Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(1):313-324. doi:10.7150/jca.64846 This issue Cite
Review
A Concise Review of MicroRNA-383: Exploring the Insights of Its Function in Tumorigenesis
1. The Central Laboratory, Shenzhen Second People's Hospital/First Affiliated Hospital of Shenzhen University Health Science Center, Shenzhen, Guangdong 518035, P.R. China.
2. Department of Physiology, School of Basic Medical Science, Southwest Medical University, Luzhou, Sichuan province 646099, P.R. China.
3. Department of Orthopedics, Shenzhen Second People's Hospital/First Affiliated Hospital of Shenzhen University Health Science Center, Shenzhen, Guangdong 518035, P.R. China.
4. Laboratory of Anesthesia and Organ Protection, Southwest Medical University, Luzhou, Sichuan province 646099, P.R. China.
#. Q.Y. and W.X. contributed equally to this review.
Received 2021-7-12; Accepted 2021-11-10; Published 2022-1-1
Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that commonly have 18-22 nucleotides and play important roles in the regulation of gene expression via directly binding to the 3'-UTR of target mRNAs. Approximately 50% of human genes are regulated by miRNAs and they are involved in many human diseases, including various types of cancers. Recently, microRNA-383 (miR-383) has been identified as being aberrantly expressed in multiple cancers, such as malignant melanoma, colorectal cancer, hepatocellular cancer, and glioma. Increasing evidence suggests that miR-383 participates in tumorigenic events including proliferation, apoptosis, invasion, and metastasis as well as drug resistance. Although downstream targets including CCND1, LDHA, VEGF, and IGF are illustrated to be regulated by miR-383, its roles in carcinogenesis are still ambiguous and the underlying mechanisms are still unclear. Herein, we review the latest studies on miR-383 and summarize its functions in human cancers and other diseases. The goal of this review is to provide new strategies for targeted therapy and further investigations.
Keywords: miR-383, Cancer, Proliferation, Apoptosis, Invasion, Metastasis
1. Introduction
Cancer is one of the most life-threatening diseases and the leading cause of death in humans [1,2]. Lung cancer is the most commonly diagnosed cancer and the leading cause of cancer-related death among males, while breast cancer is the most prevalent among females [3]. There are many risk factors that contribute to cancer development, such as gender, age, region and race. The occurrence, development, and pathogenesis of different types of cancer can vary widely. Therefore, investigating the specific molecular mechanisms of different types of cancer may provide us with new ideas to develop novel and effective therapies for cancer treatment.
MicroRNAs (miRNAs) are characterized as a class of small noncoding RNAs comprised of approximately 18-22 nucleotides [4]. The critical role of miRNAs is to regulate gene expression in a post-transcriptional manner. Through binding to the specific sites of the 3'-UTRs of targeted mRNAs, miRNAs mediate their degradation and translational repression. Approximately 50% of human genes are regulated by miRNAs, including tumor suppressor genes and oncogenes [5, 6]. Therefore, miRNAs play important regulatory roles in multiple biological progressions, such as cell proliferation, apoptosis, and carcinogenesis. Meanwhile, studies have also found that the expression of miRNAs is differ among the early, middle, and advanced stages of various cancers, thus indicating that miRNAs could be clinical or diagnostic biomarkers. Exploring the role and mechanism of miRNAs in tumors is critical for the development of unique and effective miRNA-based therapies.
MicroRNA-383 (miR-383) is located on chromosome 8p22, within the third intron of the sarcoglycan zeta (SGCZ) gene [7]. It has been reported that chromosome 8p is frequently lost or mutated and the loss of chromosome 8p is a characteristic of prostate cancer, with more than half of the loss being due to genomic alterations [8, 9]. The genomic alterations on chromosome 8p21-p23 also frequently occur in stage I squamous cell lung carcinoma [10]. Therefore, the expression of miR-383 was found to be aberrant in multiple types of cancer. In lung cancer, the expression of miR-383 was markedly lower than in the non-cancerous lung tissues, and further reduced in advanced-stage carcinomas [11]. Down-regulation of miR-383 was also observed in cancers that include glioma, hepatocellular cancer, and breast cancer (Table 1). However, in primary human malignant melanoma, the expression level of miR-383 was higher than normal epidermal melanocytes [12]. Upregulated expression and oncogenic function of miR-383 has also observed in cholangiocarcinoma, epithelial ovarian cancer, and renal cell carcinoma. These results suggest that miR-383 might play significant roles in tumor progression and its functions in tumorigenesis are controversial.
miR-383 expression in various cancers and its target genes.
| Cancer types | Expression | Target gene | Reference |
|---|---|---|---|
| Lung cancer | Decreased | /, Wnt1, E2F7, EPAS1, CIP2A | [11,13-16] |
| Glioma | Decreased | PRDX3, IGF-1R, /, VEGF, CCND1 | [17-21] |
| Testicular embryonal carcinoma | Decreased | PNUTS, IRF1 | [22,23] |
| Ovarian cancer | Decreased | /, LDHA, TRIM27 | [24-26] |
| Colorectal cancer | Decreased | PAX6, CREPT/RPRD1B, / | [27,28] |
| Hepatocellular cancer | Decreased | LDHA, APRIL, PHF8 | [29-31] |
| Medulloblastoma | Decreased | /, FOXM1, PRDX3 | [32-34] |
| Pancreatic carcinoma | Decreased | SOX11, GAB1, ROBO3 | [35-37] |
| Esophageal squamous cell carcinoma | Decreased | SP1, 5S rRNA, MALAT1 | [38-40] |
| Breast cancer | Decreased | LDHA, Gadd45g, PD-L1 | [41-43] |
| Gastric cancer | Decreased | PP2A, HDAC9, ERBB4 | [44-46] |
| Cervical cancer | Decreased | PARP2 | [47] |
| Colon cancer | Decreased | APRIL | [48] |
| Prostate cancer | Decreased | CD44 | [49] |
| MALT lymphoma | Decreased | ZEB2 | [50] |
| Pituitary adenoma | Decreased | / | [51] |
| Ependymoma | Decreased | / | [52] |
| Nasopharyngeal carcinoma | Decreased | HMGA2 | [53] |
| Cholangiocarcinoma | Increased | IRF1 | [54] |
| Epithelial ovarian cancer | Increased | CASP2 | [55] |
| Canine malignant melanoma | Increased | ATR serine/threonine kinase, CDK2 | [56] |
| Hepatocellular carcinoma | Increased | /, EIF5A2 | [57,58] |
| Renal cell carcinoma | Increased | DIO1 | [59] |
“/” means no miR-383 targeted gene.
In this paper, we review the roles and mechanisms of miR-383 in cancer cell proliferation, apoptosis, invasion and metastasis, development and differentiation, as well as its functions in other human diseases. This review aims to provide evidence or suggestions for further investigations and clinical applications regarding miR-383.
2. miR-383 in cell proliferation
Several studies have shown that miR-383 mediates cell proliferation via regulating the expression of proliferation-associated genes. Cyclin D1 is well-known in regulating cell proliferation and has been demonstrated to be overexpressed in many human cancers [60]. It has been reported that CREPT can bind to the promoter of Cyclin D1, enhancing its transcription and expression [61]. Li et al. illustrated that miR-383 directly binds to the 3'-UTR of CREPT mRNA and inhibits the expression of CREPT and cyclin D1, thereby suppressing cell growth and colony formation of colorectal cancer cells [28]. It has been demonstrated that miR-383 can directly bind to Cyclin D1 mRNA, inducing cell cycle arrest at the G0/G1 phase and inhibiting glioma cell growth [21].
A proliferating-inducing ligand (APRIL) belongs to the tumor necrosis factor super-family. As a cytokine, it stimulates cell proliferation and modulates cell apoptosis, playing important roles in tumorigenesis [62, 63]. Overexpression of miR-383 in colon cancer cells was associated with the decreased expression of APRIL and the inhibition of cell proliferation [48]. The direct binding between miR-383 and APRIL mRNA was recently identified in hepatocellular carcinoma cells. Overexpression of miR-383 also induced cell cycle arrest in G0/G1 phase and inhibited the proliferation of HepG2 and SK-Hep-1 cells [30].
