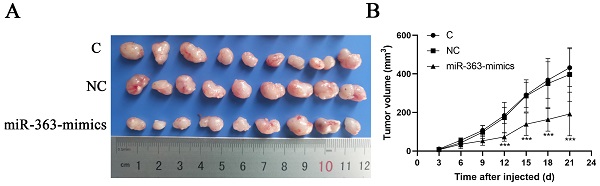
HCT-116 cells transfected with miR-363 mimics, NC or C were subcutaneously injected into either flank of nude mice. Compared with NC, miR-363 reduced the volume of tumors formed from HCT-116 cells at different time-points (P<0.001) (Fig 5A and B). The results suggested that the miR-363 might reduce tumorigenicity and tumor progression in the nude mouse model.
Effect of miR-363 on CRC cell proliferation, migration and invasion. A. miR-363 expression was determined by RT-PCR. B. CCK8 was used to measure the cell proliferation. C. Colony formation assays was performed to measure the cell proliferation. D. Wound-healing assay and E. cell invasion assay were used to test cell migration (100×) and invasion (200×). F. MMP-2 expression was detected by western blot. G. Cell apoptosis and H. cell cycle were detected by flow cytometry. n = 3. *P<0.05, **P<0.01, ***P<0.001. Take the average of normal group or C group as 1.
Discussion
Up to now, treatment failure and recurrence of CRC are still an urgent problem to be solved in clinical, and one of the main reasons for the poor prognosis of CRC patients is the late diagnosis [32, 33]. Therefore, it is of great significance to find predictive bio-markers.
Numerous studies demonstrated that miRNAs are important in regulating tumor cell proliferation, migration and invasion [34-37], and are involved in the pathogenesis of CRC [38, 39]. MiR-4323 promotes CRC cell proliferation by inhibiting HDGF [40]. MiR‑4306 inhibits proliferation, migration and invasion by targeting lncRNA-FoxD2‑AS1 in CRC [41]. miR-545 promotes colorectal cancer by inhibiting transferring in the non-normal ferroptosis signaling [42]. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization [43].
E2F3 was the target of miR-363 in CRC cell lines. The TargetScan database showed that miR-363 may bind to target sequences located at nucleotides 272-278 of E2F3 3'-UTR. B. Luciferase activity of E2F3 3'-UTR and mutant E2F3 3'-UTR were transfected with miR-363 mimics was detected by dual-luciferase reporter assay system. C. The mRNA levels of E2F3 were quantified by RT-PCR. D. E2F3 expression was measured by western blot. n = 3. ***P<0.001. Take the average of normal group or C group as 1.
MiR-363 is downregulated in various tumors and regulates tumor cell development [44, 45]. Studies reported that [46-48] miR-363 overexpression can significantly reduce the resistance of HepG2-R cells to cisplatin, decrease the migration ability of head and neck tumor cells, and inhibit the renal cancer cell proliferation and migration. Jamali Z et al [49] observed that patients with miR-363 low expression in head and neck tumors had a poor prognosis, and miR-363 role is via targeting of FBW7 ubiquitin ligase expression [50]. Aberrant miR-363 expression affects head and neck cancer infiltrate and metastasize by targeting podoplanin [45], indicating that miR-363 plays a role in the development and prognosis of tumors. miR-363 can degrade target genes by fully binding to the 3'UTR region of target genes, or incompletely binding to inhibit the translation process, forming a negative regulatory role. However, the effect of miR-363 on CRC and its mechanism remain to be further studied.
In this study, we first preformed RT-PCR in CRC cell lines and found that miR-363 level in HCT-116 and SW480 cells were markedly lower than that in NCM460 cells. Then, miR-363 mimics were transferred into HCT-116 and SW480 cells, and CCK8, wound-healing assay and cell invasion assay were used to detect cell behavior. The results showed that miR-363 overexpression inhibited CRC cell proliferation, migration and invasion. Cell cycle is divided into the early stage of DNA synthesis (G1 stage), the DNA synthesis stage (S stage) and the late DNA synthesis stage (G2 stage). The main function of G1 phase is to synthesize RNA and ribosomes to prepare material and energy for DNA replication in S phase, S phase is mainly for DNA synthesis, and G2 phase is to prepare for mitosis. Cell cycle experiments showed that miR-363-mimics blocked the cell cycle of HCT-116 and SW480 in G1 phase, suggesting that miR-363-mimics can inhibit DNA synthesis. We further used flow cytometry to detect the cell apoptosis of HCT-116 and SW480. The results showed that the apoptotic rate of HCT-116 and SW480 was increased in miR-363-mimics group compared to the control and NC group, suggesting that overexpression of miR-363 can promote CRC cell apoptosis and inhibit cell proliferation, migration and invasion of CRC.
