3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(2):541-553. doi:10.7150/jca.61437 This issue Cite
Research Paper
CircRNA_400029 promotes the aggressive behaviors of cervical cancer by regulation of miR-1285-3p/TLN1 axis
Department of Gynecology, Cancer Hospital of China Medical University, Liaoning Cancer Hospital and Institute, Shenyang, Liaoning 110042, P. R. China.
Received 2021-4-9; Accepted 2021-10-11; Published 2022-1-1
Abstract
Background: Cervical cancer (CC) is the highest incidence of female malignant tumor in China. Circular RNAs (circRNAs) has been reported to affect CC progression by altering mRNA stability at the transcriptional level or binding to miRNAs to produce competitive endogenous RNA (ceRNA). For this purpose, our study was aimed to investigate the function effects and the potential regulatory mechanism of the circRNA_400029 in CC cells.
Materials and methods: The expression levels of circRNA_400029 and miR-1285-3p were detected by real time polymerase chain reaction (RT-PCR). Similarly, the mRNA and protein levels of TLN1 (Talin 1) was detected by RT-PCR and Western blot. Cell-Counting Kit-8 (CCK-8), EdU and Flow cytometry assay were used to detect cell proliferation, cell cycle and apoptosis. Then the Transwell assays were used to test cell migration and invasion. Besides this, the functional targets were confirmed by Dual luciferase reporter assays. Tumor xenograft in nude mice checked the result in vivo.
Results: To begin with, circRNA_400029 was upregulated in CC cells and tissue. Knockdown circRNA_400029 inhibited cell proliferation, migration and invasion while induced cell apoptosis. Interestingly, miR-1285-3p targeted circRNA_400029 and down-regulated of miR-1285-3p could reverse the effects of circRNA_400029 weak-expression on progression and apoptosis of CC cells. Moreover, TLN1 was up-regulated in CC cells and identified as a direct target of miR-1285-3p. Meanwhile, we found that miR-1285-3p negatively regulated the function of TLN1. Finally, the circRNA_400029/miR-1285-3p/TLN1 axis could affect tumor growth in vivo.
Conclusion: The overexpressed circRNA_400029 promoted CC proliferation, migration and invasion while deduced apoptosis by sponging miR-1285-3p to regulate TLN1. CircRNA_400029 was a potential onco-circRNA in CC, and might be a promised therapy target.
Keywords: Cervical cancer, circRNA_400029, miR-1285-3p, TLN1, proliferation, invasion
Introduction
Cervical cancer (CC) is the most common malignant tumor in women with relatively high morbidity and mortality and remains the main cause of cancer-related deaths among women in developing countries [1]. Prevention and control of CC is difficult on account of the risk factors such as bacterial factors, virus factors, environmental factors and genetic susceptibility [2]. Among them, high risk human papillomavirus (HPV) infection is the main cause of CC [3]. However, the occurrence and development of cervical cancer is a multi-stage, involving the abnormal regulation of multiple genes in the pathophysiological process. As screening has become more widespread and treatment has improved, the mortality rate for early-stage patients has been falling [4]. However, the prognosis for patients with advanced stage is still poor.
Non-coding RNAs, including long non-coding RNAs (lncRNAs), circular RNAs (circRNAs) and microRNAs (miRNAs), do not encode proteins but interact with other RNAs, proteins and DNA to modulate their functions have become the stars of molecular biology research [5]. Recent evidence suggested that circRNA acted as putative biomarkers involved in the pathogenesis of human cancer by regulating different gene functions [6-8]. CircRNAs is a special structure of non-coding RNA with covalently closed circular structure, which is produced by the tail-head splicing of its host gene pre-messenger RNA (mRNA). It is widely expressed in eukaryotes and plays a role in specific patterns of tissue and developmental stages. The circRNA circFoxO3a was identified as a serum biomarker for squamous cervical cancer (SCC) [9]. Furthermore, a previous document uncovered that the upregulation of circRNA_PVT1 could induce invasion and metastasisi of cervical cancer cells via exosome pathway [10]. Meanwhile, knockdown of circ-0084927 restrained proliferation and facilitated apoptosis through sponging miR-634 and regulating TPD52 level in CC [11]. These outcomes showed that circRNA had a great effect on CC and could serve as a potential clinical test biomarker in CC treatment.
MicroRNAs (miRNAs) are noncoding RNA molecules containing approximately 23 nts that playing gene-regulatory roles on proteins expression and evolution in different cells [12]. Previous reports presented that miR-7 modulated multiple molecular targets and served as a potential therapeutic target in CC [13]. Up-regulated miR-216a-3p could suppress the proliferation and invasion of CC through modulated multiple molecular targets and served as a new positive regulatory factor of YAP signaling pathway. miRNA-499a promotede the cell migration and invasion and enhanced chemoresistance of CC cells by targeting SOX6 [14]. In addition, exosomal miRNA is also an important target for remodeling tumor microenvironment and molecular diagnosis [15].
In this study, over-expressed circRNAs in CC were obtained through high-throughput microarray, and the potential molecular targets were analyzed and verified. We found the circRNA_400029 was overexpressed in CC cells and tissue. It contributed the CC cell proliferation, migration and invasion while retarded apoptosis. Mechanically, it was achieved by the circRNA_400029/miR-1285-3p/TLN1 (Talin 1) axis.
Materials and methods
Cell culture and cell transfection
Four CC cell lines (SiHa, HeLa, CaSki, C-33A) and human cervical epithelial immortalized cell line H8 were obtained from were obtained from China Medical University (Shenyang, China). They were cultured with DMEM (Gibco BRL, USA) and 10% fetal bovine serum (FBS, USA) placed to incubator with 5% CO2 and 37 °C.
All oligomers used in this study including siRNA targeting circRNA_400029 junction site, miR-1285-3p mimics and inhibitors were commercially obtained from Sangon Biotech (Shanghai, China). Before transfection the SiHa and CaSki were cultured in 24-well plates with a density of 30% for 24 h, siRNAs were transfected into SiHa and CaSki cells by Lipofectamine 2000 (Invitrogen, USA). After transfection and cultured for 24 h, 48 h and 72 h, samples were collected for the next experiment.
Patients and specimens
A total of 50 CC tissues and their neighboring healthy tissues were obtained from Liaoning Province Cancer Hospital &Institute. All patients had a clear pathological diagnosis. The degrees of these CC patients were divided into stage on the basis of the AJCC TNM staging system. All patients had not received any therapy. All enrolled patients had no other tumors or systematic diseases shortening survival. These specimens were stored in -80 °C. And we got the approval of the Ethics Committee of Liaoning Province Cancer Hospital &Institute in this study and written informed consent was signed by every participant. The information of the patients were shown in Table 1.