The expression of miR-383 was also decreased in medulloblastoma, ovarian cancer, pancreatic carcinoma, and colorectal cancer (Table 1). In medulloblastoma, miR-383 regulated the transcription and translation of PRDX3. Ectopic expression of miR-383 significantly suppressed cell growth [34]. In ovarian cancer, miR-383-5p suppressed cell proliferation in vitro and inhibited tumor growth in vivo by targeting TRIM27 [26]. Overexpression of miR-383 markedly suppressed the proliferation of SW620 and HCT116 human colorectal cancer cells through targeting PAX6 [27]. Moreover, miR-383 inhibited the gastric mucosa-associated lymphoid tissue lymphoma proliferation via targeting ZEB2 [50]. In pancreatic carcinoma, miR-383 inhibited the expression of ROBO3 and suppressed cell growth [37]. In testicular embryonal carcinoma, miR-383 targeted the tumor suppressor IRF1 to reduce the expression of Cyclin D1, CDK2, and p21, which inhibited proliferation through inactivation of the pRb pathway [23, 64]. The proliferation-regulation role of the miR-383-LDHA axis has also been demonstrated in hepatocellular cancer cells such as HepG2 and SMMC-7721 cells [29]. Additionally, miR-383-5p suppresses cell proliferation via directly targeting CIP2A in lung adenocarcinoma [16]. Furthermore, miR-383 targeted VEGF to suppress glioma-exposed endothelial cells proliferation [20]. In esophageal squamous carcinoma cells, miR-383 decreased the expression of 5S rRNA and intensified the rpL11-c-Myc interaction, resulting in the attenuation of c-Myc and inhibition of cell proliferation [39]. In testicular embryonal carcinoma cells, miR-383 impaired the phosphorylation of H2AX and induced cell cycle arrest via the direct targeting of PNUTS mRNA [22]. The anti-proliferation function of miR-383 has also been reported in other types of cancer; its expression in different cancers is summarized in Table 1.
The expression of miR-383 was found to be frequently decreased in various malignant tumors; however, it was upregulated in cholangiocarcinoma tissues and acted as an oncogenic miRNA by inhibiting the expression of tumor suppressor gene IRF1 [54]. Sheng et al. also reported that the expression of miR-383 was significantly elevated in immortal human epithelial ovarian cancer cell lines and human epithelial ovarian cancer tumors. They found that miR-383 targeted CASP2, and stable knockdown of miR-383 expression was associated with the suppression of cell proliferation [55]. The upregulation of miR-383 was also found in clear cell renal cell carcinoma. MiR-383 was negatively associated with the expression of DIO1, which was reported to inhibit the proliferation of renal cancer cells [59]. Together, these data indicate that the role of miR-383 in tumor proliferation is critical, complex, and involves multiple signaling pathways (Figure 1).
3. miR-383 in cell apoptosis
A large body of evidence has uncovered the relationship between impaired cell apoptosis and cancer development. Bcl-2 family signaling consists of Bax pro-apoptosis protein, Bcl-2 anti-apoptosis protein, and BH3-only protein. It is a classical mitochondria pathway that mediates cell apoptosis. Recently, miR-383 has been found to promote medulloblastoma cell apoptosis via repressing the expression of PRDX3, up-regulating cleaved PARP expression, and reducing the expression of BCL-XL and/or BCL-2 [34]. Moreover, in human retinal pigment epithelial cells, a high glucose treatment could increase the expression of miR-383, promote reactive oxygen species (ROS) formation, downregulate Bcl-2 and Bax expression, and induces apoptosis by repressing PRDX3 [65].
Ultraviolet radiation (UV) or ionizing radiation can induce DNA damage and activate ATR and ATM, members of the phosphatidylinositol 3-kinase-related kinase (PIKK) family [66]. Studies have shown that ATR plays a significant role in DNA damage response, proliferation, and apoptosis [55,56]. In A431 melanoma cells, Stat3 decreased the expression of miR-383 and, as the direct target of miR-383, ATR expression was increased. MiR-383 also mediated ATR activity to control DNA damage and affect cell apoptosis [67]. Studies have also reported that ATR was a target for miR-383 in canine malignant melanoma, indicating that miR-383 may be involved in melanoma tumorigenesis by inhibiting DNA repair or apoptosis [68].
Gadd45a, Gadd45b, and Gadd45g constitute the Gadd45 family. Via interactions with PCNA, p21, and cdc2/Cyclin B1, they regulate cell proliferation, the cell cycle, and apoptosis [69]. Recently, Gadd45g was found as a direct target of miR-383. MiR-383 promoted apoptosis and increased the sensitive of breast cancer cells to both UV irradiation and cisplatin treatment [42]. The caspase family are key mediators in the maintenance of cell homeostasis by regulating inflammatory response and apoptosis. The expression of miR-383 was positively related to the number of apoptotic nuclei in brain infarct area of ischemic stroke in a rat model. The expression of cleaved caspase-3 and cleaved PARP were also found to increase after ischemic stroke. These results shown us that miR-383 may promote apoptosis in ischemic stroke [70]. In non-small cell lung cancer, overexpression of miR-383 induced apoptosis via targeting the Wnt/β-catenin signaling pathway [13]. In human glioma cells, miR-383 overexpression increases the rate of apoptotis of U251 and U87 cells from 8.0% and 1.9% to 36.6% and 16.9%, respectively [19].
The above studies show that, while controversial, there are apoptosis-promoting functions of miR-383. Shuai et al. found that propofol treatment significantly decreased the expression of miR-383. The reduced expression of Bcl-2 and increased expression of Bax induced by propofol was inhibited by miR-383 mimic treatment. These data indicate that miR-383 inhibited the neuron apoptosis by regulating the expression of Bcl-2 and Bax [71]. Moreover, Resveratrol treatment reduced the expression of miR-383-5p in human podocytes, and it effectively inhibited high-glucose-induced apoptosis via stimulating autophagy [72]. Furthermore, in human epithelial ovarian cell lines, the overexpression of miR-383 decreased the expression of caspase-2, indicating that miR-383 was acting as an oncogene in human epithelial ovarian cell lines [55]. Furthermore, in homocysteine-induced endothelial injury in rat coronary arteries, miR-383-3p negatively regulated the expression of IL1R2 and caspase-1. Therefore, miR-383-3p may function by decreasing cell apoptosis of coronary artery endothelial cells [73].
4. miR-383 in cancer invasion and metastasis
Cancer invasion and metastasis is a complex process that is the major obstacle to cancer treatment. The epithelial-to-mesenchymal transition (EMT) is a process characterized by a decrease in E-cadherin expression and increase of N-cadherin/Vimentin expression, which confers cells migratory and invasive properties. Recently, it has been reported that miR-383 repressed the metastasis of pancreatic carcinoma through regulating EMT. MiR-383 decreased ROBO3 expression and inhibited the Wnt/β-catenin signaling pathway, resulting in an increased expression of E-cadherin and decreased expression of Vimentin/N-cadherin [37]. In nasopharyngeal carcinoma, miR-383-3p suppressed HMGA2 expression and inhibited the invasion of NPC cells via modulating the EMT process [53].
Lactate dehydrogenase A (LDHA) is an important enzyme involved in the regulation of the glycolysis pathway and cell metabolism. Recently, its critical roles in cell proliferation, glycolysis, and invasion of cancer cells have been reported. LDHA is a direct target of miR-383, with the enhanced invasive capacity of HepG2 and SMMC-7721 cells induced by LDHA overexpression being abolished by miR-383 overexpression [29]. In Li and colleagues' study, SKOV3 ovarian cells were transfected with a miR-383 inhibitor and OVCAR3 ovarian cells was transfected with miR-383 mimics. These two cell lines have relatively high and low miR-383 expression, respectively. The transwell assays showed that the invasive ability of OVCAR3 and SKOV3 cells were decreased and increased after transfection, respectively. The authors found that the function of miR-383 in suppressing ovarian cancer cell invasion was mediated by LDHA [25].
IGF1R signaling is constitutively active in many human cancers and the IGF1R/AKT/MMP2 axis plays crucial roles in tumor invasion. In human glioma cancer cells, miR-383 expression was suppressed. MiR-383 mimic significantly reduced the invasive ability of U87MG glioma cell, while miR-383 suppression dramatically increased the A172 cell invasion. Further studies demonstrated that IGF1R was a direct target of miR-383, which regulated the IGF1R/AKT signaling pathway and MMP2 expression, thereby influencing glioma cell invasion [18]. Moreover, PARP2 expression was higher in the cervical cancerous tissue compared to the paracancerous tissues. The high expression of PARP2 was associated with high expression of PI3K, AKT, and mTOR. MiR-383 suppressed the expression of PARP2, reduced the activity of PI3K-AKT-mTOR signaling, and inhibited cell migration and invasion [47].
