Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(15):3660-3673. doi:10.7150/jca.78835 This issue Cite
Research Paper
miR-140-3p enhances the sensitivity of LUAD cells to antitumor agents by targeting the ADAM10/Notch pathway
1. Department of Thoracic Surgery, General Hospital of Northern Theater Command, Shenyang City 110011, Liaoning Province, China.
2. Department of Clinical Pharmacy, Shenyang Pharmaceutical University, Shenyang City 110011, Liaoning Province, China.
*These authors equally contributed to this work.
Received 2022-9-11; Accepted 2022-11-7; Published 2022-11-21
Abstract
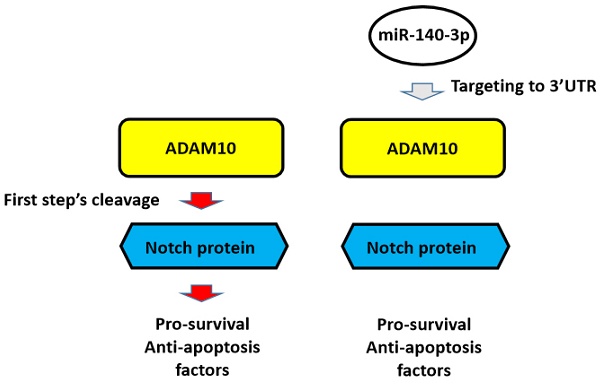
Background: The Notch pathway, which is related to the drug-resistance of lung adenocarcinoma (LUAD) type of non-small cell lung cancer (NSCLC) cells, is activated by cleavage of Notch proteins mediated by ADAMs, ADAM10 or ADAM17.
Methods: In the present study, our results demonstrated that of these two ADAMs, the expression of ADAM10 in clinical samples of the LUAD type of NSCLC was much higher than that of ADAM17, while miR-140-3p - an miRNA that could target ADAM10 - was identified by an online tool: miRDB (miRNA database). The detail function and mechanism of miR-140-3p in regulating the sensitivity of NSCLC cells to antitumor drugs was systematically explored in vitro and in vivo.
Results: In A549, a typical NSCLC LUAD cell line, miR-140-3p decreased ADAM10 expression and repressed activation of the Notch pathway by repressing cleavage of Notch proteins. The expression of miR-140-3p was negatively related to ADAM10 in clinical specimens. Nucleocytoplasmic separation/subfraction assays showed that miR-140-3p was able to inhibit the cleavage of Notch protein, and led to the accumulation of Notch intracellular domains (NICD) in the nucleus. Overexpression of miR-140-3p enhanced the sensitivity of A549 cells to antitumor agents by targeting the 3'UTR region of ADAM10 mRNA in both cultured cells and in vivo models.
Conclusion: ADAM10 plays a major role in LUAD, and miR-140-3p acts on ADAM10 and inhibits its expression and the cleavage of Notch protein, leading to the inhibition the activity of the Notch pathway, and ultimately upregulating LUAD cell sensitivity to anti- tumor drugs.
Keywords: Notch pathway, A disintegrin and metalloproteinase domain 10, miR-140-3p, chemotherapeutic agents, molecular targeted agents
Introduction
Lung cancer poses a serious threat to the health of individuals worldwide and in China, and its morbidity and mortality rank first among all malignant tumors [1,2]. Among the types of lung cancer, non-small cell lung cancer (NSCLC) is the most important and common pathological type [3-5]. Furthermore, NSCLC is the most important type of lung adenocarcinoma, making lung adenocarcinoma-related research important [3-6]. Currently, antitumor drug treatments for lung adenocarcinoma mainly include various small molecule protein kinase inhibitors and cytotoxic chemotherapeutics [7-12]. Although these drugs or drug combinations are believed to prolong the lives of patients and improve their quality of life, there are still many shortcomings: (1) the sensitivity of patients to these antitumor drugs is purely individual [13-15]; (2) antitumor drug treatment is prone to drug resistance [13-15]; and (3) the side effects of antitumor drug treatment are very serious [16]. Small molecule inhibitors or therapeutic antibodies targeting the PD-1/PD-L1 pathway have been available for a short time, and the influencing factors or biomarkers that influence the efficacy of these drugs are not yet clear [17-19]. Therefore, it is of great significance to study and develop safer and more effective anti-adenocarcinoma drug treatment strategies, to achieve the superior treatment strategies at drug treatment doses that are the same or smaller.
Increasing evidence has shown that the Notch pathway is not only an important positive regulator of NSCLC but also plays a central role in antitumor drug resistance [20-22]. It has been confirmed that in the process of anti-tumor treatment, chemotherapy drugs and radiotherapy do more harm than good to the body, but they are serious damaging factors for malignant tumor cells/tissues by targeting tumor cell components or to macromolecules [23,24]. In this process, the Notch pathway can be activated [23,24]. The activated Notch pathway can induce cell survival, anti-apoptosis-related factors, and malignant tumor cell epithelial-mesenchymal transition (EMT) [25]. In this way, malignant tumor cells respond to exogenous damage factors (i.e., stress factors) through Notch, which is involved in the cell's stress response mechanism to improve cell resistance, promote survival, and anti-apoptosis mechanisms, ultimately resulting in cell resistance to these antitumor treatments [22-25]. In this process, the activation of the Notch pathway is mainly dependent on the cleavage of the Notch protein: when the Notch protein is cleaved, the extracellular and intracellular segments of the Notch protein separate to release the intracellular products, which migrate into the nucleus and ultimately mediate the expression of factors related to cell survival and anti-apoptosis [25,26]. The activation of Notch protein is a two-step cleavage, process in which the first cleavage is completed by two ADAMs: ADAM17 and ADAM10 [27-29]. This makes ADAM17 or ADAM10 an ideal intervention target for the treatment of malignant tumors. It is of great significance to determine the expression of both in different tumor tissues, and to develop corresponding intervention strategies.
MicroRNA is a type of small noncoding RNA that can recognize and act on the 3'UTR of the target gene mRNA in a sequence-specific manner to degrade the mRNA and finally achieve post-transcriptional silencing [30-32]. Not only can miRNA be used as an important strategy for anti-tumor therapy but also the lack of expression of miRNA is also an important mechanism for the higher expression of pro-oncogenes in malignant tumor tissues [33]. In this study, the expression of two ADAMs in lung adenocarcinoma (LUAD) tissues was first determined, and it was found that ADAM10 is the main ADAM subtype that plays a major role in LUAD. IT was found that miR-140-3p is the miRNA that acts on ADAM10, miR-140-3p can inhibit the activity of the Notch pathway by acting on ADAM10, and ultimately up-regulates the sensitivity of lung adenocarcinoma cells to anti-tumor drugs.
Materials and Methods
Clinical specimens, cells lines, and agents
A total of 26 pairs of malignant tumor tissues and adjacent non-tumor tissues were collected and stored by the Department of Thoracic Surgery, General Hospital of Northern Theater Command from November 2019 to March 2021. All clinical specimens were obtained from routine surgical resection and LUAD was confirmed by pathological analysis by the General Hospital of Northern Theater Command) of the author's institution. The diagnosis, inclusion and exclusion criteria of patients were all in accordance with the clinical diagnosis and treatment guidelines for lung cancer of the Chinese Medical Association (2022 version) [34]. The patient was diagnosed according to imaging methods, including CT (Computed Tomography), MRI (Nuclear Magnetic Resonance Imaging), or PET [Positron Emission Computed Tomography]. Head CT, bone CT, and liver-gallbladder-pancreas-kidney-adrenal-color ultrasound were performed before surgery. The patients with no severe obstructive difficulty in pulmonary function; no serious heart disease (including no stent placement within half a year, and cardiac function>56%) underwent surgical operation. The pathological staging and classification of the specimens removed by surgery are determined by pathological staining and analysis (that is, confirmed as lung adenocarcinoma). The baseline information of the patients were shown as Table 1. The sample size (26 paired nontumor/tumor specimens) has adequate power to detect a pre-specified effect size (the 1-β: 0.8; α/2: 0.025; P < 0.05) of the statistical analysis by original hypothesis/ alternative hypothesis. The original hypothesis was that the expression level of the targeting gene (for example, ADAM10 or miR-140-3p) was not significantly different in the non-tumor tissues compared with the tumor tissues; whereas the alternative hypothesis was that the expression level of the targeting gene was significantly different in the non-tumor tissue compared with the tumor tissue.
The baseline information of patients involved in the present work
| Characters | Values |
|---|---|
| Age | 55 (38~75) (Median [upper and lower limit]) |
| Gender | Male (58%, 15/26) |
| Tumor stage | I (65%, 17/26) (IA and IB) |
| II (35%, 9/26) (IIA and IIB) | |
| Tumor number | 1 (92%, 24/26) |
| 2 (8%, 2/26) | |
| Tumor sites | Left lower lobe (11%, 3/26) |
| Left upper lobe (27%, 7/26) | |
| Right lower lobe (19%, 5/26) | |
| Right middle lobe (-) | |
| Right upper lobe (43%, 11/26) | |
| Tumor location | Central (15%, 8/26) |
| Peripheral (85%, 22/26) | |
| Tumor sizes | 1.4 (0.4~2.2 [cm]) (Median [upper and lower limit]) |
The expression vector containing full length sequences of ADAM10 (the wild type of ADAM10 or the ADAM10 with the mutated miR0140-3p targeted sites located in its 3' untranslated region (3'UTR), hsa-pre-miR-140 or NICD was constructed and prepared as lentivirus and stored at our lab as described in previous studies [25,35-37]. The 3'UTR region sequence of ADAM10 containing wild-type or mutant miR-140-3p action site (about 200bp upstream and downstream of the action site) was cloned into the pGL4.26 vectors to form the luciferase reporters named as Luc-1 or Luc-1Mut.