CircRNA_400029 expressions and clinicopathologic characteristics
| Characteristics | circRNA_400029 | P | X2 | |
|---|---|---|---|---|
| Low (%) | High (%) | |||
| 26 | 24 | |||
| Age | 0.59 | 0.31 | ||
| ≥45 | 12 | 11 | ||
| <45 | 14 | 13 | ||
| Histology | 0.00 | 12.86 | ||
| Adenocarcinoma | 3 | 10 | ||
| Squamous carcinoma | 23 | 14 | ||
| Tumor Size | 0.36 | 0.93 | ||
| ≥4 cm | 13 | 10 | ||
| <4 cm | 13 | 14 | ||
| HPV infection | 0.72 | 0.40 | ||
| Negative | 2 | 1 | ||
| Positive | 24 | 23 | ||
| Tumor differentiation | 0.00 | 10.05 | ||
| Moderate and high | 11 | 19 | ||
| Poor | 15 | 5 | ||
| Lymph node invasion | 0.17 | 1.36 | ||
| Yes | 10 | 11 | ||
| No | 16 | 13 | ||
| TNM stage | 0.03 | 5.59 | ||
| I | 11 | 6 | ||
| II | 15 | 18 | ||
RNA isolation and RT-PCR
The total RNA of samples was separated and extracted by TRIzol (Invitrogen, USA). cDNA was obtained by the RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Canada). PCR was running in an ABI SYBR Green Master Mix (Invitrogen, USA). The primers sequences were as follows: 5'-CACAGTGCTGGGATTACA-3' and 5'-CAGGAGAATCGCTTGAATC-3' for circRNA_400029; 5'- TCTGGGCAACAAAGTGAG-3' and 5'- CTCAACTGGTGTCGTGGA -3' for miR-1285-3p; 5'-CTCGCTTCGGCAGCACA-3' and 5'- AACGCTTCACGAATTTGCGT-3' for U6; TLN-1: 5'- TGTAGAGGAGCACGAGACGC -3' and 5'- AAGGAGACAGGGTGGGAGC -3' for TLN1. These experiments were repeated 3 times and data was analyzed with 2-ΔΔCt method.
Nuclear and cytoplasmic separation assay
Following to the manufacturer's protocol, the PARIS kit (AM1921; Thermo Fisher Scientifc, Yokohama, Japan) divide the total cellular fractions into nuclear and cytoplasmic fractions. The kit isolates the nuclear and cytoplasmic components prior to RNA extraction, and ensures that RNA is isolated from the same experimental sample.
Immunofluorescence
At room temperature, fix the cells with 4% paraformaldehyde PBS for 10 minutes, and infiltrate with 0.1% Triton-X and 1% BSA PBS for 30 minutes. DNA staining was performed using 4',6-diamino-2-phenylindole (DAPI). An ECLIPSE Ni microscope (Nikon, Japan) was used for imaging.
RNA fluorescence in situ hybridization
SiHa and CaSki cells were fixed with 4% formaldehyde for 15 min, and then washed with PBS, treated with pepsin and dehydrated with ethanol. RNA fluorescence in situ hybridization (FISH) was performed using a RiboTM Fluorescent In situ Hybridization Kit (RiboBio, China) according to the manufacturer's instructions. The FITC-labeled hsa circRNA_400029 probe were detected and observed by ECLIPSE Ni microscope (Nikon, Japan).
CCK-8 proliferation vitality assay
The cell proliferation viability were detected by the Cell-Counting Kit-8 (Dojindo Laboratories). SiHa and CaSki cells were cultivated into a 96-well plate with a density of 5×104 cells/well and incubated in 37 °C for 24, 48, 72 or 96 hours. Then the cells proliferation viability was detected by a microplate reader (Bio-Rad, CA) using spectrophotometry.
Transwell assay
Cells migration and invasion were detected by transwell cell culture chambers with or without Matrigel (Corning Life Sciences, USA). To begin with, cells were added 100 ml of no-serum medium and inoculated to the upper chamber, and 500 ml of DMEM with 10% serum was inoculated to the lower chamber. Then the cells attaching to the lower surface of the upper chamber was disposed with 4% paraformaldehyde (PFA) after 24 h of incubation. Observation under the microscope after staining with crystal violet and statistics analysis.
5-ethynyl-20-deoxyuridine (EdU) staining proliferation assay
The proliferation of CC cells was measured by using EdU assay kit (RiboBio, Guangzhou, China). In breif, the transfected CC cells were incubated with 50 μm EdU for 2 h. And the cell nuclei were dyed with 4', 6-diamidino-2phenylindole (DAPI, Sigma, USA). Finnally, the EdU-positive cells were observed by using a fluorescence microscope.
Cell cycle
Pre-chilled 1 X PBS used to re-suspended cells. 1× Annexin binding buffer diluted cells to 1×105 cells/ml. Cells were precooled with 70% ethanol at 4 °C overnight before assay.
Flow cytometry
Transfected cells were dyed by Annexin V-FITC Apoptosis Detection Kits (BD, USA). Then collected and washed using phosphate-buffered saline (PBS). Added to 1× binding buffer and resuspended cells, then mixed into the test container 500 μL cell suspension, 5 μL Annexin V-FITC, and 5 μL propidium iodide solution incubated for 15 minutes. The samples apoptosis was detected by a FACScan flow cytometer analyzer (BD, USA).
Western blot analysis
Proteinase inhibitor (Solarbio) was added and tissue lysate was extracted using RIPA reagent. The concentration of protein was measured by BCA reagent (Beyotime Institute of Biotechnology). Then proteins were separated by 10% SDS-PAGE and transferred to PVDF membrane (Millipore, USA). After incubation with 5% skimmed milk in TBST, PBS was used to washing membrane and then cultivated with the primary antibody overnight at 4°C. This study used following antibodies: anti-TLN1 antibody (1:1000, Abcam, USA) and anti-β-actin antibody (1:1000, Santa Cruz, USA). After that we used the individual antibody for western blot analysis. The signals were observed by an enhanced chemiluminescence detection reagent (GE Healthcare, USA).