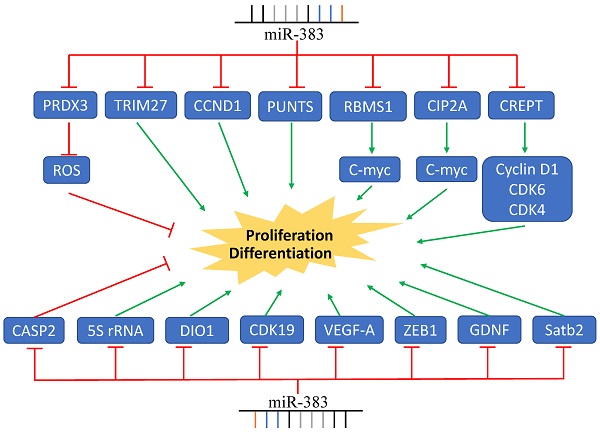
MiR-383 Regulates Cancer Cell Proliferation. MiR-383 overexpression suppressed cell proliferation via inhibition of the expression of CCND1, CREPT, VEGF-A, and others, while it increased proliferation by inhibiting the expression of CASP2. Green arrows indicate promotion and red lines indicate suppression.
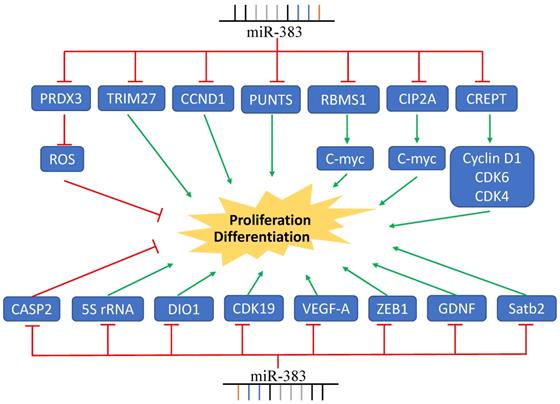
MiR-383 functions in cancer cell EMT, invasion, and metastasis. LDHA, ROBO3, PARP2, and other genes are regulated by miR-383 resulting in the suppression of cancer cell invasion and metastasis. MiR-383 also regulates the expression of IRF- and promotes the cell invasion and metastasis.
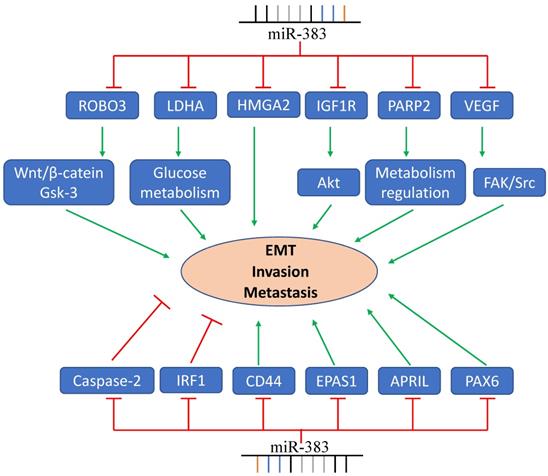
Angiogenesis plays crucial role in cancer cell metastasis, invasion, and tumor progression. Vascular endothelial growth factor (VEGF), an endothelial cell-specific mitogen, is an important mediator of angiogenesis. It has been reported that VEGF was a target gene of miR-383. In glioma-exposed endothelial cells (GECs), miR-383 overexpression decreased the expression of p-VEGFR2, p-FAK, and p-Src mediated by VEGF and inhibited the migration of GECs [20]. In human lung cancer cells, miR-383, via inhibiting the expression of EPAS1, repressed the wound healing capacity and invasive capacity of lung cancer cells [15]. Moreover, overexpression of miR-383 in SW620 and HCT116 colorectal cells evidently decreased the cell invasion through directly targeting PAX6 [27]. Transfecting HT-29 and LoVo cells with rno-miR-383 mimics significantly reduced the migratory and invasive capacity of colon cancer cells [48]. Additionally, in prostate cancer, miR-383 had a strong inhibitory effect on prostate cancer metastasis, mediated by CD44 [49].
In contrast, in a study on sorafenib's effects on lung metastasis in hepatocellular carcinoma, researchers found that the expression of miR-383 was up-regulated in lung metastatic tissue, providing new evidence regarding the role of miR-383 in metastasis [57]. It has been found that miR-383 inhibition significant repressed human epithelial ovarian cancer cell invasion through the regulation of the caspase-2 gene [55]. Interferon regulatory factor 1 (IRF1) is a tumor suppressor in cholangiocarcinoma and was reported to be a direct target of miR-383. High expression of miR-383 induced cholangiocarcinoma cell migration and invasion through repressing the expression of IRF1 [54].
These studies show that miR-383 has vital functions in tumor invasion, metastasis, and EMT (Figure 2). Up-regulating the expression of miR-383 inhibits cancer cell invasion and metastasis, while some groups also reported contrary results. Therefore, more studies about the targets and signaling pathway related to miR-383 should be investigated to elucidate the function of miR-383 on cancer cell invasion and metastasis.
5. miR-383 in development and cell differentiation
Emerging evidence indicates that microRNAs have important roles in the regulation of osteoblastic differentiation. Recently, miR-383 has been reported as a critical regulator of osteoblastic differentiation. The expression of miR-383 was significantly decreased in the osteoblastic differentiation process of bone marrow mesenchymal stem cells. MiR-383 decreased the expression of alkaline phosphatase, RUNX2, and OCN, and also suppressed matrix mineralization. STAB2 has been identified as a direct target of miR-383 in osteoblastic differentiation, with evidence suggesting that the inhibitory role of miR-383 in osteoblastic differentiation may be mediate by STAB2 [74].
It has been reported that miR-383 influences the characteristics of bone-marrow-derived mesenchymal stem cells and reduce their use in spinal cord injury. Guo et al. found that miR-383 targeted binding to the mRNA of GDNF, an identified neural growth and survival factor, which inhibited its translation in mesenchymal stem cells (MSCs). The depletion of miR-383 in MSCs increased the expression of GDNF and the therapeutic potentials of MSCs in the treatment of spinal cord injury in a rat model [75]. The authors also reported that the expression of vascular endothelial growth factor A (VEGF-A) and cyclin-dependent kinase 19 (CDK-19) were also inhibited by miR-383. MiR-383 suppression increased the proliferation of MSCs and MSC-mediated angiogenesis due to the increased expression of CDK-19 and VEGF-A, respectively. The up-regulated expression of CDK19 and VEGF-A further improved the therapeutic potential of MSCs in treating spinal cord injury (SCI) in rats [76].
MiR-383 has been found to have essential functions in spermatogenesis. It has been reported that the expression of miR-383 in primary spermatocyte was higher than in spermatid, and miR-383 expression was decreased in patients with non-obstructive azoospermia [64]. In fragile X mental retardation protein (FMRP) knockout mice testes tissue and FMRP downregulated maturation arrest (MA) in patients' testes tissue. The expression of miR-383 was decreased and associated with impaired expression of CDK4 and increased DNA damage [77]. The expression of miR-383 was increased during male germ line development, while during female germ line development, it showed a slightly increase but then decreased to a low level. MiR-383 may, through downregulating the expression of cDNMT3B, regulate germ line development in meiotic stages [78].
MiR-383 has also been reported to participate in ovarian follicular and luteal development [79]. The expression of miR-383 was significantly downregulated in TGF-β1-treated mouse ovarian granulosa cells [80]. Overexpression of miR-383 in ovarian granulosa cells resulted in the decreased expression of c-Myc and increased release of estradiol via targeting of RBMS1 expression [81]. Sun et al. found that miR-383 upregulated and transactivated miR-320, which regulated the function of granulosa cell by targeting E2F1 and SF-1 [82]. Although many studies report the specific roles of miR-383 in regulating ovarian follicle development, some research groups have found controversial results. Donadeu and Schauer aspirated the follicular fluid from dominant follicles during the ovulatory and anovulatory seasons to analyze the physiological roles of miRNAs during follicular development; however, they could not detect the expression of miR-383 in follicular fluid [83]. In a study to compare the different expressions of miRNAs in follicular fluid from dominant ovulatory, largest subordinate, and dominant anovulatory follicles, the expression of miR-383 was too low to be accurately measured [84]. In the early luteal phase of the bovine estrous cycle, the expression of miR-383 was different between granulosa cells of subordinate (SF) and dominant follicles (DF). The SFs expressed abundant miR-383, while it was not detected in granulosa cells of the DFs [85].