The NSCLC LUAD cell line A549, NSCLC large cell lung cancer (LCC) cell line H460 (NCI-H460), and NSCLC Lung squamous cell carcinoma (LSCC) cell line H520 (NCI-H520), and WI-38, a non-tumor lung derived cell line was purchased from the Type Culture Collection of the Chinese Academy of Sciences (the culture collection center of the Chinese government, Shanghai, People's Republic of China).
The antitumor agents, gemcitabine, paclitaxel, etoposide, doxorubicin, gefitinib (Cat. No.: S1025), erlotinib (Cat. No.: S7786), osimertinib (Cat. No.: S7297) or anlotinib (Cat. No.: S8726), were purchased from Selleck Corporation, Houston, TX, USA. For in vitro cell-based cell survival assays or MTT experiments. The antitumor agents (the powders of agents with purity > 98.7%) were carefully dissolved by using the organic solvents, dimethyl sulfoxide (DMSO), and then diluted by DMEM without FBS with stirring and ultrasonic solubilization [38-41]. The concentrations of these agents used in this study in DMSO or DMEM are listed in Table 2. For the subcutaneous tumor experiments in nude mice, gefitinib (4 mg) was dissolved by a mixture of organic solvents: DMSO (15 μL), PEG400 (polyethylene glycol) (60 μL), and Tween80 (40 μL) and was carefully added with physiological saline to a total volume 2 mL with stirring and ultrasonic solubilization [38-41]. This Gefitinib-containing solution was used for oral administration to nude mice.
Antibodies and Western blotting
Antibodies against ADAM10 (Cat. no ab124695), Lamin A (Cat. no ab108595), GAPDH (Cat. no ab8245) and β-actin (Cat. no ab205) were obtained from Abcam (Cambridge, MA, USA). Antibodies to detect the NICD of Notch protein was generated by our lab as described in previous studies [42-44]. A549 cells, following transfection with vectors, were harvested for total protein extraction for western blotting experiments under standard protocols. The cellular sub-fraction assays were performed following the methods described in previous studies [42-44]. Briefly, the A549 cells were transfected with plasmids and then trypsinized and harvested. The cells were disrupted using sonication, and the nuclear fraction was collected at 800 rpm, while the cytoplasmic fraction was collected after centrifugation at 12000 rpm. The above components were detected using western blotting. Lamin A was used as an to normalize nuclear components/sub-fraction, while β-Actin was used for cytoplasmic components/sub-fraction. The images of western blot were quantitatively analyzed by Image J software (National Institutes of Health [NIH], Bethesda, Maryland, USA) [45-47]. The total area and the intensity of bands of ADAM10, NICD or loading control (Lamin A for nuclear sub-fraction and β-Actin for cytoplasm sub-fraction) from in the western blot images was quantitatively identified by Image J. The expression level of ADAM10 or NICD was adjusted by β-Actin or Lamin A [45-47].
Cell culture and proliferation analysis
The MTT assay was used to evaluate the survival-inhibitory effect of antitumor drugs on A549 cells as following the methods described in previous studies [45-48]. Briefly, transfected A549 cells were seeded in 96-well plates (8000 cells per well) and then treated with the indicated concentrations of antitumor agents for 48h. The cells were then subjected to MTT assay agents for 5-6 h. Lastly, the cell samples were diluted using SDS, the absorbance was read using a multi-function microplate reader at 490 nm. The inhibition rates of the drug on A549 cells was determined bas on the 490 nm values, and a drug inhibition curve was constructed for the A549 cells, and the IC50 value of the drug effect was determined.
The concentrations of antitumor agents in the present work
| Agents | Gemcitabine | Paclitaxel | Etoposide | Doxorubicin | Gefitinib | Erlotinib | Osimertinib | Anlotinib |
|---|---|---|---|---|---|---|---|---|
| DMSO (mmol/L) | 3 | 0.3 | 3 | 1 | 10 | 10 | 10 | 10 |
| 1 | 0.1 | 1 | 0.3 | 3 | 3 | 3 | 3 | |
| 0.3 | 0.03 | 0.3 | 0.1 | 1 | 1 | 1 | 1 | |
| 0.1 | 0.01 | 0.1 | 0.03 | 0.3 | 0.3 | 0.3 | 0.3 | |
| 0.03 | 0.03 | 0.03 | 0.01 | 0.1 | 0.1 | 0.1 | 0.1 | |
| 0.01 | 0.01 | 0.01 | 0.003 | 0.03 | 0.03 | 0.03 | 0.03 | |
| 0.003 | 0.003 | 0.003 | 0.001 | 0.01 | 0.01 | 0.01 | 0.01 | |
| DMEM (μmol/L) | 3 | 0.3 | 3 | 1 | 10 | 10 | 10 | 10 |
| 1 | 0.1 | 1 | 0.3 | 3 | 3 | 3 | 3 | |
| 0.3 | 0.03 | 0.3 | 0.1 | 1 | 1 | 1 | 1 | |
| 0.1 | 0.01 | 0.1 | 0.03 | 0.3 | 0.3 | 0.3 | 0.3 | |
| 0.03 | 0.03 | 0.03 | 0.01 | 0.1 | 0.1 | 0.1 | 0.1 | |
| 0.01 | 0.01 | 0.01 | 0.003 | 0.03 | 0.03 | 0.03 | 0.03 | |
| 0.003 | 0.003 | 0.003 | 0.001 | 0.01 | 0.01 | 0.01 | 0.01 |
Quantitative polymerase chain reaction (qPCR) and luciferase assays
Quantitative polymerase chain reaction experiments were carried out according to the methods reported in previous studies and in accordance with the manufacturer's instructions [49-51]. Briefly, total RNA samples from A549 and other cells and tissue samples were extracted and then reverse transcribed into cDNA. The ABI-7500 real-time quantitative PCR instrument was used for qPCR experiments, and GAPDH was used as the internal reference to calculate the expression of each gene in each sample. Finally, the relative expression of each gene in each group was determined based on the control group (fold change of control), and a heatmap was drawn [51]. The primers of ADAM10 were Forward Sequence, 5'-GAGGAGTGTACGTGTGCCAGTT-3' and Reverse Sequence, 5'-GACCACTGAAGTGCCTACTCCA-3'. The primers of hsa-pre-miR-140-3p were: (1) RT-primer, 5'-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCCGTG-3', the Forward Sequence, 5'-AGAACCACGGGTCGTATCCA and Reverse Sequence, 5'-CAGTGCGTGTCGTGGAGT-3. Other primers were gifts from Dr. Yan Ma in Department of Gastroenterology and Hepatology, Chinese PLA General Hospital, Beijing, China [35].
For the luciferase assays, A549 cells were co-transfected with the miRNA-140-3p, Luc-1 or Luc-1Mut and harvested for luciferase assays according to the previous publication [30]. Luciferase test kit produced by Promega Company was used for the experiment. After the cells were scraped off with a scraper, the cells were mixed with the lysate, and then the cells were broken by shaking (vortex shaking for 15 min). After that, the cells were centrifuged at 4 °C at 12000 rpm for 10 min, and the supernatant was collected as the cell sample. Luciferase activity and β-Gal activity to β-Gal activity reading corrects luciferase reading. Relative luciferase activation (folds to control) of each group is determined with luciferase activity of control group as unit 1.
Subcutaneous LUAD tumor model
The in vivo antitumor activity of agents on A549 cells was examined using the subcutaneous tumor model [52,53]. The use of nude mice and the relevant protocols were reviewed and approved by the Institutional Animal Care and Use Committee of the General Hospital of Northern Theater Command, People's Liberation Army of China. The subcutaneous tumor model was performed according to the methods described in previous studies. A549, previously transfected with plasmids/vectors, were injected into nude mice (approximately 5×106 cells per injection point). The mice were administered the indicated doses of gefitinib via oral administration. The length and width of tumor tissues were measured using a Vernier caliper and the tumor volume was calculated as tumor width × tumor width × tumor length / 2. Tumor tissue was accurately weighed using a precision balance.
Ethics statement
The collection, and usage of clinical specimens, and all the methods and experimental design/protocol were reviewed and approved by the ethics review organization: the Medical Ethics Committee of General Hospital of Northern Theater Command. All experiments were performed according to the Helsinki Declaration (WHO) with the written consent from patients.
The animal experiments related information (including animal breeding, animal welfare, experimental design and methods of animal experiments) were all reviewed and approved by the Animal Ethics Committee of General Hospital of Northern Theater Command. All experiments were performed in accordance with the UK Animals (Scientific Procedures) Act, 1986 and associated guidelines.