Knockdown of E2F3 significantly inhibited HCT-116 and SW480 cell proliferation, migration and invasion. A. Expression of E2F3 was determined by RT-PCR. B. Cell proliferation was test by CCK8. C. Cell migration and invasion were detected using wound-healing assay (100×) and cell invasion assay (200×). n = 3. *P<0.05, **P<0.01, ***P<0.001. Take the average of normal group or C group as 1.
miR-363 suppresses tumor growth in vivo. A. Images of tumors from transfected with miR-363 mimics or NC after 21 days of injected. B. Volumes of tumors in vivo were detected every 3 days. n = 3. **P<0.01, ***P<0.001.
MMPs is a highly conserved class of enzymes composed of a series of zinc ion dependent proteolytic enzymes, which are widely distributed in various organisms. Under normal physiological conditions, MMPs and tissue metalloproteinases inhibitors (TIMP) co-regulates the renewal of ECM and maintain cell stability, while the imbalance of MMPs can destroy tissue barrier, promote matrix degradation, and then promote tumor invasion and metastasis. According to the substrate and fragment homology, MMPs can be divided into four categories. Gelatinase (i.e. type IV collagenase) is an important class, mainly including MMP9 and MMP2. Studies have shown that gelatinases exhibit increased activity in a variety of malignant tissues, cultured tumor cells, and oncogene transformed cells, and both in vivo and in vitro invasion assays have confirmed that the high invasive ability of tumor cells is associated with the increased activity of gelatinases, which are thus considered to be the major proteolytic enzymes involved in tumor invasion and metastasis. MMP2 and MMP9 have a lot of the same components in the degradation substrates, but MMP9 has no ability to hydrolyze collagen directly [51]. Under physiological conditions, MMP2 is ubiquitously expressed in vivo, while MMP9 is only present in neutrophil granules. Numerous studies have demonstrated that MMP-2 plays a critical role in tumor cell-mediated extracellular matrix degradation. The increase of MMP-2 activity and expression is closely related to the invasion and metastasis potential and prognosis of a variety of human malignant tumors [52], so it has become the hotspot of tumor invasion and metastasis research in recent years. Tumor invasion and metastasis is a continuous multi-step process. In addition to the degradation of extracellular matrix, the formation of tumor microvessels is also indispensable for tumor invasion and metastasis [53]. MMP2 has the ability to degrade collagen in vascular basement membrane, which is closely related to tumor angiogenesis [54], and its activation state plays a crucial role in angiogenesis. Thus, in this study, we selected MMP2, one of the genes most closely related to tumor invasion and metastasis, for detection, and the results showed that the expression of MMP2 was significantly decreased in the miR-363-mimics group compared with the control and NC groups, and the results still supported the foregoing conclusion.
Luciferase reporter assay was used to test whether E2F3 is a direct target of miR-363. RT-PCR and WB were used to detect the E2F3 level in HCT-116 and SW480 cell which miR-363 overexpressed, and the results showed that E2F3 was target of miR-363. To further confirm the hypothesis that overexpression of miR-363 suppresses cell behavior by targeting E2F3 in CRC, siRNA-E2F3 was transfected to HCT-116 and SW480 cells, and the results indicated that knockdown of E2F3 dramatically inhibited the cell migration and invasion in HCT-116 and SW480 cells.
We inoculated HCT-116 cells transfected with miR-363 mimics into nude mice to observe whether miR-363 could inhibit tumor progression. Results suggested that the miR-363 might reduce tumorigenicity and tumor progression in a nude mouse model, which further confirmed that miR-363 might suppress cell proliferation, migration and invasion by targeting E2F3 in CRC.
In this study, we only used miR-363 mimics to intervene, and observed CRC cell growth and motility. Lacking miR-363 inhibitor is one of the shortcomings of this study, and we will refine it in the next in-depth study.