6. miR-383 in other diseases
Insulin resistance and insufficient pancreatic beta cell insulin secretion are the mainly characteristics of type 2 diabetes. Valeria et al. found that the expression of miR-383 was decreased in the islets of adult db/db diabetic mice and high-fat-diet-fed mice [86]. Insulin resistance is caused by the disruption of insulin signal transduction, which requires the participation of various proteins, such as insulin, insulin receptor, PI3-K, and glucose transporters. Studies have reported that miR-383 regulated the activity of IGF-1 and IGF-1R, and stimulated the AKT signaling pathway [18, 87]. In studies regarding age-associated beta cell dysfunction, researchers have found that miR-383 expression was increased in the islets of older rats. Interestingly, expression changes of miR-383 have no effect on the insulin content, insulin secretion, cell proliferation, and apoptosis [88]. In addition, Xia et al. reported that free fatty acids can increase miR-383 expression, highlighted by the observation that the expression of DIO1 was inversely associated with miR-383 expression. The authors suggested that miR-383 may influence the different propensities to diet-induced obesity by regulating the DIO1 [89].
Stroke is the second leading cause of death in populations over the age of 60 worldwide and neuroinflammation is a main cause of it, with PPARγ playing a beneficial role in ischemia brain injury in stroke [90]. In a rat model of middle cerebral artery occlusion, Pei et al. found that the expression of PPARγ can be upregulated due to the downregulation of miR-383. These results demonstrated that miR-383 plays an important function in ameliorating injury after focal cerebral ischemia [91]. MiR-383 was increased in rats after acute ischemic stroke, and erythropoietin and cyclosporine can decrease miR-383 expression by reducing brain infarct area, indicating that miR-383 participates in the regulation of apoptosis in ischemic stroke [70]. Upregulated expression of miR-383 was also reported in a study of ischemic infarction in whole blood [92]. However, in the study of the role of miR-383-3p in coronary atherosclerosis, Lian and colleagues reported that miR-383-3p directly targeted IL1R2, demonstrating an anti-inflammatory effect against homocysteine-induced endothelial injury in rat coronary arteries. Their research indicated that miR-383-3p was helpful in preventing coronary atherosclerosis and other cardiovascular diseases [73].
MiRNAs also have essential regulatory roles in the central nervous system. It has been reported that the expression of miR-383 was higher in the marginal division (MrD) than in the hippocampus of Sprague-Dawley rats' brain [93], suggesting miR-383 plays an important role in the learning and memory function of MrD. In the propofol anesthesia-induced cognitive impairment rat model, downregulated miR-383 expression was associated with neuron apoptosis, an increased Bax/Bcl-2 ratio, and decreased expression of PSD95 and CREB. These results suggest that miR-383 protected against hippocampal neuron apoptosis and cognitive impairment [71]. The expression of miR-383 in the hypothalamus of leptin-deficient (or non-functional leptin receptor) mice was significantly higher than control C57BL/6 mice. These results indicate that miR-383 expression is modulated by leptin in the hypothalamus and may, via regulating the POMC gene, influence the central control energy homeostasis [94].
It has been reported that miR-383 also correlated with various inflammatory diseases. The expression of miR-383 was significantly lower in TNF-α-treated jurkat cells and T cells from rheumatoid arthritis patients [95]. In lipopolysaccharide (LPS)-induced RAW264.7 cells, the expression of miR-383 was upregulated, indicating that miR-383 may participate in the regulation of the immune response [96]. MiR-383 expression was higher in the additional colonic mucosal tissue of ulcerative colitis (UC) and Crohn disease (CD) patients compared to the control, and CD expressed more miR-383 compared with UC. The differential expression of miR-383 between UC and CD indicates that miR-383 may have important functions in regulating idiopathic inflammatory bowel disease [97].
In some diseases, the expression of miR-338 was downregulated. For example, the expression of miR-383-5p was significantly downregulated in db/db mice and human podocytes after resveratrol treatment and overexpression of miR-383-5p inhibited resveratrol-induced autophagy and apoptosis [72]. The expression of miR-383-5p was decreased in rat serum and liver tissue samples kept at 4°C for 12 h, and it may regulate the metabolic pathway, which responds to cold stress [98]. Dengue fever (DF) patients had a higher miR-383 expression compared to DF patients with clinical fluid accumulation. These findings suggest that the downregulation of miR-383 could be involved in the complications of DF patients [99]. Rats suffering from chronic unpredictable mild stress have upregulated expression of miR-383-5p, while the expression of miR-383 was significantly decreased after electro-acupuncture intervention [100]. Therefore, miR-383-5p might affect depression by regulating neurotrophy and neurons apoptosis. However, the expression of miR-383 was upregulated in other diseases. This includes the villi of recurrent pregnancy loss patients, which had increases in miRNA-383, suggesting that miR-383 may regulate the pathogenesis of recurrent pregnancy loss by targeting MALAT1 [101]. Moreover, miRNA-383 was significantly increased in vitiligo patients. Through regulating EDN1, TYRP1, and PRDX3 expression, miRNA-383 participates in the pathogenesis and progression of oxidative stress, autoimmunity, or ER-stress-mediated vitiligo [102].
7. Regulation of miR-383 Expression
The expression and function of miRNAs could be regulated by transcriptional regulation and epigenetic modification. In the human genome, miR-383 is locates in the chr8p22 region within the third intron of the SGCZ gene. It has been reported that the loss of heterozygosity at the chr8p22 locus leads to the downregulated expression of miR-383 in prostate cancer [49]. DNA methylation was closely related to gene silencing and methyltransferases played essential function in this process. Zhang et al. reported that a liver-specific knockout of histone methyltransferase G9a significantly increased miR-383 expression [103]. Sun and colleagues reported that the expression of miR-383 was decreased by TGF-β1 in mouse ovarian granulosa cells [80], and they further illustrated that miR-383 was transcriptionally regulated by transcription factor steroidogenic factor-1 (SF-2) [81]. Moreover, the transcription activity of miR-383 has also been reported to be downregulated by signal transducer and activator of transcription 3 (STAT3) in human skin cancer [67]. Furthermore, it has been reported that HIF-1α promoted macrophage necroptosis by downregulating miR-383 [104]. In addition, the expression of miR-383 could be regulated by some anti-cancer agents. Lv et al. demonstrated that allicin treatment increased the expression of miR-383 in gastric carcinoma [46], while Huang et al. reported that resveratrol treatment decreased the expression of miR-383 in podocytes [72]. Guo et al. also revealed that piperine could decrease miR-383 expression and inhibit proptosis in myocardial ischaemia/reperfusion injury [105].
Interestingly, competing endogenous RNAs (ceRNAs), including long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs), could also regulate the expression and function of miR-383 through influencing the interaction of miR-383 with its target transcript. For example, lncRNA-FGD5-AS1 via sponging miR-383 accelerates the malignant characteristics of esophageal squamous cell carcinoma [38]. LncRNA-TMPO-AS1 promotes tumor growth, cell migration, and invasion in pancreatic carcinoma by regulating the miR-383/SOX11 axis [35]. Additionally, circRNA-CCS, via sponging miR-383, promotes lung cancer cell growth, metastasis, and predicted poor prognosis [14]. Circ-0136666 was reported to facilitate the carcinogenesis of colorectal cancer via targeting the miR-383/CREB1 axis [106]. We summarize the ceRNAs-miR-383 networks in Table 2.
Summarization of ceRNAs-miR-383 networks.