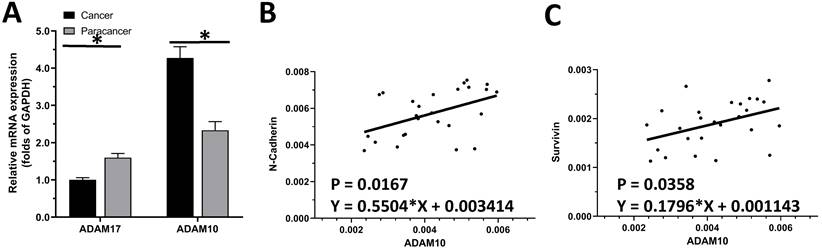
The endogenous level of ADAMs in clinical specimens. (A) The expression of ADAM10 and ADAM17 was detected in cancer (LSCC or LUAD) and paracancerous tissues, respectively. (B) Correlation analysis of N-Cadherin and ADAM10 in LSCC tissues. (C) Correlation analysis of survivin and ADAM10 in LSCC tissues.
Statistical analysis
Statistical analyses in the present work were performed by Bonferroni's correction with two-way ANOVA using SPSS Software (software version 9.0, IBM Corporation, Armonk, NY, USA). The IC50 values were calculated using an Origin software (Version No 6.1, OriginLab Corporation, Northampton, MA, USA). A P-value of <0.05 was considered to indicate statistically significant differences between two groups.
Results
The expression of ADAM10 was much higher compared with ADAM17 in LUAD clinical specimens
The expression of ADAM10 or ADAM17 in LUAD specimens was examined by qPCR. As shown in Figure 1A, the expression of ADAM17 and ADAM10 in the selected 30 pairs of clinical specimens was higher in LUAD tissues than in adjacent tissues, and the expression of ADAM10 in LUAD tissues was significantly higher than that of ADAM17. As an important control, the expression of ADAM17 is much higher in LSCC compared with LUAD (Figure 1A). This indicated that of the two ADAMs, ADAM10 is the ADAM subtype that plays a major role in LUAD.
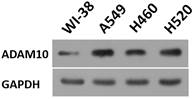
To further reveal the role of ADAM10 in LUAD, a correlation between the expression of ADAM10 with the Notch pathway members was examined. As shown in Figure 1B and C, the expression of ADAM10 was positively correlated with the expression of N-cadherin or survivin, two typical downstream factors of the Notch pathway, in LUAD clinical specimens. Moreover, the expression level of ADAM10 in lung derived cell lines was also examined by western blot. As shown in Figure 2, the expression of ADAM10 in NSCLC cells (A549, H460 and H520) was much higher compared with its expression level in WI-38, a lung derived non-tumor cell line. Therefore, these findings suggested that ADAM10 participates in the activation of Notch pathway in LUAD.
miR-140-3p targeting of ADMA10 in LUAD
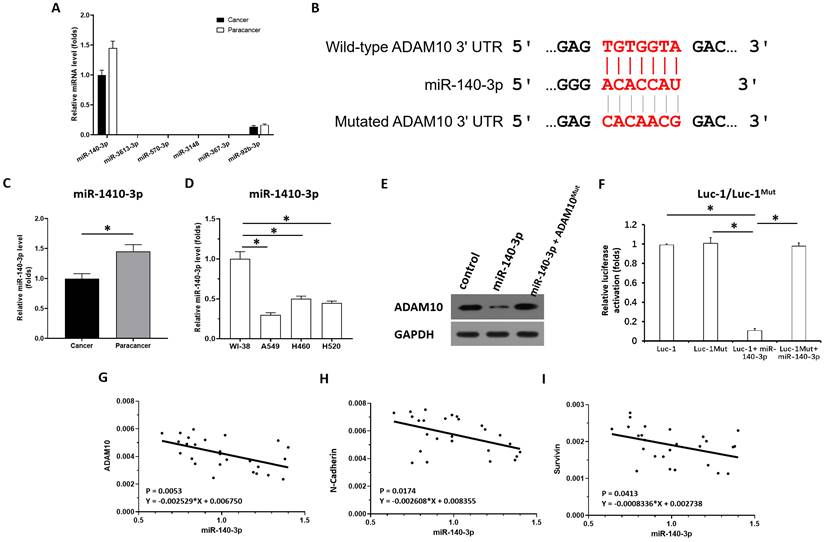
To reveal the potential mechanisms associated with the high expression of AMAM10 in LUAD, the expression level of miRNAs which were predicted by online tool miRDB potentially targeting to ADAM10 was examined in clinical specimens. As shown in Figure 3A, among the miRNAs (miR-140-3p, miR-3613-3p, miR-570-3p, miR-3148, miR-367-3p, miR-92b-3p) potentially targeting to ADAM10 with high scores, the miR-140-3p was detected in clinical specimens (the LUAD specimens and the paired non-tumor specimens); whereas miR-3613-3p, miR-570-3p, miR-367-3p, miR-3148, could not be detected in clinical specimens. Although miR-92b-3p can be detected in clinical specifications, its expression level is far lower than miR-140-3p. Therefore, miR-140-3p may be a miRNA with physiological and clinical significance that targets on ADAM10 in LUAD.
Next, as shown in Figure 3B and C, miR-140-3p could target the 3'UTR of ADAM10 in LUAD. The expression of miR-140-3p was much lower in LUAD specimens compared with the paired non-tumor tissues (Figure 3C). Moreover, the expression level of miR-140-3p in lung derived cell lines was also examined by qPCR. As shown in Figure 3D, the expression of miR-140-3p in NSCLC cells (A549, H460 and H520) was much lower compared with its expression level in WI-38, a lung derived non-tumor cell line. Overexpression of miR-140-3p downregulated the protein levels of wild-type ADAM10 (the endogenous ADAM10 expressed in A549 cells) but not the ADAM10 harboring a mutated miR-140-3p target site in the 3'UTR (ADAM10Mut) (Figure 3E) in A549 cells. Thus, miR-140-3p repressed the expression of ADAM10 in A549 cells. The effect of miR-140-3p on ADAM10 was further confirmed by the luciferase assays (Figure 3F). Moreover, the correlation between the expression of miR-140-3p and the Notch pathway was examined. As shown in Figure 3G, the expression of miR-140-3p was negatively associated with the expression of ADAM10 in LUAD clinical specimens. The expression of miR-140-3p was also negatively associated with the expression of N-cadherin or survivin, two typical downstream factors of the Notch pathway, in LUAD clinical specimens (Figure 3H and 3I). Therefore, these findings suggested that miR-140-3p could inhibit the expression of ADAM10 protein by targeting the 3'UTR of ADAM10 in LUAD.
miR-140-3p repressed the activation of the Notch pathway
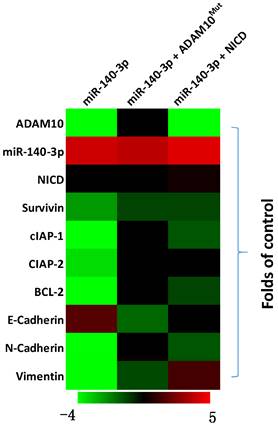
The above results indicated that miR-140-3p inhibited the expression of ADAM10 in A549 cells. To further examine the effects of miR-140-3p on the Notch pathway, the expression of Notch pathway's downstream factors was examined by the qPCR. As shown in Figure 4, overexpression of miR-140-3p in A549 cells repressed the expression of Notch pathway downstream anti-apoptosis or the pro-survival related factors (e.g., survivin, cIAPs) and EMT related factors (e.g., N-cadherin, vimentin). Co-transfection of A549 cells with miR-140-3p with ADAM10Mut or NICD almost completely abolished the inhibitory effects of miR-140-3p on the Notch pathway, while NICD did not influence the expression of ADAM10 in A549 cells. Therefore, miR-140-3p repressed the activation of Notch pathway by targeting to the 3'UTR of ADAM10 in A549 cells.
The expression level of ADAM10 in lung derived cell lines. The lung derived cell lines (WI-38 [a non-tumor cell line], A549 [a LUAD cell line], H460 [a LCC cell line], H520 [a LSCC cell line]) were cultured and harvested for western blot. The results were shown as images of western blot. GAPDH was used as the loading control.
miR-140-3p potentially targets the 3'UTR of ADAM10. (A) The miRNAs (miR-140-3p, miR-3613-3p, miR-570-3p, miR-3148, miR-367-3p, miR-92b-3p) potentially targeting to ADAM10 with high scores was examined in the clinical specimens (the LUAD specimens and the paired non-tumor specimens) via qPCR. (B) The targeting sites of miR-140-3p in the 3'UTR of ADAM10. (C) The expression levels of miR-140-3p were detected in cancer (LUAD) and para-cancerous tissue. (D) The expression level of miR-140-3p in lung derived cell lines (WI-38 [a non-tumor cell line], A549 [a LUAD cell line], H460 [a LCC cell line], H520 [a LSCC cell line]) was examined by qPCR. (E and F) The effects of miR-140-3p on the expression of ADAM10 in A549 cells were confirmed by western blotting assays (E) or luciferase assyas (F). (G) Correlation analysis of miR-140-3p and ADAM10 expression in LSCC tissues. (H) Correlation analysis of miR-140-3p and N-cadherin expression in LSCC tissues. (I) Correlation analysis of miR-140-3p and surviving expression in LSCC tissues, *P<0.05.