Dual luciferase reporter assays
CircRNA_400029 and miR-1285-3p were predicted having the binding sequences by RegRNA 2.0 [16]. Similarly, TLN1 and miR-1285-3p were also predicted having the binding sequences by TargetScan database [17]. Next to verify whether the circRNA_400029 and TLN1 as a directly target of miR-744-5p, then the wild type circRNA_400029 sequences (circRNA_400029-wt), mutant circRNA_400029 sequences (circRNA_400029-mut), wild type TLN1 3'UTR sequences (TLN1 3'UTR-wt) and mutant TLN1 3'UTR sequences (TLN1 3'UTR-mut) were cloned into the pGL-3 luciferase reporter vector (Promega, USA). The reporter vectors with miR-744-5p or miR-NC were constructed into 293T cells by Lipofectamine 2000 (Invitrogen, USA). After transfection for 36 h we detected the activity by Dual Luciferase Reporter Assay System (Promega, USA).
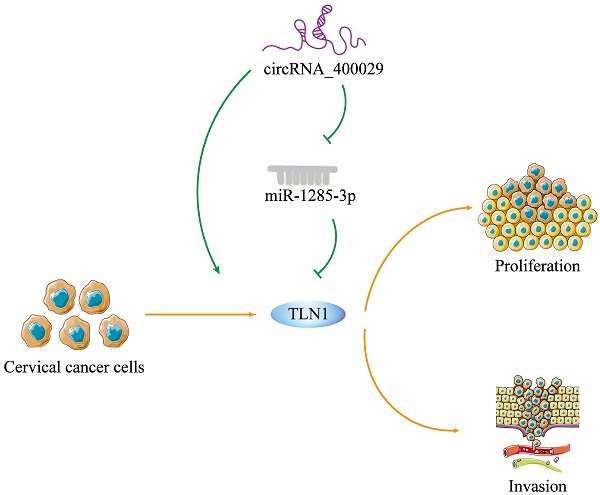
CircRNA_400029 was overexpressed in cervical cancer. A. CircRNA_400029 was overexpressed in CC cells and tissue. B. CircRNA_400029 expression in CC tissue compare to paired adjacent tissue in 50 patients. C, D. The expression level of circRNA_400029 in the subcellular fractions of CC cells was detected by RT-PCR. U6 and β-actin were used as nuclear and cytoplasmic markers, respectively. E. Immunofluorescence indicated that CircRNA_400029 was mainly localized in the cytoplasm (Yellow arrows). U6 was used as a loading control in RT-PCR; n=3, *P< 0.05.
Tumor xenograft in nude mice
Obtained four/five-week-old female BALB/c nude mice from Beijing Vital River Laboratory Animal Co., Ltd., (Beijing, China). Resuspend the cell concentration in PBS to 1×107 cells/ml. The cell suspension was then injected subcutaneously into the flanks of nude mice (0.1 ml). Measure the width and length of the tumor every 3 days. All nude mice were sacrificed three weeks after injection, xenogeneic tumors were excised and weighed. The tumor volume is calculated as (length×width2)/2. The animal experiment procedure was approved by the Animal Care and Use Committee of Liaoning Cancer Hospital.
Statistical analysis
All data were displayed as mean ± standard deviation (SD) of more than 3 experiment repeats and statistical analyses were finished by SPSS 23.0 software. The significance differences were analyzed by Student's t-test or one-way analysis of variance (ANOVA), P<0.05 considered as statistical significance.
Results
CircRNA_400029 was upregulated in cervical cancer cells and tissues
To investigate the potential role of circRNA_400029 in CC, we firstly detected the RNA level of circRNA_400029 in CC cells and tissues. RT-PCR showed that circRNA_400029 was significantly increased in CC cells, especially in SiHa and CaSki (Fig. 1A). Besides, circRNA_400029 was also up-regulated in CC tissues compared with adjacent normal tissues (Fig. 1B). These results indicated that circRNA_400029 might play an oncogenic role in CC. Subsequently, we tested the localization of circRNA_400029 in cells in order to further explore the function of circRNA_400029 in CC. The nuclear/cytoplasmic RNA fractionation in the subcellular distribution assay exhibited that circRNA_400029 was mainly located at cytoplasmic in both SiHa (Fig. 1C) and CaSki (Fig. 1D). Not surprisingly, the immunofluorescence also showed the similar results (Fig. 1E). So, we speculated that circRNA_400029 might participate in the corresponding molecular regulation and function at the post-transcriptional level. In addition, we chose the median of circRNA_400029 expression in CC samples as the cutoff value. The CC samples were classified as high- and low- groups based on the expression of circRNA_400029. We found that the expression of circRNA_400029 was closely related to histology, tumor differentiation, and TNM stage (Table 1).
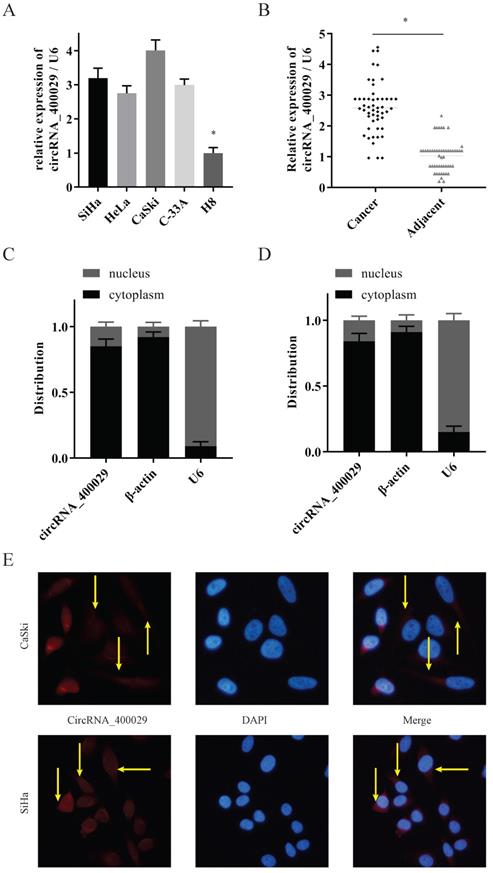
CircRNA_400029 could crosstalk with miR-1285-3p through direct binding. A. The diagram showed designed siRNAs targeting the junction site of circRNA_400029. B. RT-PCR analysis verifying the knockdown efficiency of siRNAs. C. The potential binding sites of circRNA_400029 and miR-1285-3p transcript. D, E. miR-1285-3p was weak-expressed in both CC cells and tissue. F. The expressions of circRNA_400029 and miR-1285-3p were negatively correlated. G. RT-PCR analyzed miR-1285-3p expression in CC cells treated with mimics and inhibitor. H. The mimics of miR-1285-3p inhibited the expression of circRNA_400029. I. Luciferase activity in 293T co-transfected with luciferase reporter encompassing circRNA_400029 and miR-1285-3p mimics. Results represent renilla and firefly luciferase activity ratio.
