3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(23):6931-6936. doi:10.7150/jca.63333 This issue Cite
Research Paper
Clinicopathological Significance of ATRX Expression in Nasopharyngeal Carcinoma Patients: A Retrospective Study
1. Department of Oncology, The Second Xiangya Hospital, Central South University, Changsha, Hunan, 410011, China.
2. Department of Pathology, The Second Xiangya Hospital, Central South University, Changsha, Hunan, 410011, China.
Received 2021-5-30; Accepted 2021-9-22; Published 2021-10-3
Abstract
Background: Nasopharyngeal carcinoma (NPC) is the most common head and neck squamous cell carcinoma in south China. Radiation technology improves the local control rates in early NPC. However, the distant metastases are still the main cause of treatment failure. Thus, to find biomarkers for prognosis will help to enhance the survival of NPC. ATRX is a chromatin remodeling protein localized in the nucleus. Deletion or mutation of ATRX gene has been demonstrated in a variety of malignancies. However, the significance of ATRX expression in the prognosis of NPC remains unclear.
Methods: Tumor tissues from 227 NPC patients diagnosed in the Second Xiangya Hospital of Central South University from 2011 to 2016 were selected. Immunohistochemistry was used to detect the ATRX expression level of the tumor tissue. Chi-square test was used to analyze the relationship between ATRX expression and clinical characteristics such as age, sex, T stage, N stage and clinical stage. Kaplan-Meier method was used for survival analysis, and log-rank was used to compare the difference in survival rate.
Results: There were 53 patients with negative ATRX expression, accounting for 24.2% of the total group. ATRX expression was not significantly associated with age, sex, N stage, clinical stage, and progression-free survival (PFS) (P>0.05). However, patients with negative ATRX expression had earlier T staging (P=0.045) and a higher 5-year overall survival (84.9% vs 66.9%, P=0.022).
Conclusions: Loss of ATRX expression may contribute to better prognosis in patients with NPC.
Keywords: Nasopharyngeal Carcinoma, ATRX, Staging, Survival, Prognosis
Introduction
Nasopharyngeal Carcinoma (NPC) is the most common head and neck squamous cell carcinoma in south China and southeastern Asia [1, 2]. Due to its radiosensitive behavior, radiation (RT) and chemoradiotherapy (CRT) has become the cornerstone of the treatment for early or locoregionally advanced diseases [3, 4]. However, about 25-30% of the patients will suffer from disease relapse or metastasis, and the prognosis of advanced patients is poor. The biological behavior of NPC varied dramatically in individual patients. Therefore, seeking for biomarkers for survival prediction, patient stratification and treatment adaptation is urgently needed [5].
Alpha thalassemia/mental retardation X-linked (ATRX) is a protein containing an ATPase/helicase domain, which belongs to the switch/sucrose nonfermentable (SWI/SNF) family of chromatin remodeling proteins [6]. It is found to be involved in cell cycle-dependent phosphorylation, which regulates its nuclear matrix and chromatin association and suggests its involvement in gene regulation at interphase and chromosomal segregation in mitosis [7]. It has been demonstrated that ATRX is frequently mutated in a variety of mesenchymal tumors including gliomas [8], neuroendocrine neoplasms [9], and sarcomas [10]. ATRX mutation leads to decreased ATRX protein expression, and results in tumor genome instability, higher tumor mutation burden, and thus leading to increased sensitivity to chemotherapy, radiation therapy and immunotherapy agents [11, 12]. However, the data on the incidence of ATRX expression and clinical significance in epithelial carcinomas is limited. It is reported that ATRX loss predicts good prognosis in cervical carcinoma and hepatocarcinoma [13, 14]. And ATRX mutation results in increased immune checkpoint inhibitor (ICI) sensitivity in NSCLC [12]. However, the significance of ATRX expression in the prognosis of NPC remains unknown.
In the present study, we evaluated the ATRX expression in NPC patients via immunohistochemistry and analyzed the correlation between ATRX expression and clinicopathological characteristics and patient's prognosis, and demonstrated that ATRX deficiency is correlated with earlier T stage and longer overall survival in NPC patients.