| Cancer types or Diseases | LncRNAs or circRNAs | Expression | Reference |
|---|---|---|---|
| Esophageal squamous cell carcinoma | LncRNA-FGD5-AS1 | Increased | [38] |
| Pancreatic carcinoma | lncRNA-TMPO-AS1 | Increased | [35] |
| Lung cancer | lncRNA-TMPO-AS1 | Increased | [107] |
| Glioma | lncRNA-TMPO-AS1 | Increased | [108] |
| Glioma | LINC01614 | Increased | [109] |
| Glioma | LINC00162 | Increased | [110] |
| Oral Squamous cell carcinoma | RP11-284F21.9 | Increased | [111] |
| Cervical cancer | LINC01128 | Increased | [112] |
| Hepatocellular carcinoma | LncRNA-PTTG3P | Increased | [113] |
| Liver cancer | LncRNA-HULC | Increased | [114] |
| Prostate cancer | LncRNA-SNHG1 | Increased | [115] |
| Breast cancer | LINC00096 | Increased | [116] |
| Head and neck squamous carcinoma | lncRNA-HOXC-AS | Increased | [53] |
| Head and neck squamous carcinoma | LncRNA-MIR4435-2HG | Increased | [117] |
| Nephropathy | LncRNA-PTTG3P | Increased | [118] |
| Diabetic retinopathy | LINC00162 | Increased | [119] |
| Diabetic retinopathy | lncRNA-LUADT1 | Increased | [120] |
| Diabetic retinopathy | lncRNA-AK077216 | Decreased | [121] |
| Recurrent pregnancy loss | LncRNA-MALAT1 | Decreased | [101] |
| Spinal cord injury | lncRNA-CASC9 | Decreased | [122] |
| Lung cancer | CircRNA-CCS | Increased | [14] |
| Colorectal cancer | CircRNA-0136666 | Increased | [106] |
| Medulloblastoma | CircRNA-SKA3 | Increased | [33] |
| Breast cancer | CircRNA-0001791 | Increased | [123] |
8. Conclusion
In this review, we summarized the roles of miR-383 in human diseases, which may be beneficial for further clinical applications. First, miR-383 has been reported to be dysregulated in various cancers in some studies, whereas it may present a different expression level in an identical cancer in other studies. For instance, miR-383 expression has been reported as decreased in hepatocellular cancer, whereas it was increased in two studies regarding the same type of cancer. Similarly, miR-383 was downregulated in ovarian cancer, while it was significantly upregulated in epithelial ovarian cancer. The possible explanation of the different miR-383 expression levels in the same types of cancer may due to the various detection methods, histological grade, or pathological stage. Second, the underlying mechanisms of miR-383 in the biological processes are complex and variable. We summarized the target genes and signaling pathways that were regulated by miR-383 in various tumors. However, many studies reported that the expression of miR-383 was markedly changed during tumorigenesis, while they did not explore the target genes. Therefore, other target genes and signal pathways of miR-383 should be revealed for further investigation. Third, up to now, most studies indicate that miR-383 functions as a tumor suppressor because it represses cell proliferation, decreases xenograft development, inhibits invasion and metastasis, promotes cell apoptosis, and sensitizes tumor cells to chemotherapy agents. However, it also has oncogenic functions in promoting proliferation, enhancing metastasis, and inducing tumorigenesis.
In conclusion, we summarized the aberrant expression of miR-383 in various human cancers, highlighting the functions of miR-383 in proliferation, development and differentiation, apoptosis, invasion and metastasis, as well as its roles in other human diseases. Additionally, we also reviewed the diverse target genes and signaling pathways regulated by miR-383 in cancer. This review provides some suggestions and evidence for further investigations and clinical applications. Although the relationship of miR-383 and tumorigenesis was explored, further investigations are required to explore its role in cancer biological behaviors. Future work will have help lead to strategies for cancer diagnosis and treatment.
Abbreviations
MiRNAs: microRNAs; UTR: Untranslated Region; UV: Ultraviolet radiation; EMT: Epithelial to mesenchymal transition; TNF-α: Tumor necrosis factor-α; TGF-β1: Transforming growth factor β 1; ATR: ATR serine/threonine kinase; LPS: lipopolysaccharide; ATM: ATM serine/threonine kinase; MALT lymphoma: Mucosa-associated lymphoid tissue lymphoma; ROS: Reactive Oxygen Species; CLL: Chronic lymphocytic leukemia; NSCLC: Non-Small-Cell Lung Cancer; DF: Dengue fever; UC: ulcerative colitis; CD: Crohn disease; GECs: Glioma-exposed endothelial cells; MSCs: Mesenchymal stem cells; LDHA: Lactate dehydrogenase A; VEGF: Vascular endothelial growth factor; FAK: Focal adhesion kinase; APRIL: A proliferating-inducing ligand; CDK2: Cyclin-dependent kinase 2; IRF1: Interferon regulatory factor 1; CCND1: Cyclin D1; IGF: Insulin like growth factor 1; IGF-1R: Insulin like growth factor 1 receptor; BCL-2: B cell lymphoma-2; SP1: Sp1 transcription factor; GDNF: Glial cell derived neurotrophic factor; PARP: Poly(ADP-ribose) polymerase; H2AX: H2AX variant histone; ZEB2: Zinc finger E-box binding homeobox 2; IRF1: Interferon regulatory factor 1; c-MYC: MYC proto-oncogene, bHLH transcription factor; MALAT1: Metastasis associated lung adenocarcinoma transcript 1; HMGA2: High mobility group AT-hook 2; PHF8: PHD finger protein 8; FOXM1: Forkhead box M1; SOX11: SRY-box transcription factor 11; GAB1: GRB2 associated binding protein 1; ROBO3: Roundabout guidance receptor 3; GADD45g: Growth arrest and DNA damage inducible gamma; PD-L1: CD274 molecule; PP2A: Protein phosphatase 2 phosphatase activator; HDAC9: Histone deacetylase 9; ERBB4: Erb-b2 receptor tyrosine kinase 4; PARP2: poly(ADP-ribose) polymerase 2; CD44: CD44 molecule; CASP2: Caspase 2; EIF5A2: Eukaryotic translation initiation factor 5A2; DIO1: Iodothyronine deiodinase 1; SGCZ: Sarcoglycan zeta; WNT1: Wnt family member 1; E2F7: E2F transcription factor 7; EPAS1: Endothelial PAS domain protein 1; CIP2A: Cellular inhibitor of PP2A; PRDX3: Peroxiredoxin 3; PNUTS: Protein phosphatase 1 regulatory subunit 10; RBMS1: RNA binding motif single stranded interacting protein 1; E2F1: E2F transcription factor 1; SF-1: Splicing factor 1; PPARγ: Peroxisome proliferator activated receptor gamma; PSD95: Discs large MAGUK scaffold protein 4; MMP: Matrix metalloproteinase; mTOR: Mechanistic target of rapamycin kinase; OCN: Bone gamma-carboxyglutamate protein; TRIM27: Tripartite motif containing 27; PAX6: Paired box 6; RPRD1B: Regulation of nuclear pre-mRNA domain containing 1B.
Acknowledgements
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding
This work was supported by funds from the National Natural Sciences Foundation of China (No. 82172831 to Yi Liao and No. 82003126 to Weichao Sun); China Postdoctoral Science Foundation Grant (No. 2019M663106 to Yi Liao); and Shenzhen Science and Technology Projects (No. JCYJ20190807102601647 to Qian Yi and JCYJ20210324103604013 to Weichao Sun).
Author Contributions
Qian Yi and Wei Xie performed the literature search and prepared the first draft of the manuscript; Weichao Sun wrote, reviewed, and edited the manuscript; Wei Sun supervised the review; Weichao Sun was the project administrator; and Weichao Sun and Yi Liao acquired funding. Qian Yi and Wei Xie contributed equally to this work. All of the authors have read and agreed to the published version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68:394-424
2. Rebecca L. Siege. Kimberly D. Miller.Ahmedin Jemal. Cancer Statistics,2019. CA: A Cancer journal for clinicians. 2019;69:7-34
3. Lee CH, Kuo WH, Lin CC, Oyang YJ, Huang HC, Juan HF. MicroRNA-regulated protein-protein interaction networks and their functions in breast cancer. International Journal of Molecular Sciences. 2013;14:11560-606
4. Takahashi RU, Prieto-Vila M, Hironaka A, Ochiya T. The role of extracellular vesicle microRNAs in cancer biology. Clinical Chemistry and Laboratory Medicine. 2017;55:648-56
5. Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20-51
6. Volinia S, Calin GA, Liu CG. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2257-61
7. Piovani G, Savio G, Traversa M. et al. De novo 1Mb interstitial deletion of 8p22 in a patient with slight mental retardation and speech delay. Molecular Cytogenetics. 2014;7:25
8. Kim JH, Dhanasekaran SM, Mehra R. et al. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer Research. 2007;67:8229-39
9. Taylor BS, Schultz N, Hieronymus H. et al. Integrative Genomic Profiling of Human Prostate Cancer. Cancer Cell. 2010;18:11-22
10. Kang J. Genomic alterations on 8p21-p23 are the most frequent genetic events in stage i squamous cell carcinoma of the lung. Experimental and Therapeutic Medicine. 2015;9:345-50
11. Shang Y, Zang A, Li J. et al. MicroRNA-383 is a tumor suppressor and potential prognostic biomarker in human non-small cell lung caner. Biomedicine and Pharmacotherapy. 2016;83:1175-81
12. Mueller DW, Rehli M, Bosserhoff AK. MiRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. Journal of Investigative Dermatology. 2009;129:1740-51
13. Gu B, Wang J, Song Y, Wang Q, Wu Q. microRNA-383 regulates cell viability and apoptosis by mediating Wnt/β-catenin signaling pathway in non-small cell lung cancer. Journal of Cell Biology. 2018
14. Yuan Y, Zhou X, Kang Y. et al. Circ-CCS is identified as a cancer-promoting circRNA in lung cancer partly by regulating the miR-383/E2F7 axis. Life Sciences. 2021;267:118955
15. Ma H, Liu B, Wang S, Liu J. MicroRNA-383 is a tumor suppressor in human lung cancer by targeting endothelial PAS domain-containing protein 1. Cell Biochemistry and Function. 2016;34:613-9
16. Zhao S, Gao X, Zang S, Li Y, Feng X, Yuan X. MicroRNA-383-5p acts as a prognostic marker and inhibitor of cell proliferation in lung adenocarcinoma by cancerous inhibitor of protein phosphatase 2A. Oncology Letters. 2017;14:3573-9
17. Xu Z, Zeng X, Li M, Liao J, Chen Q. MicroRNA-383 promotes reactive oxygen species-induced autophagy via downregulating peroxiredoxin 3 in human glioma U87 cells. Experimental and Therapeutic Medicine. 2021;21:439
18. He Z, Cen D, Luo X. et al. Downregulation of miR-383 promotes glioma cell invasion by targeting insulin-like growth factor 1 receptor. Medical Oncology. 2013;30:557
19. Dawei Xu, Pengju Ma, Guojun Gao, Yongkun Gui, Xiaolu Niu, Baozhe Jin. MicroRNA-383 expression regulates proliferation, migration, invasion, and apoptosis in human glioma cells. Tumor Biology. 2015;36:7743-53
20. Zhao LN, Wang P, Liu YH. et al. MiR-383 inhibits proliferation, migration and angiogenesis of glioma-exposed endothelial cells in vitro via VEGF-mediated FAK and Src signaling pathways. Cellular Signalling. 2017;30:142-53
21. Xu Z, Zeng X, Tian D. et al. MicroRNA-383 inhibits anchorage-independent growth and induces cell cycle arrest of glioma cells by targeting CCND1. Biochemical and Biophysical Research Communications. 2014;453:833-8
22. Huang H, Tian H, Duan Z, Cao Y, Zhang XS, Sun F. MicroRNA-383 impairs phosphorylation of H2AX by targeting PNUTS and inducing cell cycle arrest in testicular embryonal carcinoma cells. Cellular Signalling. 2014;26:903-11
23. Lian J, Tian H, Liu L. et al. Downregulation of microRNA-383 is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation by targeting IRF1. Cell Death and Disease. 2010;1:e94
24. Vilming Elgaaen B, Olstad OK, Haug KBF. et al. Global miRNA expression analysis of serous and clear cell ovarian carcinomas identifies differentially expressed miRNAs including miR-200c-3p as a prognostic marker. BMC Cancer. 2014;14:80
25. Han RL, Wang FP, Zhang PA, Zhou XY, Li Y. MiR-383 inhibits ovarian cancer cell proliferation, invasion and aerobic glycolysis by targeting LDHA. Neoplasma. 2017;64:244-52
26. Jiang J, Xie C, Liu Y, Shi Q, Chen Y. Up-regulation of miR-383-5p suppresses proliferation and enhances chemosensitivity in ovarian cancer cells by targeting TRIM27. Biomedicine and Pharmacotherapy. 2019;109:595-601
27. Yan F, Tu Z, Duan L, Wang D, Lin F. MicroRNA-383 suppresses cell proliferation and invasion in colorectal cancer by directly targeting paired box 6. Molecular Medicine Reports. 2018;17:6893-901
28. Jian Li, Amber R. Smith, Rebecca T. Marquez, Jun Li KL, Lan Lan, Xiaoqing Wu, Linxi Zhao, Fangli Ren YW, Yinyin Wang, Baoqing Jia, Liang Xu Z. MicroRNA-383 acts as a tumor suppressor in colorectal cancer by modulating CREPT RPRD1B expression. Molecular Carcinogenesis. 2018;57:1408-20
29. Fang Z, He L, Jia H, Huang Q, Chen D, Zhang Z. The miR-383-LDHA axis regulates cell proliferation, invasion and glycolysis in hepatocellular cancer. Iranian Journal of Basic Medical Sciences. 2017;20:187-192
30. Chen L, Guan H, Gu C, Cao Y, Shao J, Wang F. miR-383 inhibits hepatocellular carcinoma cell proliferation via targeting APRIL. Tumor Biology. 2016;37:2497-507
31. Cheng Y, Liu N, Yang CF. et al. MicroRNA-383 inhibits proliferation, migration, and invasion in hepatocellular carcinoma cells by targeting PHF8. Molecular Genetics and Genomic Medicine. 2020;8:e1272
32. Ferretti E, De Smaele E, Po A. et al. MicroRNA profiling in human medulloblastoma. International Journal of Cancer. 2009;124:568-77
33. Wang X, Xu D, Pei X. et al. Circska3 modulates foxm1 to facilitate cell proliferation, migration, and invasion while confine apoptosis in medulloblastoma via mir-383-5p. Cancer Management and Research. 2020;12:13415-26
34. Li KKW, Pang JCS, Lau KM. et al. MiR-383 is downregulated in medulloblastoma and targets peroxiredoxin 3 (PRDX3). Brain Pathology. 2013;23:413-25
35. Xue F, Song X, Zhang S. et al. Long non-coding RNA TMPO-AS1 serves as a tumor promoter in pancreatic carcinoma by regulating miR-383-5p/SOX11. Oncology Letters. 2021;21:255
36. Su QL, Zhao HJ, Song CF, Zhao S, Tian ZS, Zhou JJ. MicroRNA-383 suppresses pancreatic carcinoma development via inhibition of GAB1 expression. European Review for Medical and Pharmacological Sciences. 2019;23:10729-39
37. Han S, Cao C, Tang T. et al. ROBO3 promotes growth and metastasis of pancreatic carcinoma. Cancer Letters. 2015;366:61-70
38. Gao J, Zhang Z, Su H, Zong L, Li Y. Long noncoding rna fgd5-as1 acts as a competing endogenous rna on microRNA-383 to enhance the malignant characteristics of esophageal squamous cell carcinoma by increasing sp1 expression. Cancer Management and Research. 2020;12:2265-78
39. Wang X, Ren Y, Wang Z. et al. Down-regulation of 5S rRNA by miR-150 and miR-383 enhances c-Myc-rpL11 interaction and inhibits proliferation of esophageal squamous carcinoma cells. FEBS Letters. 2015;589:3989-97
40. Zhao X, Chen D, Cai Y, Zhang F, Xu J. RBPvsMIR: A computational pipeline to identify competing miRNAs and RNA-binding protein pairs regulating the shared transcripts. Genes. 2018;9:426
41. Zhang J, Kong X, Shi Q, Zhao B. MicroRNA-383-5p acts as a potential prognostic biomarker and an inhibitor of tumor cell proliferation, migration, and invasion in breast cancer. Cancer Biomarkers. 2020;27:423-32
42. Zhao L, Gu H, Chang J. et al. MicroRNA-383 regulates the apoptosis of tumor cells through targeting Gadd45g. PLoS ONE. 2014;9:e110472
43. Azarbarzin S, Hosseinpour-Feizi MA, Banan Khojasteh SM, Baradaran B, Safaralizadeh R. MicroRNA -383-5p restrains the proliferation and migration of breast cancer cells and promotes apoptosis via inhibition of PD-L1. Life Sciences. 2021;267:118939
44. Li X, Yuan J, Cao Q, Xie A, Chen J. MicroRNA-383-5p inhibits the proliferation and promotes the apoptosis of gastric cancer cells by targeting cancerous inhibitor of PP2A. International Journal of Molecular Medicine. 2020;46:397-405
45. Xu G, Li N, Zhang Y, Zhang J, Xu R, Wu Y. Microrna-383-5p inhibits the progression of gastric carcinoma via targeting hdac9 expression. Brazilian Journal of Medical and Biological Research. 2019;52:e8341