In conclusion, our research showed that miR-363 was significantly decreased in CRC cells, and miR-363 suppresses cell proliferation, migration and invasion by targeting E2F3 in CRC cells.
Acknowledgements
Funding
This project was supported by the PhD‑initiated funding project of Hubei University of Chinese Medicine (No. 10110428), Hubei Province Administration of Traditional Chinese Medicine Young Talents Program (No. ZY2023Q041), Hubei Provincial Science and Technology Planning Project (No.2020BCB015), Key project of Scientific Research of Hubei Provincial Education Department (No. D20152004) and Key discipline Project of TCM of Hubei Provincial Administration of TCM (No. 5432-10100204).
Author contributions
WY and XY contributed to conception of the study. WY, XY, YZ and YL designed and performed the experiments, analyzed the data and drafted the manuscript. WY, XY, YL and WL analyzed the data and provided technical support. All authors have read and approved the final manuscript. WY, XY, YZ, YL and WL confirm the authenticity of all the raw data.
Ethics approval and consent to participate
The study strictly conformed to the international health and medical research guidelines for animal welfare and approved by the animal ethics committee of Wuhan Myhalic Biotechnological Co., Ltd (HLK-20210501-01).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Health Commission of the People's Republic of China. Chinese Criteria for Diagnosis and Treatment of colorectal Cancer (2020 edition). Chinese Journal of Practical Surgery. 2020;40(6):601-625
2. Zhu J, Tan Z, Hollis-Hansen K. et al. Epidemiological trends in colorectal cancer in China: an ecological study. Dig Dis Sci. 2017;62(1):235-243
3. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297
4. Irwandi RA, Vacharaksa A. The role of microRNA in periodontal tissue: A review of the literature. ARCH ORAL BIOL. 2016;72:66-74
5. Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. CURR GENOMICS. 2010;11:537-561
6. Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J PHYSIOL BIOCHEM. 2011;67:129-139
7. Long J, He Q, Yin Y, Lei X, Li Z, Zhu W. The effect of miRNA and autophagy on colorectal cancer. Cell Prolif. 2020Oct;53(10):e12900
8. Jung G, Hernández-Illán E, Moreira L, Balaguer F, Goel A. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. 2020;17(2):111-130
9. Wang H. MicroRNAs and Apoptosis in Colorectal Cancer. Int J Mol Sci. 2020;21(15):5353
10. Wan J, Yang J, Qiao C. et al. MicroRNA-362 Inhibits Cell Proliferation and Invasion by Directly Targeting SIX1 in Colorectal Cancer. Yonsei Med J. 2019;60(5):414-422
11. Huang C, Liu J, Xu L. et al. MicroRNA-17 promotes cell proliferation and migration in human colorectal cancer by downregulating SIK1. Cancer Manag Res. 2019;11:3521-3534
12. Tsuji S, Kawasaki Y, Furukawa S. et al. The miR-363-GATA6-Lgr5 pathway is critical for colorectal tumourigenesis. Nat Commun. 2014;5:3150
13. Zhou P, Huang G, Zhao Y. et al. MicroRNA-363-mediated downregulation of S1PR1 suppresses the proliferation of hepatocellular carcinoma cells. Cell Signal. 2014;26:1347-1354
14. Wang SH, Zhang WJ, Wu XC. et al. The lncRNA MALAT1 functions as a competing endogenous RNA to regulate MCL-1 expression by sponging miR-363-3p in gallbladder cancer. J Cell Mol Med. 2016;20:2299-2308
15. Zhang R, Li Y, Dong X, Peng L, Nie X. MiR-363 sensitizes cisplatin-induced apoptosis targeting in Mcl-1 in breast cancer. Med Oncol. 2014;31:347
16. Wang J, Liang H, Ge H, Guo X, Gu D, Yuan Y. MicroRNA-363-3p inhibits hepatocarcinogenesis by targeting HMGA2 and is associated with liver cancer stage. Mol Med Rep. 2019;19(2):935-942
17. Johnson J, Thijssen B, McDermott U, Garnett M, Wessels LF, Bernards R. Targeting the RB-E2F pathway in breast cancer. Oncogene. 2016;35(37):4829-35
18. Sun Q, Zhang J, Cao W. et al. Dysregulated miR-363 affects head and neck cancer invasion and metastasis by targeting podoplanin. INT J BIOCHEM CELL B. 2013;45:513
19. Pidíkova P, Reis R, Herichova I. miRNA Clusters with Down-Regulated Expression in Human Colorectal Cancer and Their Regulation. Int J Mol Sci. 2020;21(13):4633
20. Yan XR, Wang LB, Chen DM, Li YK, Yang YX.Effect of miR-363 expression on cell proliferation, migration activity in human colorectal cancer. Chinese Journal of Cancer Prevention. 2014;21(7):509-513.