The effects of miR-140-3p on the Notch pathway in A549 cells. A549 cells were transfected with vectors and harvested for qPCR experiments. The related expression level (fold-change with respect to the control group) of miR-140-3p (hsa-pre-miR-140), AMDA10 or Notch pathway related factors are shown as a heat-map.
The specificity of miR-140-3p on the Notch pathway. The effects of miR-140-3p on the cleavage of Notch protein shown in cellular sub-fraction assays using A549 cells. The accumulation of NICD in the nuclear sub-fraction of A549 cells reflected the cleavage of Notch protein. Lamin A was used as the indicator of nuclear proteins; whereas the β-Actin was used as the indicator of cytoplasmic proteins. The results were shown as the images of western blot (A) or the quantitative results (B and C). *P<0.05.
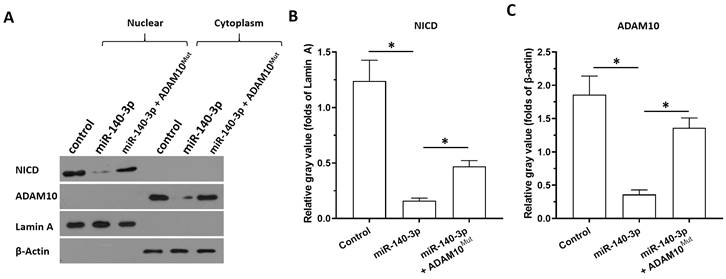
Next, the specificity of miR-140-3p on the ADAM10/Notch pathway was further examined using cellular sub-fraction assays. The cleaving of Notch protein was directly reflected by the accumulation of NICD in nuclear sub-fraction of A549 cells. As shown in Figure 5, ADAM10 was detected in the cytoplasm sub-fraction and the NICD was detected in the nuclear sub-fraction of A549 cells. Overexpression of miR-140-3p decreased the expression of ADAM10 in the cytoplasmic fraction and decreased the accumulation of NICD in the nuclear fraction of A549 cells. Therefore, miR-140-3p inhibited the cleavage of Notch protein and led to the nuclear accumulation of NICD to inhibit the activation of Notch pathway.
miR-140-3p enhanced the sensitivity of AC cells to antitumor agents
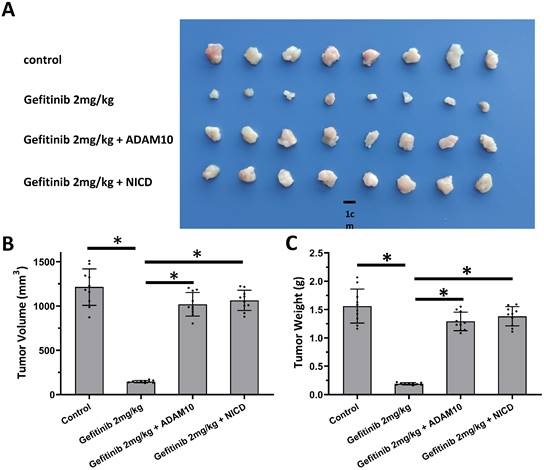
The above results revealed the effects of miR-140-3p on the ADAM10/Notch pathway. To confirm the role of ADAM10/Notch pathway, MTT and the nude mice model was used. As shown in Table 3, overexpression of ADAM10 or NICD enhanced the resistance of A549 cells to antitumor agents, and the IC50 values of these antitumor agents on A549 were increased. Overexpression of NICD or ADAM10 also enhanced the resistance of A549 cells to gefitinib, a typical molecular targeted agent used for LUAD treatment, in the nude mice model (Figure 6). These results confirmed the roles of ADAM10/Notch in LUAD cells.
Overexpression of NIVD or ADAM10 enhanced the resistance of LUAD cells to antitumor agents
| Agents | control | NICD | ADAM10 |
|---|---|---|---|
| IC50 values (μmol/L) | |||
| Gemcitabine | 0.30±0.12 | 1.77±0.87 | 1.66±0.28 |
| Paclitaxel | 0.10±0.01 | 0.62±0.37 | 0.59±0.66 |
| Etoposide | 0.44±0.05 | 2.34±0.48 | 1.94±0.80 |
| Doxorubicin | 0.29±0.04 | 1.62±0.43 | 1.81±0.41 |
| Gefitinib | 1.79±0.35 | 10.68±0.31 | 9.94±1.11 |
| Erlotinib | 1.55±0.94 | 9.08±0.74 | 10.42±0.63 |
| Osimertinib | 1.08±0.47 | 12.28±1.01 | 11.03±0.95 |
| Anlotinib | 0.92±0.55 | 10.49±0.72 | 8.06±1.33 |
Table notes: NICD, the intra-cellular domain of Notch protein; AMDA10, A disintegrin and metalloproteinase domain 10.
To further examine the effects of miR-140-3p on LUAD's sensitivity to antitumor drugs, the MTT assays were performed. As shown in Table 3, the antitumor agents, including the cytotoxic chemotherapies (gemcitabine, paclitaxel, etoposide or doxorubicin) or the molecular targeted agents (gefitinib, erlotinib, osimertinib or anlotinib) inhibited the survival of A549 cells in a dose-dependent manner. Overexpression of miR-140-3p enhanced the sensitivity of A549 cells to these agents: the IC50 values of these agents in A549 cells decreased in the presence of miR-140-3p (Table 3). Transfection of AMAM10Mut or NICD repressed the effects of miR-140-3p on the sensitivity of A549 cells to the antitumor agents (Table 3). To further confirm the effect of miR-140-3p on LUAD cells, H460 (LCC cell line) and H520 (LSCC cell line), two typical cell lines of the other two types of NSCLC were used. Overexpression of miR-140-3p enhanced the sensitivity of H460 (Table 4) or H520 (Table 5) cells to these agents: the IC50 values of these agents in (Table 4) or H520 (Table 5) cells decreased in the presence of miR-140-3p. Transfection of AMAM10Mut or NICD repressed the effects of miR-140-3p on the sensitivity of (Table 4) or H520 (Table 5) cells to the antitumor agents. Therefore, miR-140-3p enhanced the sensitivity of A549 cells to antitumor agents by targeting ADAM10.
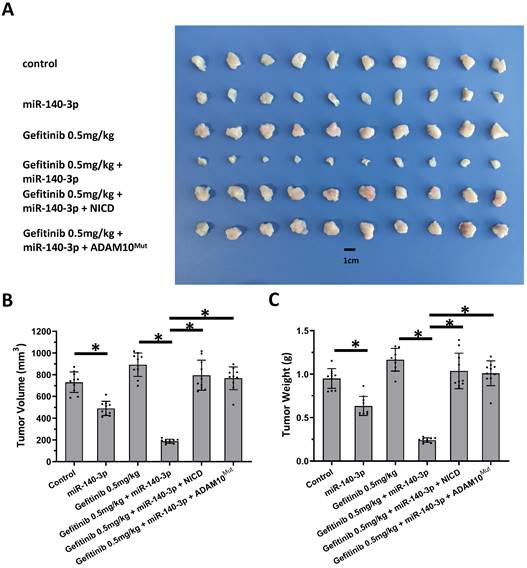
Next, the nude mice model was used in combination with gefitinib chosen as the representative drugs. As shown in Figure 7, oral administration of gefitinib inhibited the subcutaneous growth of A549 cells in a dose-dependent manner. Overexpression of miR-140-3p enhanced the in vivo antitumor activation of gefitinib on A549 cells. Moreover, as shown in Figure 8, overexpression of miR-140-3p in A549 cells exerted a certain anti-tumor activity, and gefitinib at a dose of 0.5 mg/kg was selected. Gefitinib at this dose had no significant anti-tumor activity (Figure 8). However, exposure to miR-140-3p significantly up-regulated the antitumor activity of 0.5 mg/kg gefitinib (Figure 8). Thus, the simultaneous transfection of NICD or ADAM10Mut inhibited the effects of miR-140-3p (Figure 8). These results indicated that miR-140-3p enhanced the in vitro and in vivo antitumor activation of antitumor agents by targeting the 3'UTR of ADAM10 in A549 cells (Figure 9).
miR-140-3p enhanced the sensitivity of LUAD cells A549 to antitumor agents by targeting to the 3'UTR of ADAM10
| Agents | control | miR-140-3p | miR-140-3p + AMDA10Mut | miR-140-3p + NICD |
|---|---|---|---|---|
| IC50 values (μmol/L) | ||||
| Gemcitabine | 0.42±0.15 | 0.07±0.01 | 0.59±0.10 | 0.50±0.11 |
| Paclitaxel | 0.11±0.01 | 0.03±0.00 | 0.15±0.01 | 0.13±0.00 |
| Etoposide | 0.45±0.10 | 0.12±0.06 | 0.50±0.07 | 0.57±0.34 |
| Doxorubicin | 0.37±0.22 | 0.06±0.02 | 0.48±0.20 | 0.44±0.06 |
| Gefitinib | 1.99±0.65 | 0.61±0.23 | 2.21±0.78 | 2.41±0.87 |
| Erlotinib | 1.74±0.29 | 0.36±0.08 | 1.80±0.86 | 1.65±0.26 |
| Osimertinib | 1.22±0.62 | 0.25±0.05 | 1.24±0.23 | 1.00±0.32 |
| Anlotinib | 1.05±0.30 | 0.14±0.03 | 0.90±0.21 | 1.37±0.25 |
Table notes: NICD, the intra-cellular domain of Notch protein; AMDA10Mut, A disintegrin and metalloproteinase domain 10 with mutated miR-140-3p's targeting sites in 3'UTR.