Materials and Methods
Patients enrollment and clinical data
Patients who were diagnosed with NPC at the Second Xiangya Hospital of Central South University (Hunan, China) between 2011 and 2016 were retrospectively reviewed. And patients without complete clinical data or unavailable paraffin blocks were excluded. Finally, a total of 227 patients were enrolled in this study. Two pathologists (HH Wang and SQ Fan) independently reviewed all H&E staining and IHC staining slides. And all the clinical and pathological characteristics were obtained from electronic medical records system of the hospital. Approval for using the patient material in this study was obtained from the Ethics Committee of the Second Xiangya Hospital of Central South University. All procedures were conducted in accordance with the declaration of Helsinki. Tissue samples were obtained from the Department of Pathology of the Second Xiangya Hospital of Central South University.
Immunohistochemical staining and semi-quantitative scores
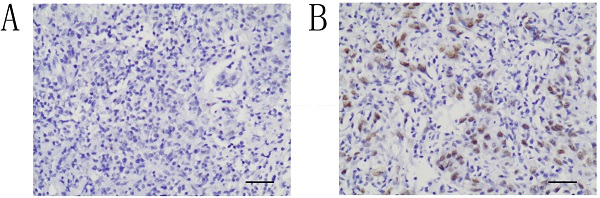
Immunohistochemical staining for ATRX was performed with 4-μm-thick sections from paraffin blocks using the MaxVision HRP polymer anti-rat IHC kit. Each section was deparaffinized and rehydrated, and high-temperature antigen retrieval was achieved for all antibodies by heating the samples in 0.01 M citrate buffer in a domestic microwave oven at full power (1000 W) for 15 mins. Besides internal positive control, positive control slides were included in every experiment. Methanol containing 3% H2O2 was applied for fifteen minutes to inactivate endogenous peroxidase. The slides were incubated with a rabbit monoclonal antibody against human, a 1:200 dilution of the primary antibody to ATRX (Sigma, Sigma-Aldrich, St. Louis, MO, USA) at 4 °C overnight. Then, all the slides were rinsed with PBS three times for 5 mins each. After the incubation with secondary antibody, the visualization signal was conducted with 3,3′-diaminobenzidine tetrachloride. Subsequently, all slides were counterstained with hematoxylin. The specificity of the antibody was determined with matched IgG isotype antibody as a negative control. Tissue samples were evaluated independently by HHW and SQF who were blinded to the clinicopathological data, at 200× magnification light microscopy. Nucleus staining of the tumor cells was assessed. Representative microscopic photos are shown in Figure 1. According to the proportion of nucleus staining of the tumor cells, the ATRX expression was classified as negative and positive. ATRX staining loss of tumor nucleus was defined as ATRX negative while ATRX positive was defined as that ATRX is expressed in at least 1% of nuclei in the tumor cells. The concordance rate between the two pathologist is 97%.
Immunohistochemical study of ATRX expression in NPC tissues. The intensity of nucleus staining was graded as negative (A) or positive (B) (A-B, ×200).
Statistical analysis
Statistical analysis was performed using SPSS software V.22 (IBM, Armonk, USA). The χ² test was used to evaluate the association between ATRX expression and clinicopathological parameters including Age, Gender, T stage, N stage and clinical stage according to AJCC 8th staging system. Progression-free survival (PFS) and overall survival (OS) curves were plotted using Kaplan-Meier method, and the log-rank test was used to compare the differences. P<0.05 was considered as statistically significant.
Results
Patient characteristics
The clinicopathological characteristics of the patients are summarized in Table 1. A total of 227 patients were enrolled, among which 138 (60.8%) patients were less than 50 years old, with a median age of 48 years. 156 (68.7%) patients were men, with a male-to-female ratio of 2.2:1. All patients were diagnosed with NPC and treated with chemoradiation therapy. 26 (11.5%) were T1 stage, 88 (38.8%) were T2 stage, 68 (30.0%) were T3 stage and 45 (19.8%) were T4 stage. 15 (6.6%) were N0 stage, 37 (16.3%) were N1 stage, 144 (63.4%) were N2 stage and 31 (13.7%) were N3 stage. According to the 8th AJCC staging system, 24 (10.6%) were stage II, 132 (58.1%) were stage III and 71 (31.3%) were stage IV. A total of 103 (45.4%) patients received concurrent chemoradiation, and 124 (54.6%) received sequential chemoradiation. Among all the patients, 145 (63.9%) received cisplatin-based chemotherapy, 68 (30.0%) received nedaplatin-based chemotherapy, while 14 (6.1%) received other agents.