21. Hu F, Min J, Cao X. et al. MiR-363-3p inhibits the epithelial-to-mesenchymal transition and suppresses metastasis in colorectal cancer by targeting Sox4. Biochem Biophys Res Commun. 2016;474(1):35-42
22. Dong J, Geng J, Tan W. MiR-363-3p suppresses tumor growth and metastasis of colorectal cancer via targeting SphK2. Biomed Pharmacother. 2018;105:922-931
23. Pourdavoud P, Pakzad B, Mosallaei M. et al. MiR-196: emerging of a new potential therapeutic target and biomarker in colorectal cancer. Mol Biol Rep. 2020;47(12):9913-9920
24. Liao Y, Du W. Rb-independent E2F3 promotes cell proliferation and alters expression of genes involved in metabolism and inflammation. FEBS Open Bio. 2017;7(10):1611-1621
25. Gong C, Liu H, Song R. et al. ATR-CHK1-E2F3 signaling transactivates human ribonucleotide reductase small subunit M2 for DNA repair induced by the chemical carcinogen MNNG. Biochim Biophys Acta. 2016;1859(4):612-26
26. Olsson AY, Feber A, Edwards S. et al. Role of E2F3 expression in modulating cellular proliferation rate in human bladder and prostate cancer cells. Oncogene. 2007;26:1028-1037
27. Trikha P, Sharma N, Pena C. et al. E2F3 in tumor macrophages promotes lung metastasis. Oncogene. 2016;35(28):3636-46
28. Yao J, Wang C, Dong X. et al. lncRNA SNHG22 sponges miR-128-3p to promote the progression of colorectal cancer by upregulating E2F3. Int J Oncol. 2021;59(3):71
29. Chang SW, Yue J, Wang BC, Zhang XL. miR-503 inhibits cell proliferation and induces apoptosis in colorectal cancer cells by targeting E2F3. Int J Clin Exp Pathol. 2015;8(10):12853-60
30. Jiang Z, Shu M, Wang YY, Pu LS. Studies on the effect of Mir-363 on proliferation and apoptosis of HepG2 cells by targeting regulation of E2F3 expression. Journal of Practical Liver Disease. 2018;21(6):825-828
31. Sheng L, He P, Yang X, Zhou M, Feng Q. miR-612 negatively regulates colorectal cancer growth and metastasis by targeting AKT2. CELL DEATH DIS. 2015;6:e1808
32. Weitz J, Koch M, Debus J, Höhler T, Galle PR, Büchler MW. Colorectal cancer. Lancet. 2005;365(9454):153-65
33. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394(10207):1467-1480
34. Endo-Munoz L, Cumming A, Sommerville S, Dickinson I, Saunders NA. Osteosarcoma is characterised by reduced expression of markers of osteoclastogenesis and antigen presentation compared with normal bone. Br J Cancer. 2010;103:73-81
35. Wang T, Xu H, Qi M, Yan S, Tian X. miRNA dysregulation and the risk of metastasis and invasion in papillary thyroid cancer: a systematic review and meta-analysis. Oncotarget. 2017
36. Wu F, Li J, Guo N, Wang XH, Liao YQ. MiRNA-27a promotes the proliferation and invasion of human gastric cancer MGC803 cells by targeting SFRP1 via Wnt/beta-catenin signaling pathway. AM J CANCER RES. 2017;7:405-416
37. Li J, Li Q, Huang H. et al. Overexpression of miRNA-221 promotes cell proliferation by targeting the apoptotic protease activating factor-1 and indicates a poor prognosis in ovarian cancer. INT J ONCOL. 2017
38. Wang L, Sun GT. Effects of micrornA-152-3P on proliferation, migration and invasion of colon cancer cells and their mechanisms. Anhui Med Pharm. 2022;26(9):1835-1839 +1912