Activation of Notch pathway by Notch protein's cleavage enhanced the resistance of A549 cells. A549 cells which were transfected with expression vectors (control, NICD or ADAM10) were injected into the nude mice to form the subcutaneous tumor tissues. Mice received 2mg/kg dose of gefitinib via oral administration. The results shown are images of tumor tissues (A), the tumor volumes (B), and the tumor weights (C). *P<0.05.
Overexpression of miR-140-3p enhanced the sensitivity A549 cells to Gefitinib. A549 cells which were transfected with vectors (control or miR-140-3p) were injected into the nude mice to form the subcutaneous tumor tissues. Mice received the indicated doses of gefitinib via oral administration. The results are shown as images of tumor tissues (A), the tumor volumes (B), and the tumor weights (C). *P<0.05.
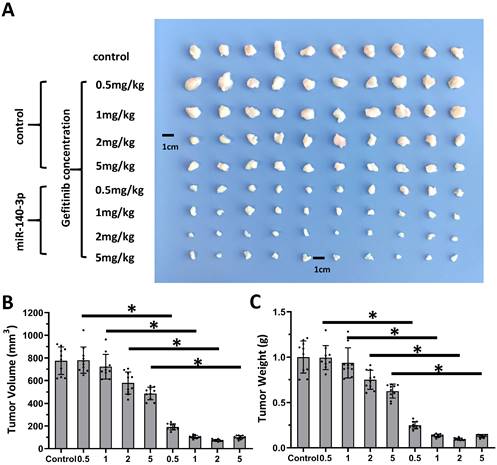
Overexpression of miR-140-3p enhanced the sensitivity A549 cells to Gefitinib. A549 cells which were transfected with vectors (control, miR-140-3p, miR-140-3p + NICD or miR-140-3p + ADAM10Mut) were injected into nude mice to form the subcutaneous tumor tissues. Mice received the indicated 0.5 mg/kg dose of gefitinib via oral administration. The results are shown as images of tumor tissues (A), tumor volumes (B) and tumor weights (C). *P<0.05.
miR-140-3p enhanced the sensitivity of LCC cells H460 to antitumor agents by targeting to the 3'UTR of ADAM10
| Agents | control | miR-140-3p | miR-140-3p + AMDA10Mut | miR-140-3p + NICD |
|---|---|---|---|---|
| IC50 values (μmol/L) | ||||
| Gemcitabine | 0.35±0.25 | 0.06±0.02 | 0.50±0.08 | 0.38±0.29 |
| Paclitaxel | 0.23±0.05 | 0.01±0.00 | 0.20±0.00 | 0.25±0.26 |
| Etoposide | 0.46±0.19 | 0.06±0.03 | 0.44±0.16 | 0.40±0.13 |
| Doxorubicin | 0.30±0.08 | 0.12±0.04 | 0.30±0.05 | 0.35±0.11 |
| Gefitinib | 2.35±0.21 | 1.03±0.56 | 2.15±0.68 | 1.56±0.70 |
| Erlotinib | 1.87±0.45 | 0.65±0.48 | 1.91±0.41 | 1.95±0.75 |
| Osimertinib | 1.50±0.42 | 0.32±0.07 | 1.73±0.21 | 1.61±0.18 |
| Anlotinib | 1.23±0.16 | 0.35±0.24 | 1.00±0.62 | 0.98±0.24 |
Table notes: NICD, the intra-cellular domain of Notch protein; AMDA10Mut, A disintegrin and metalloproteinase domain 10 with mutated miR-140-3p's targeting sites in 3'UTR.
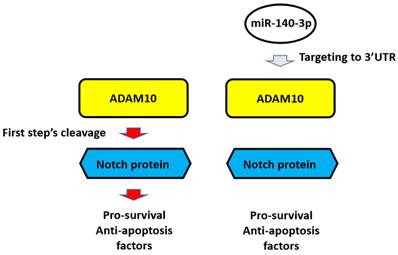
The proposal model of miR-140-3p and its downstream pathway in LUAD in the present work. In A549 cells, Notch protein can be cleaved by ADAM10 and the NICD of Notch protein translocated into the nuclear to mediate the pro-survival or anti-apoptosis rated factors' expression. These factors participate in the resistance of A549 cells to antitumor drugs. Overexpression of miR-140-3p inhibited the expression of ADAM10 by targeting to ADAM10's 3'UTR and in turn inhibited the activation of Notch pathway via inhibiting the cleavage of Notch protein by ADAM10.
The compensatory role of ADAM17 and ADAM10 in inducing LUAD cells to resist tumor drug resistance
The above results mainly focused on the effects of miR-140-3p on the ADAM10/Notch pathway. To confirm the role of ADAM10/Notch pathway and the compensatory role of ADAM17 and ADAM10 in inducing LUAD cells to resist tumor drug resistance, MTT method was used. As shown in Table 6, Inhibition of ADAM17 expression by siRNA can increase the sensitivity of A549 cells to anti-tumor drugs; ADAM10 was overexpressed in A549 cells. It can completely inhibit the effect of ADAM17. Similarly, inhibition of ADAM10 expression by siRNA could increase the sensitivity of A549 cells to antitumor drugs; ADAM17 was overexpressed in A549 cells. It can completely inhibit the effect of ADAM10. These results can reflect the compensatory role of ADAM10-ADAM17 in the anti-cancer drug resistance of A549 cells.
miR-140-3p enhanced the sensitivity of LSCC cells H520 to antitumor agents by targeting to the 3'UTR of ADAM10
| Agents | control | miR-140-3p | miR-140-3p + AMDA10Mut | miR-140-3p + NICD |
|---|---|---|---|---|
| IC50 values (μmol/L) | ||||
| Gemcitabine | 0.32±0.08 | 0.08±0.01 | 0.34±0.10 | 0.56±0.44 |
| Paclitaxel | 0.15±0.06 | 0.04±0.01 | 0.11±0.00 | 0.16±0.08 |
| Etoposide | 0.69±0.45 | 0.10±0.00 | 0.50±0.04 | 0.57±0.10 |
| Doxorubicin | 0.41±0.25 | 0.05±0.03 | 0.37±0.12 | 0.31±0.03 |
| Gefitinib | 1.77±0.39 | 0.76±0.20 | 1.60±0.58 | 1.74±0.21 |
| Erlotinib | 1.66±0.33 | 0.26±0.12 | 1.10±0.39 | 1.85±0.17 |
| Osimertinib | 1.65±0.51 | 0.18±0.11 | 1.77±0.63 | 1.75±0.43 |
| Anlotinib | 1.24±0.30 | 0.17±0.09 | 1.06±0.86 | 1.21±0.75 |
Table notes: NICD, the intra-cellular domain of Notch protein; AMDA10Mut, A disintegrin and metalloproteinase domain 10 with mutated miR-140-3p's targeting sites in 3'UTR.
Discussion
The Notch protein plays a central role in the Notch pathway. There are four sub-types of Notch protein (Notch receptors): NOTCH-1, NOTCH-2, NOTCH-3, and NOTCH-4 characterized by the extra-cellular domains and intracellular domains [54]. Among these four types of Notch receptors, the NICD is highly conserved, however, the Notch extracellular domains (EICD) of the four types of Notch receptor shows high sequence variation [55]. Therefore, in this study, transfecting the expression vector for NICD in A549 cells, or detecting the expression of NICD in the nucleus in the cellular sub-fraction experiment, could avoid the compensatory effect between the four Notch proteins. The expression of NICD was visually observed by the effect of Adam10 on the first step of cleavage of NOTCH proteins in the nucleus. Using miRNA to lower the expression of ADAM10 inhibited the NOTCH pathway, and ultimately up-regulated the sensitivity of malignant tumor cells against tumor drugs. Yang et al.2019 showed that repressing the expression of ADAM17 in HCC cells by miR-3163 enhanced the sensitivity of HCC cells to molecular targeted agents [28]. Conversely, studies investigating γ-secretase, which mediates the second step of Notch cleavage is also of great significance [25]. The results of Zhao et al. 2021 showed that miR-27-3p can act on the catalytic subunit PSEN-1 of γ-secretase to inhibit the cleavage of Notch, and ultimately up-regulate the sensitivity of triple-negative breast cancer (TNBC) cells to Olaparib [25]. Studies by Jia et al. 2021 reviewed the small molecule inhibitors that act on γ-secretase in detail [26,56]. γ-secretase has complex structure consisting of an intramembrane aspartate-lyase with multi-subunits, including the PSEN-1, the nicastrin subunit (NCSTN), the anterior pharynx-defective subunit (APH-1), and the presenilin enhancer subunit (PEN-2) [26,56]. When targeting PSEN1 alone, the effects of other subunits on PSEN-1 also need to be considered.