MiR-1285-3p targeted circRNA_400029 and negative regulated the expression of circRNA_400029
Then two siRNAs sequences were designed for circRNA_400029 junction sites to inhibit circRNA_400029 expression (Fig. 2A). Next, we transfected SiHa and CaSki cells with circRNA_400029 siRNAs. Among them, siRNA- circRNA_400029#1 had a better silence efficiency (Fig. 2B). CircRNA_400029 was predicted containing the binding site of miR-1285-3p by RegRNA 2.0 (Fig. 2C). Moreover, the expression of miR-1285-3p in CC cells and tissues was explored via RT-PCR. As expected, miR-1285-3p was weak-expressed in cells and tissues (Fig. 2D, 2E). The RT-PCR observations indicated that a negatively regulative association between circRNA_400029 and miR-1285-3p (r=0.77, P<0.01, Fig. 2F). Next, the mimics and the inhibitor of miR-1285-3p were transfected into CC cells to knockdown or up-regulated the expression of miR-1285-3p (Fig. 2G). Interestingly, the expression level of circRNA_400029 was detected in CC cells transfected with mimics of miR-1285-3p and the results showed that up-regulation of miR-1285-3p significantly inhibited the expression of circRNA_400029 (Fig. 2H). To confirm that, we constructed the pGL3 luciferase reporter plasmid containing the wild type circRNA_400029 sequences (circRNA_400029-wt) and mutant circRNA_400029 sequences (circRNA_400029-mut). Dual luciferase reporter assay revealed that miR-1285-3p decreased the luciferase expression of circRNA_400029-wt in 293T cells whereas the circRNA_400029-mut was not affected (Fig. 2I).
CircRNA_400029 functioned as ceRNA of TLN1 via sponging miR-1285-3p
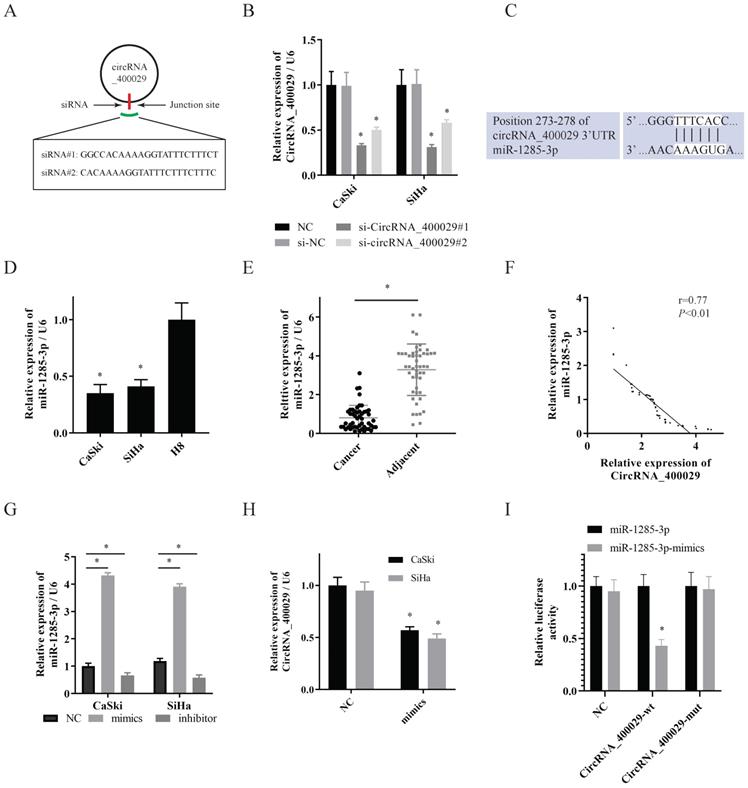
To uncover the potential target genes mediated by miR-1285-3p in CC cell, we identified TLN1 as one of the target gene of miR-1285-3p through the bioinformatics TargetScan database (Fig. 3A). The RT-PCR and western-blotting observations showed a substantial increase of TLN1 was observed in CC cell lines than in H8 (Fig. 3B, 3C). Subsequently, we also detected the expression of TLN1 CC and adjacent tissues. Consistent with circRNA_400029, TLN1 was also overexpressed in CC (Fig. 3D) and negatively correlated with miR-1285-3p expression (r=0.85, P<0.01, Fig. 3E). In addition, over-expressed miR-1285-3p could inhibit TLN1 mRNA and protein level (Fig. 3F, 3G). Based on this result, Dual-luciferase reporter assays observed that miR-1285-3p mimics transfection distinctly declining the luciferase signal of reporters comprising TLN1-WT, but no impact was detected on the luciferase signal of reporters comprising TLN1-MUT (Fig. 3H). The siRNA of circRNA_400029 could inhibited the protein level of TLN1 (Fig. 3I).
The biological function of circRNA_400029/miR-1285-3p/TLN1 axis in vitro
To further explore the functional effects of circRNA_400029 in CC cell lines, we chose the SiHa and CaSki cell lines for further phenotype experiments. Firstly, CCK-8 assay indicated that proliferation of CC cells transfected with si-circRNA_400029 was significant inhibited. On this basis, both si-circRNA_400029 and the inhibitor of miR-1285-3p were transfected; the cell proliferation ability was restored. It suggested that the effect of circRNA_400029 on CC proliferation was realized through miR-1285-3p (Fig. 4A, 4B). We conducted an EdU staining to further verify this conjecture. As expected, the fluorescence intensity of CC cells decreased after circRNA_400029 knockdown. The fluorescence intensity of CC cells recovered after siRNA and inhibitor co-transfection. This further supports the role of circRNA_400029 in CC cell proliferation (Fig. 4C). Moreover, the migration and invasion of SiHa and CaSki cells transfected with si-circRNA_400029 or miR-1285-3p inhibitor were detected by Transwell assay. The results revealed that down-regulation of circRNA_400029 suppressed CC cells migration and invasion (Fig. 4D). Furthermore, flow cytometry assessed cell cycle progression. Results confirmed that si-circRNA_400029 significantly increased the proportion of CC cells in S phase but decreased the proportion in G2/M phase. However, it could be recovered by miR-1285-3p inhibitor (Fig. 4E). Nevertheless, flow cytometry assay showed that transfection of si-circRNA_400029 result in an obvious increase of apoptosis rates in both SiHa and CaSki cells. And with the decreased of miR-1285-3p expression, apoptosis decreased (Fig. 4F).