Correlations between ATRX expression and clinicopathological factors
NPC patients with ATRX expression, defined as ATRX positive staining in at least 1% of nuclei in the tumor cells, consist of 76.7% (174/227) of the cohort. And 23.3% (53/227) of the cohort had ATRX loss. The association between ATRX expression and the clinicopathological factors are shown in Table 2. ATRX loss was significantly correlated with earlier T stage (p=0.045). Statistical significance was not found in sex, age, ECOG, smoking history, hemoglobin level, treatment mode, chemotherapy agent type, N stage and AJCC stages.
Impact of ATRX expression on the prognosis of NPC patients
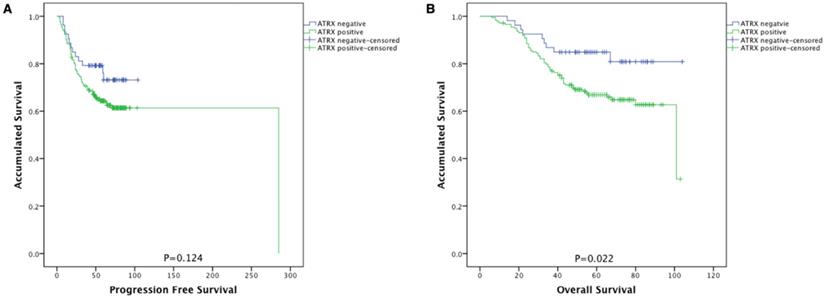
As shown in Figure 2, NPC patients with ATRX loss had significantly longer overall survival than those with ATRX expression (84.9% vs 66.9%, P=0.022). No prognostic significance between ATRX expression and progression free survival (PFS) was observed (73.1% vs 65.9%, p=0.124).
Baseline characteristics of patients with NPC
| Clinicopathological characteristics | Number (%) |
|---|---|
| Age | |
| ≤50 | 138 (60.8%) |
| >50 | 89 (39.2%) |
| Gender | |
| Male | 156 (68.7%) |
| Female | 71 (31.3%) |
| ECOG | |
| 0 | 186 (81.9%) |
| 1 | 41 (18.1%) |
| Smoking history | |
| No | 93 (40.9%) |
| Yes | 134 (59.1%) |
| HGB (M/F) (g/L) | |
| <120/110 | 9 (4.0%) |
| ≥120/110 | 218 (96.0%) |
| Treatment mode | |
| Concurrent chemoradiation | 103 (45.4%) |
| Sequential chemoradiation | 124 (54.6%) |
| Chemotherapy agent type | |
| Cisplatin | 145 (63.9%) |
| Nedaplatin | 68 (30.0%) |
| Others | 14 (6.1%) |
| Total platin dose (mg/m2) | |
| ≥300 | 164 (72.2%) |
| <300 | 63 (27.8%) |
| ECOG | |
| 0 | 185 (81.5%) |
| 1 | 42 (18.5%) |
| Smoking history | |
| No | 93 (41.0%) |
| Yes | 134 (59.0%) |
| HGB (M/F) (g/L) | |
| <120/110 | 15(6.6%) |
| ≥120/110 | 212 (93.4%) |
| Treatment mode | |
| Concurrent chemoradiation | 103 (45.4%) |
| Sequential chemoradiation | 124 (54.6%) |
| Chemotherapy agent type | |
| Cisplatin | 145 (63.9%) |
| Nedaplatin | 68 (30.0%) |
| Others | 14 (6.1%) |
| T stages | |
| T1 | 26 (11.5%) |
| T2 | 88 (38.8%) |
| T3 | 68 (30.0%) |
| T4 | 45 (19.8%) |
| N stages | |
| N0 | 15 (6.6%) |
| N1 | 37 (16.3%) |
| N2 | 144 (63.4%) |
| N3 | 31 (13.7%) |
| AJCC stages | |
| II | 24 (10.6%) |
| III | 132 (58.1%) |
| IV | 71 (31.3%) |
Kaplan-Meier survival curves of patients with NPC stratified by ATRX expression. (A) Progression-free survival and (B) overall survival according to ATRX expression in NPC patients. ATRX negative (n=53) and ATRX positive (n=174).