39. Hong S, Yan Z, Wang H, Ding L, Song Y, Bi M. miR-663b promotes colorectal cancer progression by activating Ras/Raf signaling through downregulation of TNK1. Hum Cell. 2020;33(1):104-115
40. Xia C, Li Q, Cheng X, Wu T, Gao P. miR-4323 targets hepatoma-derived growth factor (HDGF) to suppress colorectal cancer cell proliferation. Pathol Res Pract. 2021;225:153544
41. Ye J, Liu J, Tang T, Xin L, Bao X, Yan Y. miR-4306 inhibits the malignant behaviors of colorectal cancer by regulating lncRNA FoxD2-AS1. Mol Med Rep. 2021;24(4):723
42. Zheng S, Hu L, Song Q. et al. miR-545 promotes colorectal cancer by inhibiting transferring in the non-normal ferroptosis signaling. Aging (Albany NY). 2021;13(24):26137-26147
43. Xu C, Fan L, Lin Y. et al. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes. 2021;13(1):1980347
44. Sun Q, Zhang J, Cao W. et al. Dysregulated miR-363 affects head and neck cancer invasion and metastasis by targeting podoplanin. INT J BIOCHEM CELL B. 2013;45:513-520
45. Song B, Yan J, Liu C, Zhou H, Zheng Y. Tumor Suppressor Role of miR-363-3p in Gastric Cancer. MED SCI MONITOR. 2015;21:4074-4080
46. Ou Y, Zhai D, Wu N. et al. Downregulation of miR-363 increases drug resistance in cisplatin-treated HepG2 by dysregulating Mcl-1. Gene. 2015;572( 1):116-122
47. Chapman BV, Wald AI, Akhtar P. et al. MicroRNA-363 targets myosin 1B to reduce cellular migration in head and neck cancer. BMC Cancer. 2015;15:861
48. Li Y, Chen D, Li Y. Oncogenic cAMP responsive element binding protein 1 is overexpressed upon loss of tumor suppressive miR-10b-5p and miR-363-3p in renal cancer. Oncol Rep. 2016;35(4):1967-1978
49. Jamali Z, Asl AN, Attaran R. et al. MicroRNAs as prognostic molecular signatures in human head and neck squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 2015;51(4):321-331
50. Yu X, Wang J, Liu J. et al. A multimodal Pepstatin A peptide-based nanoagent for the molecular imaging of P-glycoprotein in the brains of epilepsy rats. Biomaterials. 2016;76:173-186
51. Klein T, Bischoff R. Physiology and pathophysiology of matrix metalloproteases. Amino Acids. 2011;41(2):271-290
52. Davies B, Waxman J, Wasan H. et al. Levels of matrix metalloproteinase in bladder cancer correlate with tumor grade and invasion. Cancer Res. 1993;53:5365-5369
53. Bode W, Fernandez-Catalan C, Tschesche H. et al. Structural properties of matrix metalloproteinases. Cell Mol Life Sci. 1999;55(4):639
54. Nguyen M, Arkell J, Jackson CJ. Human endothelial gelatinases and angiogenesis. Int J Biochem Cell Biol. 2001;33(10):960-970
Author contact
![]() Corresponding author: Dr. Yunhai Li, Clinical College of Chinese Medicine, Hubei University of Chinese Medicine, No. 188, Tanhualin, Wuchang District, Wuhan, China. Email: 155350975com; Dr. Wenliang Lv, Clinical College of Chinese Medicine, Hubei University of Chinese Medicine, No. 188, Tanhualin, Wuchang District, Wuhan, China. Email: lvwenliang66com.
Corresponding author: Dr. Yunhai Li, Clinical College of Chinese Medicine, Hubei University of Chinese Medicine, No. 188, Tanhualin, Wuchang District, Wuhan, China. Email: 155350975com; Dr. Wenliang Lv, Clinical College of Chinese Medicine, Hubei University of Chinese Medicine, No. 188, Tanhualin, Wuchang District, Wuhan, China. Email: lvwenliang66com.

 Global reach, higher impact
Global reach, higher impact