Furthermore, the effects of miR-1285-3p and TLN1 on CC proliferation and migration were verified by the same method. CCK-8 assays displayed that an apparently reduced cell viability in si-TLN1 group, while a substantially augmented cell viability in si-TLN1 + miR-1285-3p inhibitor group (Fig. 5A, 5B). Similarly, the knockdown of TLN1 also reduced fluorescence intensity of CC cells. With the decrease of miR-1285-3p expression, the fluorescence intensity of the cells recovered (Fig. 5C). Moreover, the migration or invasion ability of CC cells were restrained when carrying si-TLN1 transfection, whereas strengthen when carrying both si-TLN1 and miR-1285-3p inhibitor transfection (Fig. 5D).
Similarly, we also performed flow cytometry assessed to evaluate cell cycle progression and apoptosis. Results indicated that si-TLN1 increased S phase cells, but decreased G2/M phase proportion. And miR-1285-3p inhibitor could reverse this phenomenon (Fig. 5E). Besides, si-TLN1could induce cell apoptosis. And with the decreased of miR-1285-3p expression, apoptosis decreased, too (Fig. 5F).
TLN1 was overexpressed in CC and identified as a target of miR-1285-3p. A. Predicted binding sites for miR-1285-3p on the TLN1 transcript. B, C. TLN1was overexpressed in CC cells compare to it in H8 by RT-PCR and western-blot. D. TLN1 was overexpressed in CC tissue. E. TLN1 was negatively correlated with the expression of miR-1285-3p. F, G. CC cells were transfected with the mimics of miR-1285-3p Reduced TLN1 expression was shown by RT-RCR and western-blot. H. Luciferase activities were measured in 293T cells co-transfected with luciferase reporter containing TLN1 and the mimics of miR-1285-3p. I. The expression of TLN1 was decreased with the knocked down of circRNA_400029. Data are shown as mean ± SD, n = 3. The data statistical significance is assessed by Student's t-test. *P < 0.05.
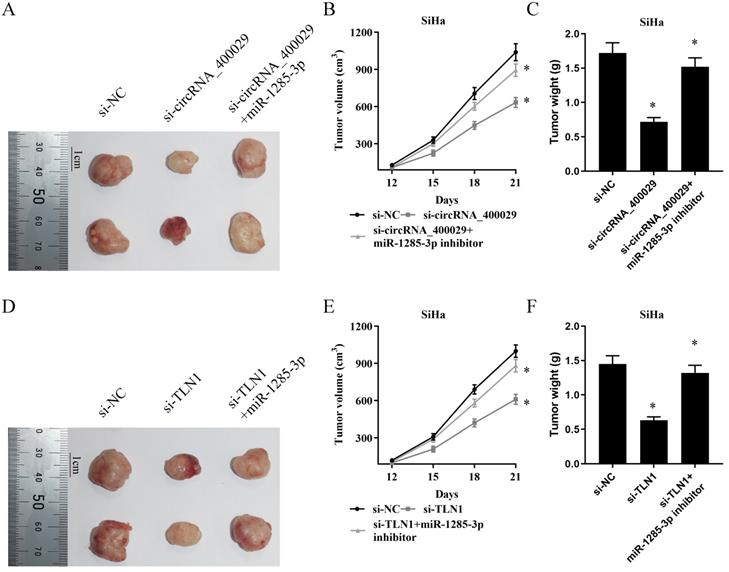
CircRNA_400029 reinforces the proliferative and invasive. A, B. Cell proliferation assessed in circRNA_400029 knockdown and circRNA_400029 knockdown + miR-1285-3p inhibitor by CCK-8. A: SiHa; B: CaSki. C. Cell proliferation assessed in circRNA_400029 knockdown and circRNA_400029 knockdown + miR-1285-3p inhibitor by Edu. up: SiHa; down: CaSki (si-NC contained more merge cells than si-circRNA_400029 and si-circRNA_400029 + miR-1285-3p inhibitor. si-circRNA_400029 + miR-1285-3p inhibitor contained more merge cell than si-circRNA_400029, marked with yellow arrows). D. Transwell assays were used to evaluate the involvement of circRNA_400029 for invasion in circRNA_400029 knockdown and circRNA_400029 knockdown + miR-1285-3p inhibitor. up: SiHa; down: CaSki. E. The cell cycle progression of CC cells transfected with si-NC, si-circRNA_400029, si-circRNA_400029 + miR-1285-3p inhibitor was identified by flow cytometry assay. F. Effects of si-circRNA_400029, si-circRNA_400029 + miR-1285-3p inhibitor on CC cell apoptosis. Data are shown as mean ± SD, n = 3. The data statistical significance is assessed by Student's t-test. *P < 0.05.
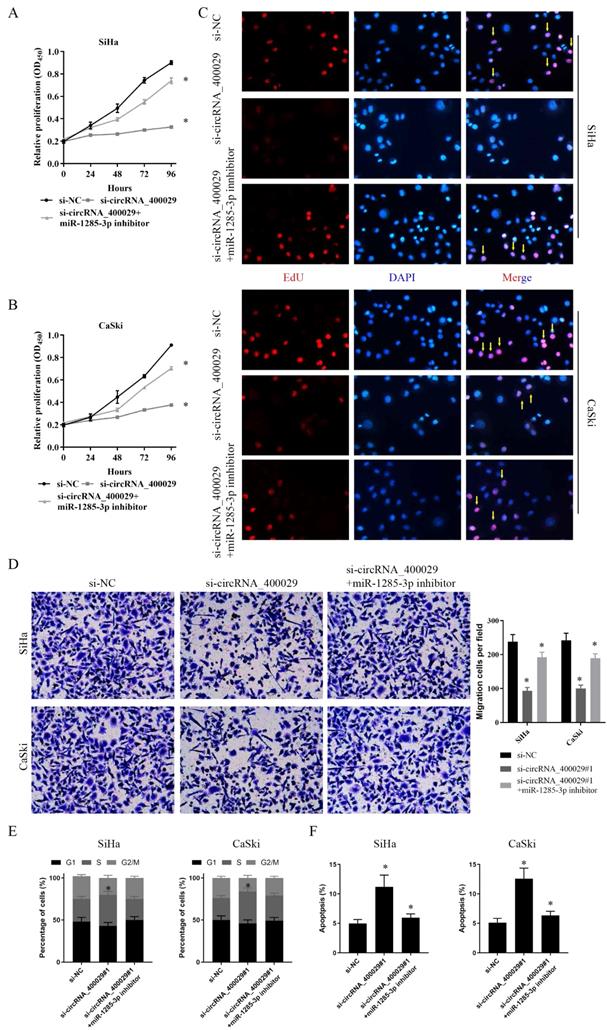
TLN1 could crosstalk with miR-1285-3p through direct binding. A, B. Cell proliferation assessed in TLN1 knockdown and TLN1 knockdown + miR-1285-3p inhibitor by CCK-8. A: SiHa; B: CaSki. C. Cell proliferation assessed in TLN1 knockdown and TLN1 knockdown + miR-1285-3p inhibitor by Edu. up: SiHa; down: CaSki (si-NC contained more merge cells than si-TLN1 and si-TLN1 + miR-1285-3p inhibitor. si-TLN1 + miR-1285-3p inhibitor contained more merge cell than si-TLN1, marked with yellow arrows). D. Transwell assays were used to evaluate the involvement of TLN1 for invasion in TLN1 knockdown and TLN1 knockdown + miR-1285-3p inhibitor. up: SiHa; down: CaSki. E. The cell cycle progression of CC cells transfected with si-NC, si-TLN1, si-TLN1 + miR-1285-3p inhibitor was identified by flow cytometry assay. F. Effects of si-TLN1, si-TLN1 + miR-1285-3p inhibitor on CC cell apoptosis. Data are shown as mean ± SD, n = 3. The data statistical significance is assessed by Student's t-test. *P < 0.05.