Correlation between ATRX expression and clinicopathological factors in NPC
| Clinicopathological factors | N | ATRX expression | ||
|---|---|---|---|---|
| negative | positive | P value | ||
| Gender | 0.630 | |||
| Male | 156 | 35 (66.0%) | 121 (69.5%) | |
| Female | 71 | 18 (34.0%) | 53 (30.5%) | |
| Age | ||||
| ≤50 | 138 | 28 (52.8%) | 110 (63.2%) | 0.175 |
| >50 | 89 | 25 (47.2%) | 64 (36.4%) | |
| ECOG | 0.559 | |||
| 0 | 186 | 46 (84.9%) | 140 (81.4%) | |
| 1 | 41 | 9 (15.1%) | 32 (18.6%) | |
| Smoking history | 0.671 | |||
| No | 93 | 24 (43.4%) | 69 (40.1%) | |
| Yes | 134 | 31 (56.6%) | 103 (59.9%) | |
| HGB (M/F) (g/L) | 0.370 | |||
| <120/110 | 9 | 1 (1.9%) | 8 (4.7%) | |
| ≥120/110 | 218 | 54 (98.1%) | 164 (95.3%) | |
| Treatment mode | 0.759 | |||
| Concurrent chemoradiation | 103 | 26 (47.2%) | 77 (44.8%) | |
| Sequential chemoradiation | 124 | 29 (52.8%) | 95 (55.2%) | |
| Chemotherapy agent type | 0.291 | |||
| Cisplatin | 145 | 32 (58.5%) | 113 (65.7%) | |
| Nedaplatin | 68 | 21 (37.7%) | 47 (27.3%) | |
| Others | 14 | 2 (3.8%) | 12 (7.0%) | |
| T stages | 0.045 | |||
| T1-2 | 114 | 33 (62.3%) | 81 (46.6%) | |
| T3-4 | 113 | 20 (37.7%) | 93 (53.4%) | |
| N stages | 0.670 | |||
| N0-N1 | 52 | 11 (20.8%) | 41 (23.6%) | |
| N2-N3 | 175 | 42 (79.2%) | 133 (76.4%) | |
| AJCC stages | 0.383 | |||
| II-III | 156 | 39 (73.6%) | 117 (67.2%) | |
| IV | 71 | 14 (26.4%) | 57 (32.8%) | |
Discussion
In the present study, we investigated the expression of ATRX in NPC and its correlation with clinicopathological characteristics and prognosis of NPC patients. ATRX protein expression was absent in 24.2% of NPC patients, and ATRX loss is correlated with earlier T stage and longer OS in NPC patients.
ATRX is a member of the SWI/SNF chromatin remodeling superfamily which is involved in remodeling and stabilizing genome [15]. It is reported that SWI/SNF genes are found to be mutated in almost 25% of malignancies, which have prognostic significance [16]. Moreover, expression changes of specific SWI/SNF chromatin remodeling proteins are also implicated as prognostic biomarkers [16]. However, the consequences of mutations or expression alterations on prognoses are highly context-specific [16]. ATRX mutation is mostly seen in mesenchymal malignances, including glioma, sarcoma, and neuroendocrine tumors, while the report on the incidence and prognostic significance on epithelial malignancies is rare. Loss of ATRX results in impaired genome damage repair system, which leads to genetic instability, microsatellite instability (MSI), and more aggressive phenotype [17]. At the same time, it also sensitizes cancer cells to treatment agents, including DNA-damaging agents, radiotherapy [11, 18], and immune checkpoint inhibitors [12, 19]. The prognostic significance of ATRX mutation on cancer patients varies between researches. It is reported that ATRX mutation is a poor prognostic factor in pancreatic neuroendocrine tumors (pNETs) [20], hepatic angiosarcomas [21], and leiomyosarcoma [22, 23], while it is found to be a good prognostic factor in glioma [24], and cervical cancer [13]. Such complexities without universal good prognosis may suggest that mutations affecting chromatin remodeling genes or expression altering chromatin remodeling proteins do not directly determine the aggressiveness or prognosis of malignancies [16]. Besides, this discrepancy may be resulted from patient selection and treatment modality. In the present study, we firstly reported the incidence of ATRX deficiency in NPC patients, and found that ATRX deficiency predicts good prognosis in NPC patients. However, the underlying mechanism is not fully understood, and it may be related with the following aspects. Firstly, since the ATRX protein plays significant epigenetic roles in maintaining telomere length via depositing histones at heterochromatin and telomeric DNA, tumors with ATRX mutations or loss exhibit altered telomeres and telomere dysfunction [25, 26]. Moreover, ATRX mutations increase tumor mutational burden (TMB), PD-L1 expression, as well as manifested interferon gamma (IFN-γ) signaling [12, 19]. Given that radiotherapy or chemoradiotherapy is the primary treatment for NPC, and the potential role of ATRX mutation in glioma patients treated with RT has been reported. Collectively, the mechanism of ATRX mutation conferring survival advantages in NPC is worth further investigation.