46. Lv Q, Xia Q, Li J, Wang Z. Allicin suppresses growth and metastasis of gastric carcinoma: the key role of microRNA-383-5p-mediated inhibition of ERBB4 signaling. Bioscience, Biotechnology and Biochemistry. 2020;84:1997-2004
47. Teng P, Jiao Y, Hao M, Tang X. microRNA-383 suppresses the PI3K-AKT-MTOR signaling pathway to inhibit development of cervical cancer via down-regulating PARP2. Journal of Cellular Biochemistry. 2018;119:5243-52
48. Cui Y, Chen LG, Yao HB, Zhang J, Ding KF. Upregulation of microrna-383 inhibits the proliferation, migration and invasion of colon cancer cells. Oncology Letters. 2018;15:1184-1190
49. Bucay N, Sekhon K, Yang T. et al. MicroRNA-383 located in frequently deleted chromosomal locus 8p22 regulates CD44 in prostate cancer. Oncogene. 2017;36:2667-79
50. Song Y, Jiang K, Su S, Wang B, Chen G. Clinical manifestations and epigenetic mechanisms of gastric mucosa associated lymphoid tissue lymphoma and long-term follow-up following Helicobacter pylori eradication. Experimental and Therapeutic Medicine. 2018;15:553-61
51. Wu S, Gu Y, Huang Y. et al. Novel Biomarkers for Non-functioning Invasive Pituitary Adenomas were Identified by Using Analysis of microRNAs Expression Profile. Biochemical Genetics. 2017;55:253-67
52. Costa FF, Bischof JM, Vanin EF. et al. Identification of micrornas as potential prognostic markers in ependymoma. PLoS ONE. 2011;6:e25114
53. Gao C, Lu W, Lou W, Wang L, Xu Q. Long noncoding RNA HOXC13-AS positively affects cell proliferation and invasion in nasopharyngeal carcinoma via modulating miR-383-3p/HMGA2 axis. Journal of Cellular Physiology. 2018:12809-20
54. Wan P, Chi X, Du Q. et al. miR-383 promotes cholangiocarcinoma cell proliferation, migration, and invasion through targeting IRF1. Journal of Cellular Biochemistry. 2018;119:9720-9
55. Liu J, Dou Y, Sheng M. Inhibition of microRNA-383 has tumor suppressive effect in human epithelial ovarian cancer through the action on caspase-2 gene. Biomedicine and Pharmacotherapy. 2016;83:1286-94
56. Noguchi S, Mori T, Hoshino Y, Yamada N, Maruo K, Akao Y. MicroRNAs as tumour suppressors in canine and human melanoma cells and as a prognostic factor in canine melanomas. Veterinary and Comparative Oncology. 2013;11:113-23
57. Shi Y, Huang A. Effects of sorafenib on lung metastasis in rats with hepatocellular carcinoma: the role of microRNAs. Tumor Biology. 2015;36:8455-63
58. Tu C, Chen W, Wang S. et al. MicroRNA-383 inhibits doxorubicin resistance in hepatocellular carcinoma by targeting eukaryotic translation initiation factor 5A2. Journal of Cellular and Molecular Medicine. 2019:1-10
59. Boguslawska J, Wojcicka A, Piekielko-Witkowska A, Master A, Nauman A. MiR-224 targets the 3′UTR of type 1 5′-iodothyronine deiodinase possibly contributing to tissue hypothyroidism in renal cancer. PLoS ONE. 2011;6:e24541
60. Qie S, Diehl JA. Cyclin D1, cancer progression, and opportunities in cancer treatment. Journal of Molecular Medicine. 2016;94:1313-26
61. Lu D, Wu Y, Wang Y. et al. Crept accelerates tumorigenesis by regulating the transcription of cell-cycle-related genes. Cancer Cell. 2012;21:92-104
62. Hahne M, Kataoka T, Schröter M. et al. APRIL, a new ligand of the tumor necrosis factor family, stimulates tumor cell growth. Journal of Experimental Medicine. 1998;188:1185-90
63. Mhawech-Fauceglia P, Allal A, Odunsi K, Andrews C, Herrmann FR, Huard B. Role of the tumour necrosis family ligand APRIL in solid tumour development: Retrospective studies in bladder, ovarian and head and neck carcinomas. European Journal of Cancer. 2008;44:2097-100
64. Lian J, Zhang X, Tian H. et al. Altered microRNA expression in patients with non-obstructive azoospermia. Reproductive Biology and Endocrinology. 2009;7:13
65. Jiang Y, Sang Y, Qiu Q. MicroRNA-383 mediates high glucose-induced oxidative stress and apoptosis in retinal pigment epithelial cells by repressing peroxiredoxin 3. American Journal of Translational Research. 2017;9:2374-83
66. Awasthi P, Foiani M, Kumar A. ATM and ATR signaling at a glance. Journal of Cell Science. 2016;129:1285
67. Liao XH, Zheng L, He HP. et al. STAT3 regulated ATR via microRNA-383 to control DNA damage to affect apoptosis in A431 cells. Cellular Signalling. 2015;27:2285-95
68. Ushio N, Rahman MM, Maemura T. et al. Identification of dysregulated microRNAs in canine malignant melanoma. Oncology Letters. 2019;17:1080-8
69. Salvador JM, Brown-Clay JD, Fornace AJ. Gadd45 in stress signaling, cell cycle control, and apoptosis. Advances in Experimental Medicine and Biology. 2013;793:1-19
70. C.-M. Y, K.-H. Y, C.G. W, et al. EPO-cyclosporine combination therapy reduced brain infarct area in rat after acute ischemic stroke: Role of innate immune-inflammatory response, micro-RNAs and MAPK family signaling pathway. American Journal of Translational Research. 2017;9:1651-66
71. Wang X, Ding G, Lai W, Liu S, Shuai J. MicroRNA-383 upregulation protects against propofol-induced hippocampal neuron apoptosis and cognitive impairment. Experimental and Therapeutic Medicine. 2018;15:3181-8
72. Huang SS, Ding DF, Chen S. et al. Resveratrol protects podocytes against apoptosis via stimulation of autophagy in a mouse model of diabetic nephropathy. Scientific Reports. 2017;7:45692
73. Lian Z, Lv FF, Yu J, Wang JW. The anti-inflammatory effect of microRNA-383-3p interacting with IL1R2 against homocysteine-induced endothelial injury in rat coronary arteries. Journal of Cellular Biochemistry. 2018;119:6684-94
74. Tang J, Zhang Z, Jin X, Shi H. miR-383 negatively regulates osteoblastic differentiation of bone marrow mesenchymal stem cells in rats by targeting Satb2. Bone. 2018;114:137-43
75. Wei GJ, An G, Shi ZW. et al. Suppression of MicroRNA-383 enhances therapeutic potential of human bone-marrow-derived mesenchymal stem cells in treating spinal cord injury via GDNF. Cellular Physiology and Biochemistry. 2017;41:1435-44
76. Wei G-J, An G, Shi Z-W. et al. Comprehensive Effects of Suppression of MicroRNA-383 in Human Bone-Marrow-Derived Mesenchymal Stem Cells on Treating Spinal Cord Injury. Cellular Physiology and Biochemistry. 2018;47:129-39
77. Tian H, Cao YX, Zhang XS. et al. The targeting and functions of miRNA-383 are mediated by FMRP during spermatogenesis. Cell Death and Disease. 2013;4:e614
78. Rengaraj D, Lee BR, Lee SI, Seo HW, Han JY. Expression patterns and mirna regulation of dna methyltransferases in chicken primordial germ cells. PLoS ONE. 2011;6:e19524
79. Donadeu FX, Schauer SN, Sontakke SD. Involvement of miRNAs in ovarian follicular and luteal development. Journal of Endocrinology. 2012;215:323-34
80. Yao G, Yin M, Lian J. et al. MicroRNA-224 is involved in transforming growth factor-β-mediated mouse granulosa cell proliferation and granulosa cell function by targeting Smad4. Molecular Endocrinology. 2010;24:540-51
81. Yin M, Lü M, Yao G. et al. Transactivation of microRNA-383 by Steroidogenic Factor-1 Promotes Estradiol Release from Mouse Ovarian Granulosa Cells by Targeting RBMS1. Molecular Endocrinology. 2012;26:1129-43
82. Yin M, Wang X, Yao G. et al. Transactivation of microRNA-320 by microRNA-383 regulates granulosa cell functions by targeting E2F1 and SF-1 proteins. Journal of Biological Chemistry. 2014;289:18239-57
83. Donadeu FX, Schauer SN. Differential miRNA expression between equine ovulatory and anovulatory follicles. Domestic Animal Endocrinology. 2013;45:122-5
84. S N Schauer, S D Sontakke, E D Watson CLE, Donadeu, FX. Involvement of miRNAs in equine follicle development. Reproduction. 2013;146:273-82
85. Salilew-Wondim D, Ahmad I, Gebremedhn S. et al. The expression pattern of microRNAs in granulosa cells of subordinate and dominant follicles during the early luteal phase of the bovine estrous cycle. PLoS ONE. 2014;9:e106795