There is a compensatory effect between ADAM10 and ADAM17 in the induction of anti-tumor drug resistance in A549 cells
| Agents | control | siADAM17 | siADAM10 | siADAM17+ADAM10 | siADAM10+ADAM17 |
|---|---|---|---|---|---|
| IC50 values (μmol/L) | |||||
| Gemcitabine | 0.50±0.27 | 0.31±0.14 | 0.11±0.00 | 0.56±0.31 | 0.45±0.13 |
| Paclitaxel | 0.15±0.02 | 0.07±0.02 | 0.02±0.00 | 0.24±0.09 | 0.34±0.15 |
| Etoposide | 0.42±0.06 | 0.18±0.05 | 0.06±0.01 | 0.58±0.21 | 0.58±0.27 |
| Doxorubicin | 0.49±0.24 | 0.23±0.10 | 0.13±0.07 | 0.52±0.30 | 0.63±0.25 |
| Gefitinib | 2.15±0.32 | 0.71±0.54 | 0.38±0.10 | 2.40±0.96 | 1.94±0.77 |
| Erlotinib | 1.80±0.43 | 0.97±0.46 | 0.40±0.26 | 1.97±0.52 | 1.68±0.73 |
| Osimertinib | 1.33±0.67 | 0.59±0.12 | 0.25±0.09 | 1.66±0.85 | 1.86±0.33 |
| Anlotinib | 1.29±0.63 | 0.48±0.09 | 0.21±0.05 | 1.35±0.91 | 1.51±0.64 |
Table notes: NICD, the intra-cellular domain of Notch protein; AMDA10Mut, A disintegrin and metalloproteinase domain 10 with mutated miR-140-3p's targeting sites in 3'UTR.
Currently, an important part of the anti-tumor treatment strategy of NSCLC, especially LUAD, is treatment with a variety of anti-tumor drugs—mainly various cytotoxic chemotherapeutics and molecular targeted drugs as protein kinase inhibitors or the combined use of these drugs [57,58]. The present study investigated four cytotoxic chemotherapeutic agents and four molecular targeted drugs. Overexpression of miR-140-3p in A549 cells up-regulated the anti-tumor activity of the selected drugs by acting on ADAM10. These findings were essentially consistent with our expectations: the activation of the Notch pathway up-regulates the resistance of cells to anti-tumor treatment strategies, so there should be no selective resistance to cytotoxic drugs or molecular targeted drugs. On this basis, the four selected cytotoxic chemotherapy drugs, gemcitabine, paclitaxel, etoposide or doxorubicin, all act on biological macromolecules such as DNA and microtubules, and kill malignant tumor cells by causing macromolecular damage [59-62]. Among the selected four molecularly-targeted drugs, gefitinib, erlotinib and osimertinib, all mainly act on the EGFR, while anlotinib is a multi-target protein kinase inhibitor that targets the VEGFR [63,64]. Gefitinib is considered the first-generation of small molecule protein kinase inhibitors acting on EGFR, while erlotinib and osimertinib are the second- and third-generation agents acting on small molecule protein kinase inhibitors [63,64]; all these drugs were used in this study and killed A549 cells in a dose-dependent manner. These drugs mainly act on the EGFR and the difference in their activity is mainly determined by the higher mutational status of the EGFR [64]. In the future, it may be of great significance to detect EGFR mutations in clinical specimens and prepare corresponding patients derived cells (PDCs). It is worth mentioning that traditional molecular targeted drugs, as represented by Gefitinib, all act on the EGFR, and related research and development strategies should continue to exploit this potential. Currently, new molecular targeted drugs, represented by anlotinib act on VEGFR-related targets [63,64], have been approved for marketing for NSCLC treatment, which will provide additional therapeutic options for NSCLC patients.
In addition to AC, NSCLC also includes LSCC [65]. At present, the research on LUAD is relatively in-depth, while studies on LSCC are relatively lagging behind, with several reports on LUAD-related molecular targeted drugs but few studies on LSCC [51]. Zhou et al. reported that VEGFR and other anlotinib targets are widely expressed in LSCC and their expression is not significantly different from the expression in LUAD [51], in addition, anlotinib can also effectively kill LSCC cells [51]. This study focuses on A549, the most common and recognized cell line model of LUAD, although, we will study the role of ADAMs in LSCC further in the future. The results of this study show that ADAM10 plays a major role in LUAD among AMAMs, while ADAM17 may play a major role in LSCC. It is worth mentioning that Qingchun Zhao's research group used a research strategy of old drugs for new applications, and found that terfenadine can act as a small molecule inhibitor of ADAM17, and can inhibit the activity of the ADAM17/Notch pathway in NSCLC cells, ultimately overcoming NSCLC cells resistance and improve sensitivity to oncology drugs [26,37,66-69]. Similar to terfenadine, ZIDI-8, a superior compound, was discovered through structural optimization and transformation [26,37,66-69]. Since ADAM17 is not expressed at a lower level than ADAM10 in LUAD cells, an effect of these drugs on ADAM10 cannot be ruled out. In addition to LSCC and LUAD, NSCLC also includes large cell lung cancer, which also merits research attention. In the future, the expression level of ADAMs will also be tested in tissue specimens and cell lines of large cell lung cancer.
The miRNA-140-3p has been considered as an important regulator of NSCLC cells. Wan et al., 2021 reported that miRNA-140-3p represses the proliferation of lung adenocarcinoma cells by targeting thymidylate synthetase [70]; Hu et al., 2022 also reported that miR-140-3p inhibits the progression of NSCLC by targeting Janus kinase 1 [71]. One miRNA may have multiple targets, so it plays a variety of physiological functions [72-74]. This makes the results of miR-140-3p targeting on other targets reported in these articles do not affect the finding that miR-140-3p can target on ADAM10 in this study. Our study focused on the expression level of ADAM17 and ADAM10 in lung adenocarcinoma related specimens and the expression level of miRNA acting on ADAM10, and finally found that miR-140-3p in lung adenocarcinoma tissues has more clinical significance compared with other miRNAs. This study explored and preliminarily clarified that ADAM10 may play a major role in NSCLC compared with ADAM17, and miR-140-3p is not only one of the possible mechanisms of ADAM10 overexpression in NSCLC, but also can potentially be used as a new therapeutic strategy targeting ADAM10. The miR-140-3p could modulate the sensitivity of NSCLC cells by targeting other proteins. Wu et al., 2020 also showed that miR-140-3p could enhance the sensitivity of NSCLC cells to cisplatin by targeting Wnt/β-catenin signaling to attenuate the stem cell-like properties [75]. It is worth mentioning that this study mainly focuses on the downstream pathway of miR-140-3p (ADAM10/Notch), and the upstream signal pathway of miR-140-3p is also worth studying, and there have been some research reported: Wei et al., 2022 reported that Circ_0020123 enhances the cisplatin resistance in NSCLC cells by sponging miR-140-3p/HOXB5 [76]; Wang et al., 2022 revealed that Hypoxia-induced PVT1 promotes lung cancer chemoresistance by PVT1/miR-140-3p/ATG5 axis [77]. Such research is of great significance to better understand the molecular mechanism of miR-140-3p. The methylation analysis of miR-140-3p promoter region can also be attempted [78,79].
Acknowledgements
We deeply appreciated for the advice and supporting from Dr. Debin Ma in Department of Respiratory and Critical Care Medicine, Northern Theater General Hospital.