Mutations in ATRX are thought to result in loss of ATRX protein which mediates loss of function (LOF), and therefore in the present study, immunohistochemistry (IHC) is used as a surrogate to detect mutations. In previous research in glioma, pancreatic neuroendocrine tumors, and neuroblastoma have demonstrated that ATRX mutations are associated with absence of ATRX protein. However, in another research, Chami et al. [27] reported that using IHC as a surrogate may miss ATRX mutations in neuroblastoma, which indicated detecting ATRX mutation by IHC protein expression as a surrogate still needs further validation. Furthermore, the consistency between ATRX protein absence and ATRX mutation in NPC is worth further investigation.
Although this study identified the prognostic significance of ATRX loss in NPCs, there are several limitations. First, confirmation of ATRX mutations by other techniques, such as sequencing from fresh tissue was not included. Additionally, this study is a retrospective study, and due to a limited sample size, there is a need for validation in larger cohorts with clinical, molecular alterations, conjointly with protein expression/activation.
In conclusion, we evaluated the clinicopathological significance of ATRX expression in 227 patients with NPC. ATRX loss was significantly associated with good prognosis in patients.
Abbreviations
NPC: nasopharyngeal carcinoma; ATRX: alpha thalassemia/mental retardation X-linked; SWI/SNF: switch/sucrose nonfermentable; ICI: immune checkpoint inhibitor; PFS: progression-free survival; OS: overall survival; MSI: microsatellite instability; pNETS: pancreatic neuroendocrine tumors; TMB: tumor mutational burden; IFN-γ: interferon gamma; LOF: loss of function; IHC: immunohistochemistry.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81802476, 81802620, 82172656), the Natural Science Foundation of Hunan Province (2020JJ5807, 2021JJ40882), and the Scientific Research Project of Hunan Provincial Health Commission (20201019).
Author Contributions
T.H., X.L.L., C.H.H. conceived and designed the study. H.H.W. and S.Q.F. contributed to data acquisition. Data analysis and manuscript drafting were performed by S.S.W. and Y.C.X. Figures and tables were created by Y.H.F. and Y.Q.F. All authors were involved in the writing or review of the manuscript and approved the final version.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Her C. Nasopharyngeal cancer and the Southeast Asian patient. Am Fam Physician. 2001;63:1776-82
2. Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. Lancet. 2016;387:1012-24
3. Lee AW, Lin JC, Ng WT. Current management of nasopharyngeal cancer. Semin Radiat Oncol. 2012;22:233-44
4. Blanchard P, Lee A, Marguet S, Leclercq J, Ng WT, Ma J. et al. Chemotherapy and radiotherapy in nasopharyngeal carcinoma: an update of the MAC-NPC meta-analysis. Lancet Oncol. 2015;16:645-55
5. Yang L, Hong S, Wang Y, Chen H, Liang S, Peng P. et al. Development and External Validation of Nomograms for Predicting Survival in Nasopharyngeal Carcinoma Patients after Definitive Radiotherapy. Sci Rep. 2015;5:15638
6. Gibbons RJ, Picketts DJ, Villard L, Higgs DR. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome). Cell. 1995;80:837-45
7. Berube NG, Smeenk CA, Picketts DJ. Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum Mol Genet. 2000;9:539-47
8. Xie Y, Tan Y, Yang C, Zhang X, Xu C, Qiao X. et al. Omics-based integrated analysis identified ATRX as a biomarker associated with glioma diagnosis and prognosis. Cancer Biol Med. 2019;16:784-96
9. Arakelyan J, Zohrabyan D, Philip PA. Molecular profile of pancreatic neuroendocrine neoplasms (PanNENs): Opportunities for personalized therapies. Cancer. 2021;127:345-53