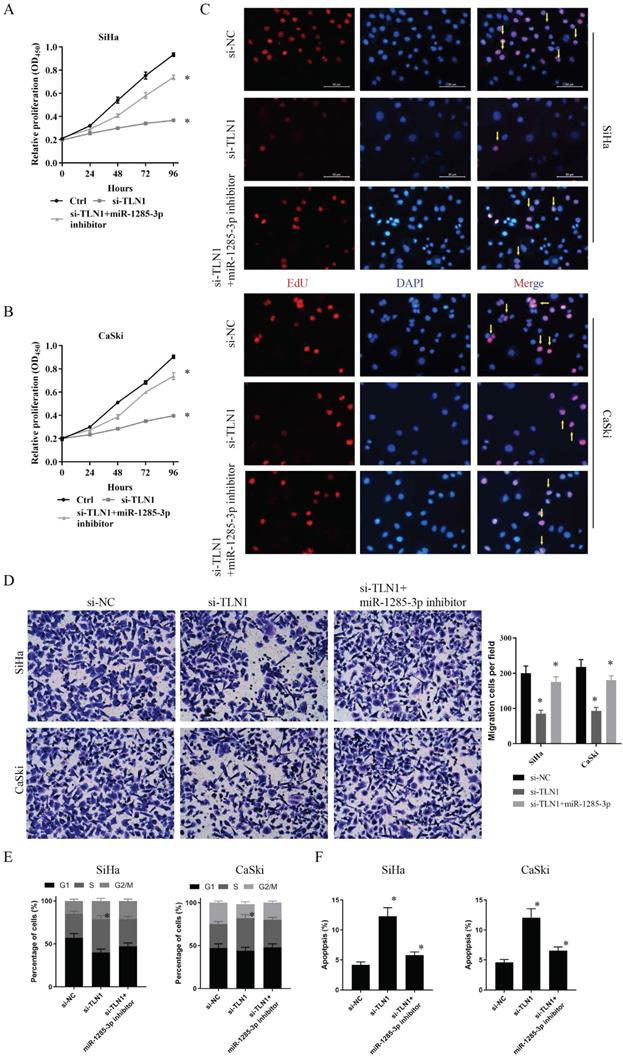
In vivo tumor lumps of xenograft mouse models composed of circRNA_400029, miR-1285-3p and TLN1. A. Representative images of the tumor lumps of each group at the endpoint of the experiment. B. The tumor growth curves of in vivo tumor volumes. C. The mean tumor weight of each group. Data are mean ± SD. of the tumor volumes, n = 5, *P < 0.05.
These data suggested that, as competitive endogenous RNA (ceRNA), circRNA_400029 was combined with miR-1285-3p to regulate the expression of TLN1, thus affecting CC cells proliferation, migration and invasion.
The biological function of circRNA_400029/miR-1285-3p/TLN1 axis in vivo
To clarify the biological functions of circRNA_400029/miR-1285-3p/TLN1 axis in vivo. Different SiHa cells were subcutaneously injected into nude mice.
In total, there were six groups: Group 1. si-NC was injected with SiHa cells; Group 2. si-circRNA_400029 was injected with SiHa cells; Group 3. si-circRNA_400029 + miR-1285-3p inhibitor was injected with SiHa cells; Group 4. si-NC was injected with SiHa cells; Group 5. si-TLN1 was injected with SiHa cells; Group 6. si-TLN1 + miR-1285-3p inhibitor was injected with SiHa cells (n = 5 per group). We measured the tumor volumes every 3 days. Moreover, the tumor volumes in the si-circRNA_400029 and si-TLN1 groups were smaller than in si-NC group. The mice were sacrificed at the end of the experiment, and the tumor volume and weight of each group were measured (Fig. 6A, 6D).
The mean tumor volume at the time of death in mice injected with si-circRNA_400029 cells was 633.21 ± 41.00 (mean value ± SD) mm3 and the mean tumor volume of mice injected with si-NC cells was 1038.91 ± 69.47 mm3. And the inhibitor of miR-1285-3p could reverse this phenomenon. The volume of si-circRNA_400029 + miR-1285-3p group of tumors was 893.30 ± 52.61 mm3 (Fig. 6B). Similarly, si-circRNA_400029 also limited tumor weight (0.72 ± 0.06 g vs si-NC: 1.72 ± 0.15 g), whereas miR-1285-3p inhibitor rescued this condition (1.52 ± 0.13 g, Fig. 6C). We get similar results in TLNN1 and miR-1285-3p: si-TLN1 decreased the volume and weight of tumors (610.81 ± 51.20 mm3, 0.63 ± 0.05 g, vs si-NC: 999.57 ± 50.29 mm3, 1.45 ± 0.12 g, Fig. 6E, 6F). The miR-1285-3p inhibitor could increase the volume and weight of tumors (880.27 ± 50.79 mm3, 1.32 ± 0.11 g, Fig. 6E, 6F).
Discussion
CC was the second most common gynecological malignant, in recent years, its onset age has a tendency to become younger [18]. More and more researches have been devoted to elucidating the genetic and epigenetic mechanisms involved in the progression of CC [19], but the pathogenesis of CC is still not fully understood. Current studies have shown that circRNA was considered as a biomarker related to clinical testing and could contribute to the poor prognosis of various tumors due to its stable structure [20]. CircRNA as an essential part of the human transcriptome, the type is various [21]. Sequence studies confirm circRNA abnormal expression in different cancer by apparent after modification, transcription [22] and its regulation and regulating tumor related gene expression of the tumor cell proliferation and invasion have an vital impact in the process of development [23, 24]. Meanwhile, it has been reported that circRNA can be used as an anti-tumor target for the therapy of cancer and has certain clinical application value [25].