Funding
This work is supported by the “Doctor initial-scientific fund of Liaoning Province Science Foundation Program [ID: 2021-BS-033]” from Chinese Government (Liaoning Province).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chen WQ, Zheng RS, Baade DP. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34
3. Thai AA, Solomon BJ, Sequist LV. et al. Lung cancer. Lancet. 2021;398:535-554
4. Rosell R, Gonzalez-Cao M. Cemiplimab monotherapy in advanced non-squamous and squamous non-small cell lung cancer. Lancet. 2021;397:557-559
5. Ludmir EB, McCaw ZR, Grossberg AJ, Wei LJ, Fuller CD. Quantifying the benefit of non-small-cell lung cancer immunotherapy. Lancet. 2019;394:1904
6. Ma D, Jia H, Qin M. et al. MiR-122 Induces Radiosensitization in Non-Small Cell Lung Cancer Cell Line. Int J Mol Sci. 2015;16:22137-22150
7. Zhang J, Tang S, Zhang C. et al. Investigation of PALB2 Mutation and Correlation With Immunotherapy Biomarker in Chinese Non-Small Cell Lung Cancer Patients. Front Oncol. 2022;11:742833
8. Zhang J, Liu Z, Zhao W. et al. Discovery of Small Molecule NSC290956 as a Therapeutic Agent for KRas Mutant Non-Small-Cell Lung Cancer. Front Pharmacol. 2022;12:797821
9. Ma DB, Liu XY, Jia H. et al. A Novel Small-Molecule Inhibitor of SREBP-1 Based on Natural Product Monomers Upregulates the Sensitivity of Lung Squamous Cell Carcinoma Cells to Antitumor Drugs. Front Pharmacol. 2022;13:895744
10. Roskoski R Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2022 update. Pharmacol Res. 2022;175:106037
11. Roskoski R Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2020 update. Pharmacol Res. 2020;152:104609
12. Chen R, Qian Z, Xu X. et al. Exosomes-transmitted miR-7 reverses gefitinib resistance by targeting YAP in non-small-cell lung cancer. Pharmacol Res. 2021;165:105442
13. Zhu H, Liu H, Zhu JH. et al. Efficacy of ginseng and its ingredients as adjuvants to chemotherapy in non-small cell lung cancer. Food Funct. 2021;12:2225-2241
14. Cui Q, Hu Y, Cui Q. et al. Osimertinib Rechallenge with Bevacizumab vs. Chemotherapy Plus Bevacizumab in EGFR-Mutant NSCLC Patients With Osimertinib Resistance. Front Pharmacol. 2022;12:746707
15. Lim SM, Kim CG, Lee JB. et al. Patritumab Deruxtecan: Paving the Way for EGFR-TKI-Resistant NSCLC. Cancer Discov. 2022;12:16-19
16. Sano K, Nakadate K, Hanada K. Minocycline prevents and repairs the skin disorder associated with afatinib, one of the epidermal growth factor receptor-tyrosine kinase inhibitors for non-small cell lung cancer. BMC Cancer. 2020;20:279
17. Genova C, Dellepiane C, Carrega P. et al. Therapeutic Implications of Tumor Microenvironment in Lung Cancer: Focus on Immune Checkpoint Blockade. Front Immunol. 2022;12:799455
18. Long K, Gu L, Li L. et al. Small-molecule inhibition of APE1 induces apoptosis, pyroptosis, and necroptosis in non-small cell lung cancer. Cell Death Dis. 2021;12:503
19. Meng L, Xu J, Ye Y. et al. The Combination of Radiotherapy with Immunotherapy and Potential Predictive Biomarkers for Treatment of Non-Small Cell Lung Cancer Patients. Front Immunol. 2021;12:723609
20. Zhang Y, Li D, Feng F. et al. Progressive and Prognosis Value of Notch Receptors and Ligands in Hepatocellular Carcinoma: A Systematic Review and Meta-analysis. Sci Rep. 2017;7:14809
21. Sharif A, Shaji A, Chammaa M. et al. Notch Transduction in Non-Small Cell Lung Cancer. Int J Mol Sci. 2020;21:5691
22. Li X, Wang Y, Li X. et al. The Impact of NOTCH Pathway Alteration on Tumor Microenvironment and Clinical Survival of Immune Checkpoint Inhibitors in NSCLC. Front Immunol. 2021;12:638763
23. Jia H, Yang Q, Wang T. et al. Rhamnetin induces sensitization of hepatocellular carcinoma cells to a small molecular kinase inhibitor or chemotherapeutic agents. Biochim Biophys Acta. 2016;1860:1417-1430
24. Kang J, Kim E, Kim W. et al. Rhamnetin and cirsiliol induce radiosensitization and inhibition of epithelial-mesenchymal transition (EMT) by miR-34a-mediated suppression of Notch-1 expression in non-small cell lung cancer cell lines. J Biol Chem. 2013;288:27343-27357
25. Zhao M, Sun B, Wang Y. et al. miR-27-3p Enhances the Sensitivity of Triple-Negative Breast Cancer Cells to the Antitumor Agent Olaparib by Targeting PSEN-1, the Catalytic Subunit of Γ-Secretase. Front Oncol. 2021;11:694491
26. Jia H, Liu M, Wang X. et al. Cimigenoside functions as a novel γ-secretase inhibitor and inhibits the proliferation or metastasis of human breast cancer cells by γ-secretase/Notch axis. Pharmacol Res. 2021;169:105686
27. Sezin T, Selvakumar B, Scheffold A. The role of A Disintegrin and Metalloproteinase (ADAM)-10 in T helper cell biology. Biochim Biophys Acta Mol Cell Res. 2022;1869(4):119192. doi: 10.1016/j.bbamcr.2021.119192
28. Nigam K, Srivastav RK. Notch signaling in oral pre-cancer and oral cancer. Med Oncol. 2021;38(12):139
29. Yang B, Wang C, Xie H. et al. MicroRNA-3163 targets ADAM-17 and enhances the sensitivity of hepatocellular carcinoma cells to molecular targeted agents. Cell Death Dis. 2019;10:784
30. Li B, Feng F, Jia H. et al. Rhamnetin decelerates the elimination and enhances the antitumor effect of the molecular-targeting agent sorafenib in hepatocellular carcinoma cells via the miR-148a/PXR axis. Food Funct. 2021;12:2404-2417
31. Cui Y, Lyu X, Ding L. et al. Global miRNA dosage control of embryonic germ layer specification. Nature. 2021;593:602-606
32. Liu J, Yang C, Huang XM. et al. Knockdown of FBI-1 Inhibits the Warburg Effect and Enhances the Sensitivity of Hepatocellular Carcinoma Cells to Molecular Targeted Agents via miR-3692/HIF-1α. Front Oncol. 2021;11:796839
33. Du FX, Sun HW, Sun F. et al. Knockdown of TANK-Binding Kinase 1 Enhances the Sensitivity of Hepatocellular Carcinoma Cells to Molecular-Targeted Drugs. Front Pharmacol. 2022;13:924523
34. Oncology Branch of Chinese Medical Association, Journal of Chinese Medical Association Guidelines for Clinical Diagnosis and Treatment of Lung Cancer of the Chinese Medical Association (2022 Edition) [J]. National Medicine Journal of China, 2022, 102 (23): 1706-1740. DOI: 10.3760/cma. j.cn112137-20220413-00795 (In Chinese).
35. Ma Y, Chai N, Jiang Q. et al. DNA methyltransferase mediates the hypermethylation of the microRNA 34a promoter and enhances the resistance of patient-derived pancreatic cancer cells to molecular targeting agents. Pharmacol Res. 2020;160:105071
36. Wang JH, Zeng Z, Sun J. et al. A novel small-molecule antagonist enhances the sensitivity of osteosarcoma to cabozantinib in vitro and in vivo by targeting DNMT-1 correlated with disease severity in human patients. Pharmacol Res. 2021;173:105869
37. Zhang Y, Li D, Jiang Q. et al. Novel ADAM-17 inhibitor ZLDI-8 enhances the in vitro and in vivo chemotherapeutic effects of Sorafenib on hepatocellular carcinoma cells. Cell Death Dis. 2018;9:743
38. Zou XZ, Zhou XH, Feng YQ. et al. Novel inhibitor of OCT1 enhances the sensitivity of human esophageal squamous cell carcinoma cells to antitumor agents. Eur J Pharmacol. 2021;907:174222
39. Feng YQ, Li BA, Feng F. et al. Novel mTOR Inhibitor Enhances the Sensitivity of Hepatocellular Carcinoma Cells to Molecular Targeting Agents. Onco Targets Ther. 2020;13:7165-7176
40. Jie Y, Liu G, E M. et al. Novel small molecule inhibitors of the transcription factor ETS-1 and their antitumor activity against hepatocellular carcinoma. Eur J Pharmacol. 2021;906:174214
41. Wang Y, Liu S, Chen Q. et al. Novel small molecular inhibitor of Pit-Oct-Unc transcription factor 1 suppresses hepatocellular carcinoma cell proliferation. Life Sci. 2021;277:119521
42. Ma H, Yao Y, Wang C. et al. Transcription factor activity of estrogen receptor α activation upon nonylphenol or bisphenol A treatment enhances the in vitro proliferation, invasion, and migration of neuroblastoma cells. Onco Targets Ther. 2016;9:3451-3463
43. Yang Q, Feng F, Zhang F. et al. LINE-1 ORF-1p functions as a novel HGF/ETS-1 signaling pathway co-activator and promotes the growth of MDA-MB-231 cell. Cell Signal. 2013;25:2652-2660
44. Lu Y, Feng F, Yang Y. et al. LINE-1 ORF-1p functions as a novel androgen receptor co-activator and promotes the growth of human prostatic carcinoma cells. Cell Signal. 2015;25:479-489