86. Guay C, Menoud V, Regazzi R. et al. Identification of particular groups of microRNAs that positively or negatively impact on beta cell function in obese models of type 2 diabetes. Diabetologia. 2013;56:2203-12
87. Chakraborty C, Doss CGP, Bandyopadhyay S, Agoramoorthy G. Influence of miRNA in insulin signaling pathway and insulin resistance: Micro-molecules with a major role in type-2 diabetes. Wiley Interdisciplinary Reviews: RNA. 2014;5:697-712
88. Tugay K, Guay C, Marques AC. et al. Role of microRNAs in the age-associated decline of pancreatic beta cell function in rat islets. Diabetologia. 2016;59:161-9
89. Tang X, Le G-W, Chen L-M. et al. Role of miR-383 and miR-146b in different propensities to obesity in male mice. Journal of Endocrinology. 2017;234:201-16
90. Mandrekar-Colucci S, Colleen Karlo J, Landreth GE. Mechanisms underlying the Rapid Peroxisome proliferator-activated receptor-γ-mediated Amyloid clearance and reversal of cognitive deficits in a murine model of Alzheimer's disease. Journal of Neuroscience. 2012;32:10117-28
91. Pei L, Meng S, Yu W, Wang Q, Song F, Ma L. Inhibition of MicroRNA-383 Ameliorates Injury after Focal Cerebral Ischemia via Targeting PPARγ. Cellular Physiology and Biochemistry. 2016;39:1339-46
92. Takuma A, Abe A, Saito Y. et al. Gene expression analysis of the effect of ischemic infarction in whole blood. International Journal of Molecular Sciences. 2017;18:2335
93. Shu SY, Qing D, Wang B. et al. Comparison of microRNA expression in hippocampus and the marginal division (MrD) of the neostriatum in rats. Journal of Biomedical Science. 2013;20:9
94. Derghal A, Djelloul M, Airault C. et al. Leptin is required for hypothalamic regulation of miRNAs targeting POMC 3'UTR. Frontiers in Cellular Neuroscience. 2015;9:172
95. Lai NS, Yu HC, Tung CH, Huang KY, Huang H Bin, Lu MC. The role of aberrant expression of T cell miRNAs affected by TNF-α in the immunopathogenesis of rheumatoid arthritis. Arthritis Research and Therapy. 2017;19:261
96. Fan G, Jiang X, Wu X. et al. Anti-Inflammatory Activity of Tanshinone IIA in LPS-Stimulated RAW264.7 Macrophages via miRNAs and TLR4-NF-κB Pathway. Inflammation. 2016;39:375-84
97. Lin J, Zhang X, Zhao Z. et al. Novel MicroRNA Signature to Differentiate Ulcerative Colitis from Crohn Disease: A Genome-Wide Study Using Next Generation Sequencing. MicroRNA. 2017;5:222-9
98. Zhen L, Guo W, Peng M. et al. Identification of cold-responsive miRNAs in rats by deep sequencing. Journal of Thermal Biology. 2017;66:114-24
99. Tambyah PA, Ching CS, Sepramaniam S, Ali JM, Armugam A, Jeyaseelan K. microRNA expression in blood of dengue patients. Annals of Clinical Biochemistry. 2015;53:466-76
100. Duan DM, Dong X, Tu Y, Liu P. A microarray study of chronic unpredictable mild stress rat blood serum with electro-acupuncture intervention. Neuroscience Letters. 2016;627:160-7
101. Wang Y, Liu HZ, Liu Y, Wang HJ, Pang WW, Zhang JJ. Downregulated MALAT1 relates to recurrent pregnancy loss via sponging miRNAs. Kaohsiung Journal of Medical Sciences. 2018;34:503-10
102. Mansuri MS, Singh M, Begum R. miRNA signatures and transcriptional regulation of their target genes in vitiligo. Journal of Dermatological Science. 2016;84:50-8
103. Lu H, Lei X, Zhang Q. Liver-specific knockout of histone methyltransferase G9a impairs liver maturation and dysregulates inflammatory, cytoprotective, and drug-processing genes. Xenobiotica. 2019;49:740-52
104. Karshovska E, Wei Y, Subramanian P. et al. HIF-1α (hypoxia-inducible factor-1α) promotes macrophage necroptosis by regulating miR-210 and miR-383. Arteriosclerosis, Thrombosis, and Vascular Biology. 2019;40:583-96
105. Guo X, Hu S, Liu JJ. et al. Piperine protects against pyroptosis in myocardial ischaemia/reperfusion injury by regulating the miR-383/RP105/AKT signalling pathway. Journal of Cellular and Molecular Medicine. 2021;25:244-58
106. Li Y, Zang H, Zhang X, Huang G. Circ_0136666 facilitates the progression of colorectal cancer via mir-383/creb1 axis. Cancer Management and Research. 2020;12:6795-806
107. Mu X, Wu H, Liu J. et al. Long noncoding RNA TMPO-AS1 promotes lung adenocarcinoma progression and is negatively regulated by miR-383-5p. Biomedicine and Pharmacotherapy. 2020;125:109989
108. Liu G, Yang H, Cao L, Han K, Li G. Lncrna tmpo-as1 promotes proliferation and invasion by sponging mir-383-5p in glioma cells. Cancer Management and Research. 2020;12:12001-9
109. Wang H, Wu J, Guo W. Sp1-mediated upregulation of lncrna linc01614 functions a cerna for mir-383 to facilitate glioma progression through regulation of adam12. OncoTargets and Therapy. 2020;13:4305-18
110. Xu Z, Chen Q, Zeng X, Li M, Liao J. Lnc-NLC1-C inhibits migration, invasion and autophagy of glioma cells by targeting miR-383 and regulating PRDX-3 expression. Oncology Letters. 2021;22:640
111. Shao B, Fu X, Li X, Li Y, Gan N. RP11-284F21.9 promotes oral squamous cell carcinoma development via the miR-383-5p/MAL2 axis. Journal of Oral Pathology and Medicine. 2020;49:21-9
112. Hu Y, Ma Y, Liu J, Cai Y, Zhang M, Fang X. LINC01128 expedites cervical cancer progression by regulating miR-383-5p/SFN axis. BMC Cancer. 2019;19:1157
113. Zhou Q, Zhang W, Wang Z, Liu S. Long non-coding RNA PTTG3P functions as an oncogene by sponging miR-383 and up-regulating CCND1 and PARP2 in hepatocellular carcinoma. BMC cancer. 2019;19:731
114. Li P, Li Y, Ma L. Long noncoding RNA highly upregulated in liver cancer promotes the progression of hepatocellular carcinoma and attenuates the chemosensitivity of oxaliplatin by regulating miR-383-5p/vesicle-associated membrane protein-2 axis. Pharmacology Research and Perspectives. 2021 9
115. Huang G, Guo X, Yang H. Long noncoding RNA SNHG1 promotes human prostate cancer progression by sponging miR-383-5p. Anti-Cancer Drugs. 2021;32:286-95
116. Tian Y, Xia S, Ma M, Zuo Y. LINC00096 promotes the proliferation and invasion by sponging miR-383-5p and regulating RBM3 expression in triple-negative breast cancer. OncoTargets and Therapy. 2019;12:10569-78
117. Wang S, Chen X, Qiao T. Long non-coding RNA MIR4435-2HG promotes the progression of head and neck squamous cell carcinoma by regulating the miR-383-5p/RBM3 axis. Oncology Reports. 2021;45:99
118. Bi M, Shi J, Zhao Y, Li CM. LncRNA PTTG3P induced aberrant glycosylated IgA1 production and B cell growth in IgA nephropathy. Environmental Science and Pollution Research. 2021
119. Fan WX, Wen XL, Zheng JF. et al. LINC00162 participates in the pathogenesis of diabetic nephropathy via modulating the miR-383/HDAC9 signalling pathway. Artificial Cells, Nanomedicine and Biotechnology. 2020;48:1047-54
120. Dai R, Sun Z, Qian Y, Zhang B, Han Y, Deng G. LncRNA LUADT1 inhibits cell apoptosis in diabetic retinopathy by regulating miR-383/peroxiredoxin 3 axis. Archives of Physiology and Biochemistry. 2020:1-6
121. Zhang X, Shi E, Yang L, Fu W, Hu F, Zhou X. LncRNA AK077216 is downregulated in diabetic retinopathy and inhibited the apoptosis of retinal pigment epithelial cells by downregulating miR-383. Endocrine Journal. 2019;66:1011-6
122. Guan C, Wang Y. LncRNA CASC9 attenuates lactate dehydrogenase-mediated oxidative stress and inflammation in spinal cord injury via sponging miR-383-5p. Inflammation. 2021;44:923-33
123. Ameli-Mojarad M, Ameli-Mojarad M, Nourbakhsh M, Nazemalhosseini-Mojarad E. Circular RNA hsa_circ_0005046 and hsa_circ_0001791 May Become Diagnostic Biomarkers for Breast Cancer Early Detection. Journal of Oncology. 2021;2021:2303946
Author contact
![]() Corresponding authors: 414464705com (W. S.); weichaosunshinecom (Wc. S.); science0528com (Y. L.)
Corresponding authors: 414464705com (W. S.); weichaosunshinecom (Wc. S.); science0528com (Y. L.)

 Global reach, higher impact
Global reach, higher impact