CircRNA_400029, also called circ_0092337, was located chr 16: 89628179- 89628539, and derived from ribosomal protein L13 (RPL13). Li et al reported that circRNA_400029 could be used as plasma circRNA in molecular detection of gastric cancer [26]. Our study confirmed that circRNA_400029 was highly expressed in both cervical cancer tissues and cells. We found and demonstrated that circRNA_400029 could promote CC proliferation, invasion and inhibit apoptosis. It suggested that circRNA_400029 might play an important role in the progression of CC. CircRNA has been reported to induce cervical cancer progression not only by altering chromatin modifications and mRNA stability at the transcriptional level, but also by binding to miRNAs to serve as ceRNA or promoting protein stabilization at post-transcriptional levels [27]. To explore whether circRNA_400029 and miRNAs can provide new insights on the mechanisms of CC proliferation and invasion, FISH clearly showed that circRNA_400029 is localized in the cytoplasm, supporting the hypothesis that circRNA_400029 regulates gene expression at the post-transcriptional level through the ceRNA mechanism. Afterwards, we used miRanda and RNAhybrid algorithm to analyze the microarray data. The prediction was combined with the RegRNA2.0 database. We found that miR-1285-3p may be a potential target of circRNA_400029. CircRNA_400029 could inhibit the expression of miR-1285-3p. miR-1285-3p antagonized the effects of circRNA_400029 on the proliferation and invasion of CC cells. The dual luciferase reporter assays confirmed the direct relationship between the two factors. Bioinformatics analysis revealed that miR-1285-3p was a potential tumor suppressor gene for breast cancer [28]. Previous reports presented that miR-1285-3p modulated multiple molecular targets and served as a sponge in ovarian cancer [29], oral squamous cell carcinoma [30], and cervical cancer [31]. Upregulated the expression of miR-1285-3p also inhibited cell proliferation and escape from apoptosis of colorectal cancer [32]. Our study confirmed that the expression level of circRNA_400029 was negatively correlated with miR-1285-3p in CC tissues and cells. Luciferase reporter gene assay confirmed that miR-1285-3p could complement and bind to miR-1285-3p.This suggested that circRNA_400029 acted as a sponge for miR-1285-3p in CC, thus promoting the progression of cervical cancer.
We identified TLN1 as an unreported potential target of miR-1285-3p using an online predictive database. The direct binding relationship between TLN1 and miR-1285-3p was confirmed by luciferase reporter gene assay. In CC cells, TLN1 was positively regulated by circRNA_400029 and negatively regulated by miR-1285-3p -mimic, suggesting that TLN1 was also a target gene of miR-1285-3p regulated by circRNA_400029. TLN1 gene encoded a cytoskeletal protein that was concentrated in areas of cell-substratum and cell-cell contacts. It played a significant role in the assembly of actin filaments and in spreading and migration of various cell types, including fibroblasts and osteoclasts. It codistributed with integrins in the cell surface membrane in order to assist in the attachment of adherent cells to extracellular matrices and of lymphocytes to other cells. Overexpression of TLN1 activated the progression of gastric cance through PTK2-PXN-VCL-E-Cadherin-CAPN2-MAPK1signaling axis [33]. Besides this, TLN1 overexpression promoted hepatocellular carcinoma epithelial-mesenchymal transition and induced metastasis [34]. In addition, the copy number alterations of TLN1 was contributed the tumorigenesis of neoplasias [35]. It was considered as a potential therapeutic target for cancer treatment [36]. In this study, we confirmed that circRNA_400029 silencing can inhibit the proliferation, metastasis and invasion ability of CC cells, as well as inhibit the change of cell cycle, and miR-1285-3p inhibitor can eliminate the biological function changes mediated by circRNA_400029 silencing. Consistent with in vitro results, down-regulation of circRNA_400029 inhibited xenograft growth in mice in vivo. Meanwhile, TLN1 interference can reverse circRNA_400029 mediated changes in cell activity, and TLN1 interference combined with miR-1285-3p inhibitor can enhance this reversal effect.
In conclusion, our study revealed that circRNA_400029 plays a positive role in regulating the growth of cervical cancer. The underlying mechanism is circRNA_400029 as a competitive endogenous RNA, through circRNA_400029/ miR-1285-3p/ TLN1 axis regulates the expression of TLN1, further affecting tumor growth, invasion and metastasis and changing cell cycle. These results provide a new idea for the mechanism between circRNAs and cervical cancer.
Acknowledgements
Author Contributions
Ma Y performed the majority of experiments and analyzed the data and drafted the manuscript; Wang DB designed the research; Liu J, Yang Z conducted the molecular biology assays and assisted in writing the manuscript; Chen P collected and analyzed the data.
Institutional review board statement
The study was reviewed and approved by the Faculty of Science Ethics Committee at Liaoning Cancer Hospital & Institute (Cancer Hospital of China Medical University, 20191227).
Informed consent
Informed consent was written by patients before surgery at the Liaoning Province Cancer Hospital & Institute.
Funding
The Key Research and Development Project of Liaoning Province [2018225037].