10. Liu W, Tong H, Zhang C, Zhuang R, Guo H, Lv C. et al. Integrated genomic and transcriptomic analysis revealed mutation patterns of de-differentiated liposarcoma and leiomyosarcoma. BMC Cancer. 2020;20:1035
11. Koschmann C, Calinescu AA, Nunez FJ, Mackay A, Fazal-Salom J, Thomas D. et al. ATRX loss promotes tumor growth and impairs nonhomologous end joining DNA repair in glioma. Sci Transl Med. 2016;8:328ra28
12. Hou T, Jiang S, Wang Y, Xie Y, Zhang H, Feng Y. et al. Alpha Thalassemia/Intellectual Disability X-Linked Deficiency Sensitizes Non-Small Cell Lung Cancer to Immune Checkpoint Inhibitors. Front Oncol. 2020;10:608300
13. Scholl SM, Beal J, de Koning L, Girard E, Popovic M, de la Rochefordiere A. et al. Genetic markers and phosphoprotein forms of beta-catenin pbeta-Cat552 and pbeta-Cat675 are prognostic biomarkers of cervical cancer. EBioMedicine. 2020;61:103049
14. Liang J, Zhao H, Diplas BH, Liu S, Liu J, Wang D. et al. Genome-Wide CRISPR-Cas9 Screen Reveals Selective Vulnerability of ATRX-Mutant Cancers to WEE1 Inhibition. Cancer Res. 2020;80:510-23
15. Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD. et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351-60
16. Mittal P, Roberts CWM. The SWI/SNF complex in cancer - biology, biomarkers and therapy. Nat Rev Clin Oncol. 2020;17:435-48
17. Terra Md S, Xie Md Ph DH, Boland Md JM, Mansfield Md AS, Molina Md Ph DJ, Roden Md AC. Loss of ATRX expression predicts worse prognosis in pulmonary carcinoid tumors. Hum Pathol. 2019;94:78-85
18. Nunez FJ, Mendez FM, Kadiyala P, Alghamri MS, Savelieff MG, Garcia-Fabiani MB. et al. IDH1-R132H acts as a tumor suppressor in glioma via epigenetic up-regulation of the DNA damage response. Sci Transl Med. 2019 11
19. Ge Y, Wei F, Du G, Fei G, Li W, Li X. et al. The association of sex-biased ATRX mutation in female gastric cancer patients with enhanced immunotherapy-related anticancer immunity. BMC Cancer. 2021;21:240
20. Marinoni I, Kurrer AS, Vassella E, Dettmer M, Rudolph T, Banz V. et al. Loss of DAXX and ATRX are associated with chromosome instability and reduced survival of patients with pancreatic neuroendocrine tumors. Gastroenterology. 2014;146:453-60 e5
21. Liau JY, Tsai JH, Yang CY, Lee JC, Liang CW, Hsu HH. et al. Alternative lengthening of telomeres phenotype in malignant vascular tumors is highly associated with loss of ATRX expression and is frequently observed in hepatic angiosarcomas. Hum Pathol. 2015;46:1360-6
22. Liau JY, Tsai JH, Jeng YM, Lee JC, Hsu HH, Yang CY. Leiomyosarcoma with alternative lengthening of telomeres is associated with aggressive histologic features, loss of ATRX expression, and poor clinical outcome. Am J Surg Pathol. 2015;39:236-44
23. Makinen N, Aavikko M, Heikkinen T, Taipale M, Taipale J, Koivisto-Korander R. et al. Exome Sequencing of Uterine Leiomyosarcomas Identifies Frequent Mutations in TP53, ATRX, and MED12. PLoS Genet. 2016;12:e1005850
24. Wiestler B, Capper D, Holland-Letz T, Korshunov A, von Deimling A, Pfister SM. et al. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013;126:443-51
25. Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C. et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425
26. Huang H, Liu Q, Zhu L, Zhang Y, Lu X, Wu Y. et al. Prognostic Value of Preoperative Systemic Immune-Inflammation Index in Patients with Cervical Cancer. Sci Rep. 2019;9:3284
27. Chami R, Marrano P, Teerapakpinyo C, Arnoldo A, Shago M, Shuangshoti S. et al. Immunohistochemistry for ATRX Can Miss ATRX Mutations: Lessons From Neuroblastoma. Am J Surg Pathol. 2019;43:1203-11
Author contact
![]() Corresponding author: Tao Hou, E-mail: houtaoedu.cn; Telephone: +8613786122515; Fax: 073185295888.
Corresponding author: Tao Hou, E-mail: houtaoedu.cn; Telephone: +8613786122515; Fax: 073185295888.

 Global reach, higher impact
Global reach, higher impact