45. Chen Y, Feng F, Gao X. et al. MiRNA153 Reduces Effects of Chemotherapeutic Agents or Small Molecular Kinase Inhibitor in HCC Cells. Curr Cancer Drug Targets. 2015;15:176-187
46. Feng F, Lu YY, Zhang F. et al. Long interspersed nuclear element ORF-1 protein promotes proliferation and resistance to chemotherapy in hepatocellular carcinoma. World J Gastroenterol. 2013;19:1068-1078
47. Shao Z, Li Y, Dai W. et al. ETS-1 induces Sorafenib-resistance in hepatocellular carcinoma cells via regulating transcription factor activity of PXR. Pharmacol Res. 2018;135:188-200
48. Feng F, Jiang Q, Cao S. et al. Pregnane X receptor mediates sorafenib resistance in advanced hepatocellular carcinoma. Biochim Biophys Acta Gen Subj. 2018;1862:1017-1030
49. Wang C, Ding S, Sun B. et al. Hsa-miR-4271 downregulates the expression of constitutive androstane receptor and enhances in vivo the sensitivity of non-small cell lung cancer to gefitinib. Pharmacol Res. 2020;161:105110
50. Du Y, Shi X, Ma W. et al. Phthalates promote the invasion of hepatocellular carcinoma cells by enhancing the interaction between Pregnane X receptor and E26 transformation specific sequence 1. Pharmacol Res. 2021;169:105648
51. Zhou W, Gao Y, Tong Y. et al. Anlotinib enhances the antitumor activity of radiofrequency ablation on lung squamous cell carcinoma. Pharmacol Res. 2021;164:105392
52. Yang H, Zhang MZ, Sun HW. et al. A Novel Microcrystalline BAY-876 Formulation Achieves Long-Acting Antitumor Activity Against Aerobic Glycolysis and Proliferation of Hepatocellular Carcinoma. Front Oncol. 2021;11:783194
53. Jiang Q, Ma Y, Han J. et al. MDM2 Binding Protein Induces the Resistance of Hepatocellular Carcinoma Cells to Molecular Targeting Agents via Enhancing the Transcription Factor Activity of the Pregnane X Receptor. Front Oncol. 2021;11:715193
54. Shivapriya PM, Singh A, Pandey P. et al. Pathways in small cell lung cancer and its therapeutic perspectives. Front Biosci (Landmark Ed). 2021;26:1668-1678
55. Shen W, Huang J, Wang Y. Biological Significance of NOTCH Signaling Strength. Front Cell Dev Biol. 2021;9:652273
56. Jia H, Wang Z, Zhang J. et al. γ-Secretase inhibitors for breast cancer and hepatocellular carcinoma: From mechanism to treatment. Life Sci. 2021;268:119007
57. Tabbò F, Pisano C, Mazieres J. et al. How far we have come targeting BRAF-mutant non-small cell lung cancer (NSCLC). Cancer Treat Rev. 2022;103:102335
58. Boch T, Frost N, Sommer L. et al. Pathologic responses in oligometastatic NSCLC patients treated with neoadjuvant immune checkpoint blockade with and without chemotherapy followed by surgery. Lung Cancer. 2022;164:46-51
59. Zhu XH, Lang HD, Wang XL. et al. Synergy between dihydromyricetin intervention and irinotecan chemotherapy delays the progression of colon cancer in mouse models. Food Funct. 2019;10:2040-2049
60. Wu HC, Chen YF, Cheng MJ. et al. Different types of components obtained from Monascus purpureus with neuroprotective and anti-inflammatory potentials. Food Funct. 2021;12:8694-8703
61. Chen YX, Wang CJ, Xiao DS. et al. eIF3a R803K mutation mediates chemotherapy resistance by inducing cellular senescence in small cell lung cancer. Pharmacol Res. 2021;174:105934
62. Chen Y, Wang L, Liu T. et al. Inhibitory effects of Panax ginseng glycoproteins in models of doxorubicin-induced cardiac toxicity in vivo and in vitro. Food Funct. 2021;12:10862-10874
63. Luo M, Sun H, Jiang Q. et al. Novel Nanocrystal Injection of Insoluble Drug Anlotinib and its Antitumor Effects on Hepatocellular Carcinoma. Front Oncol. 2021;11:777356
64. Dong RF, Zhu ML, Liu MM. et al. EGFR mutation mediates resistance to EGFR tyrosine kinase inhibitors in NSCLC: From molecular mechanisms to clinical research. Pharmacol Res. 2021;167:105583
65. Wang X, Liu H, Zhai D. et al. Multiple Primary Lung Tumors of Different Pathological Types Including Squamous Cell Carcinoma, Adenocarcinoma, and Mixed Squamous Cell and Glandular Papilloma: A Case Report. Onco Targets Ther. 2022;15:13-19
66. Lu HY, Chu HX, Tan YX. et al. Novel ADAM-17 inhibitor ZLDI-8 inhibits the metastasis of hepatocellular carcinoma by reversing epithelial-mesenchymal transition in vitro and in vivo. Life Sci. 2020;244:117343
67. Lu HY, Zu YX, Jiang XW. et al. Novel ADAM-17 inhibitor ZLDI-8 inhibits the proliferation and metastasis of chemo-resistant non-small-cell lung cancer by reversing Notch and epithelial mesenchymal transition in vitro and in vivo. Pharmacol Res. 2019;148:104406
68. Li DD, Zhao CH, Ding HW. et al. A novel inhibitor of ADAM17 sensitizes colorectal cancer cells to 5-Fluorouracil by reversing Notch and epithelial-mesenchymal transition in vitro and in vivo. Cell Prolif. 2018;51:e12480
69. An L, Li DD, Chu HX. et al. Terfenadine combined with epirubicin impedes the chemo-resistant human non-small cell lung cancer both in vitro and in vivo through EMT and Notch reversal. Pharmacol Res. 2017;124:105-115
70. Wan SZ, Liu ZM, Chen Y. et al. MicroRNA-140-3p represses the proliferation, migration, invasion and angiogenesis of lung adenocarcinoma cells via targeting TYMS (thymidylate synthetase). Bioengineered. 2021;12:11959-11977
71. Hu C, Zou Y, Jing LL. miR-140-3p inhibits progression of non-small cell lung cancer by targeting Janus kinase 1. J Biosci. 2020;45:48
72. Ding Y, Li M, Tayier T. et al. Bioinformatics analysis of lncRNA-associated ceRNA network in melanoma. J Cancer. 2021;12:2921-2932
73. Hua YT, Xu WX, Li H. et al. Emerging roles of MiR-133a in human cancers. J Cancer. 2021;12:198-206
74. Zhang Y, Huang S, Yang G. et al. The Role of miRNAs during Endoplasmic Reticulum Stress Induced Apoptosis in Digestive Cancer. J Cancer. 2021;12:6787-6795
75. Wu SM, Wang HR, Pan YP. et al. miR-140-3p enhances cisplatin sensitivity and attenuates stem cell-like properties through repressing Wnt/β-catenin signaling in lung adenocarcinoma cells. Exp Ther Med. 2020;20:1664-1674
76. Wei D, Zeng J, Rong F. et al. Circ_0020123 enhances the cisplatin resistance in non-small cell lung cancer cells partly by sponging miR-140-3p to regulate homeobox B5 (HOXB5). Bioengineered. 2022;13:5126-5140
77. Wang JY, Dong ZY, Sheng ZY. et al. Hypoxia-induced PVT1 promotes lung cancer chemoresistance to cisplatin by autophagy via PVT1/miR-140-3p/ATG5 axis. Cell Death Discov. 2022;8:104
78. Ran Q, Xu D, Wang Q. et al. Hypermethylation of the Promoter Region of miR-23 Enhances the Metastasis and Proliferation of Multiple Myeloma Cells via the Aberrant Expression of uPA. Front Oncol. 2022;12:835299
79. Wang J, Huo C, Yin J. et al. Hypermethylation of the Promoter of miR-338-5p Mediates Aberrant Expression of ETS-1 and is correlated with Disease Severity Of Astrocytoma Patients. Front Oncol. 2021;11:773644
80. He W, Gong S, Wang X. et al. DNA methylation integratedly modulates the expression of Pit-Oct-Unt transcription factors in esophageal squamous cell carcinoma. J Cancer. 2021;12:1634-1643
Author contact
![]() Corresponding authors: Dr. and prof. Shiguang Xu (E-mail: xsg13309884079com); Department of Thoracic Surgery, General Hospital of Northern Theater Command, No.83, Wenhua Road, Shenhe District, Shenyang City 110016, Liaoning Province, China. Qiong Wu (E-mail: wuqiongxiongwaicom), Department of Thoracic Surgery, General Hospital of Northern Theater Command, No.83, Wenhua Road, Shenhe District, Shenyang City, 110011, China. Dr. and prof. Yingshi Zhang (E-mail: zhangyingshi526com), Department of Clinical Pharmacy, Shenyang Pharmaceutical University, No.103, Wenhua Road, Shenhe District, Shenyang City, 110011, Liaoning Province, China.
Corresponding authors: Dr. and prof. Shiguang Xu (E-mail: xsg13309884079com); Department of Thoracic Surgery, General Hospital of Northern Theater Command, No.83, Wenhua Road, Shenhe District, Shenyang City 110016, Liaoning Province, China. Qiong Wu (E-mail: wuqiongxiongwaicom), Department of Thoracic Surgery, General Hospital of Northern Theater Command, No.83, Wenhua Road, Shenhe District, Shenyang City, 110011, China. Dr. and prof. Yingshi Zhang (E-mail: zhangyingshi526com), Department of Clinical Pharmacy, Shenyang Pharmaceutical University, No.103, Wenhua Road, Shenhe District, Shenyang City, 110011, Liaoning Province, China.

 Global reach, higher impact
Global reach, higher impact