Data availability statement
The data used to support the findings of this study are available from the corresponding author upon request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA: a cancer journal for clinicians. 2020;70:7-30
2. Fakhr E, Modic Ž, Cid Arregui A. Recent Developments in Immunotherapy of Cancers Caused by Human Papillomaviruses. Immunology. 2021;163:33-45
3. Simms KT, Hanley SJB, Smith MA, Keane A, Canfell K. Impact of HPV vaccine hesitancy on cervical cancer in Japan: a modelling study. The Lancet Public health. 2020;5:e223-e34
4. Simbar M, Ghazanfarpour M, Abdolahian S. Effects of training based on the health belief model on Iranian women's performance about cervical screening: A systematic review and meta-analysis. Journal of education and health promotion. 2020;9:179
5. Yamamoto T, Saitoh N. Non-coding RNAs and chromatin domains. Current opinion in cell biology. 2019;58:26-33
6. Wang F, Fan M, Cai Y, Zhou X, Tai S, Yu Y. et al. Circular RNA circRIMS1 Acts as a Sponge of miR-433-3p to Promote Bladder Cancer Progression by Regulating CCAR1 Expression. Molecular therapy Nucleic acids. 2020;22:815-31
7. Wang H, Song X, Wang Y, Yin X, Liang Y, Zhang T. et al. CircCNTNAP3-TP53-positive feedback loop suppresses malignant progression of esophageal squamous cell carcinoma. Cell death & disease. 2020;11:1-13
8. Huang B, Zhou D, Huang X, Xu X, Xu Z. Silencing circSLC19A1 Inhibits Prostate Cancer Cell Proliferation, Migration and Invasion Through Regulating miR-326/MAPK1 Axis. Cancer management and research. 2020;12:11883-95
9. Tang X, Liu S, Ding Y, Guo C, Guo J, Hua K. et al. Serum Circular FoxO3a Serves as a Novel Prognostic Biomarker in Squamous Cervical Cancer. Cancer management and research. 2020;12:2531-40
10. Wang H, Wei M, Kang Y, Xing J, Zhao Y. Circular RNA circ_PVT1 induces epithelial-mesenchymal transition to promote metastasis of cervical cancer. Aging. 2020;12:20139-51
11. Shi P, Zhang X, Lou C, Xue Y, Guo R, Chen S. Hsa_circ_0084927 Regulates Cervical Cancer Advancement via Regulation of the miR-634/TPD52 Axis. Cancer management and research. 2020;12:9435-48
12. Pu M, Chen J, Tao Z, Miao L, Qi X, Wang Y. et al. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci. 2019;76:441-51
13. Lin CL, Ying TH, Yang SF, Wang SW, Cheng SP, Lee JJ. et al. Transcriptional Suppression of miR-7 by MTA2 Induces Sp1-Mediated KLK10 Expression and Metastasis of Cervical Cancer. Molecular therapy Nucleic acids. 2020;20:699-710
14. Chen Y, Song Y, Mi Y, Jin H, Cao J, Li H. et al. microRNA-499a promotes the progression and chemoresistance of cervical cancer cells by targeting SOX6. Apoptosis: an international journal on programmed cell death. 2020;25:205-16
15. Nahand JS, Vandchali NR, Darabi H, Doroudian M, Banafshe HR, Moghoofei M. et al. Exosomal microRNAs: novel players in cervical cancer. Epigenomics. 2020;12:1651-60
16. Chang TH, Huang HY, Hsu JB, Weng SL, Horng JT, Huang HD. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC bioinformatics. 2013;14(Suppl 2):S4
17. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015 4
18. Shelar S, Shim E-H, Brinkley GJ, Kundu A, Carobbio F, Poston T. et al. Biochemical and epigenetic insights into L-2-hydroxyglutarate, a potential therapeutic target in renal cancer. Clinical Cancer Research. 2018;24:6433-46
19. Laengsri V, Kerdpin U, Plabplueng C, Treeratanapiboon L, Nuchnoi P. Cervical Cancer Markers: Epigenetics and microRNAs. Laboratory medicine. 2018;49:97-111
20. Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T. et al. CircRNAs in cancer metabolism: a review. Journal of hematology & oncology. 2019;12:1-10
21. Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Molecular cell. 2018;71:428-42
22. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast cancer (Tokyo, Japan). 2018;25:1-7
23. Song T, Xu A, Zhang Z, Gao F, Zhao L, Chen X. et al. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR-8075. Journal of cellular physiology. 2019;234:14296-305
24. Jin J, Chen A, Qiu W, Chen Y, Li Q, Zhou X. et al. Dysregulated circRNA_100876 suppresses proliferation of osteosarcoma cancer cells by targeting microRNA-136. Journal of cellular biochemistry. 2019;120:15678-87
25. Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. The EMBO journal. 2019;38:e100836
26. Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W. et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. Journal of molecular medicine (Berlin, Germany). 2018;96:85-96
27. Dong P, Xu D, Xiong Y, Yue J, Ihira K, Konno Y. et al. The Expression, Functions and Mechanisms of Circular RNAs in Gynecological Cancers. Cancers. 2020;12:1472
28. Peng Z, Xu B, Jin F. Circular RNA hsa_circ_0000376 Participates in Tumorigenesis of Breast Cancer by Targeting miR-1285-3p. Technology in cancer research & treatment. 2020;19:1533033820928471
29. Zhang Y, Ruan F. LncRNA LEF1-AS1 Promotes Ovarian Cancer Development Through Interacting with miR-1285-3p. Cancer management and research. 2020;12:687-94
30. Li C, Ye J, Zhang Z, Gong Z, Lin Z, Ding M. Long non-coding RNA RBM5-AS1 promotes the aggressive behaviors of oral squamous cell carcinoma by regulation of miR-1285-3p/YAP1 axis. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2020;123:109723
31. Feng Y, Ma J, Fan H, Liu M, Zhu Y, Li Y. et al. TNF-α-induced lncRNA LOC105374902 promotes the malignant behavior of cervical cancer cells by acting as a sponge of miR-1285-3p. Biochemical and biophysical research communications. 2019;513:56-63
32. Villanova L, Barbini C, Piccolo C, Boe A, De Maria R, Fiori ME. miR-1285-3p Controls Colorectal Cancer Proliferation and Escape from Apoptosis through DAPK2. International journal of molecular sciences. 2020;21:2423
33. Yan H, Guo M, Zou J, Xiao F, Yi L, He Y. et al. Promotive effect of Talin-1 protein on gastric cancer progression through PTK2-PXN-VCL-E-Cadherin-CAPN2-MAPK1 signaling axis. Journal of clinical laboratory analysis. 2020: e23555.
34. Tang H, Liu Y, Cheng W, He Z, Zhou N. microRNA-96-5p induces the epithelial-mesenchymal transition to promote the metastasis of hepatocellular carcinoma by post-transcriptionally downregulating Talin 1. International journal of clinical and experimental pathology. 2019;12:1897-908
35. Sarhadi VK, Lahti L, Scheinin I, Ellonen P, Kettunen E, Serra M. et al. Copy number alterations and neoplasia-specific mutations in MELK, PDCD1LG2, TLN1, and PAX5 at 9p in different neoplasias. Genes, chromosomes & cancer. 2014;53:579-88
36. Malla RR, Vempati RK. Talin: A Potential Drug Target for Cancer Therapy. Current drug metabolism. 2020;21:25-32
Author contact
![]() Corresponding author: Dan-bo Wang, Department of Gynecology, Liaoning Province Cancer Hospital & Institute (Cancer Hospital of China Medical University), No. 44 Xiaoheyan Road, Dadong District, Shenyang City, Liaoning Province 110042, China. E-mail: wangdanbocom.
Corresponding author: Dan-bo Wang, Department of Gynecology, Liaoning Province Cancer Hospital & Institute (Cancer Hospital of China Medical University), No. 44 Xiaoheyan Road, Dadong District, Shenyang City, Liaoning Province 110042, China. E-mail: wangdanbocom.

 Global reach, higher impact
Global reach, higher impact