Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(21):6519-6530. doi:10.7150/jca.60743 This issue Cite
Research Paper
miR-328-3p promotes migration and invasion by targeting H2AFX in head and neck squamous cell carcinoma
1. Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Changsha, China.
2. Otolaryngology Major Disease Research Key Laboratory of Hunan Province, Changsha, China.
3. Clinical Research Center for Pharyngolaryngeal Diseases and Voice Disorders in Hunan Province, Changsha, China.
4. National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China.
Received 2021-3-22; Accepted 2021-8-24; Published 2021-9-9
Abstract
Migration and invasion are the initial step in the metastatic process, while metastasis is responsible for the poor prognosis of head and neck squamous cell carcinoma (HNSCC). Since miRNA has been found as an important regulator of gene expression at the post-transcriptional level in various diseases including carcinoma, exploring the role of miRNA in cancer metastasis will facilitate the target therapy of advanced HNSCC. MiR-328-3p has been reported to be an onco-miRNA or a tumor suppressor in several cancers. However, the role of miR-328-3p in HNSCC migration and invasion remains undefined. In this study, we first demonstrated that miR-328-3p enhanced migration and invasion of HNSCC in vitro, accompanying with a promotion of epithelial-mesenchymal transition (EMT) and mTOR activity. Meanwhile, we confirmed that miR-328-3p directly targeted the 3'UTR of H2A histone family, member X (H2AFX), which served as a tumor suppressor in migration and invasion of HNSCC. Moreover, H2AFX could partially reverse the migration and invasion of HNSCC caused by miR-328-3p. Overall, our results indicated that miR-328-3p enhanced migration and invasion of HNSCC through targeting H2AFX and activated the mTOR pathway.
Keywords: Head and neck squamous cell carcinoma, miR-328-3p, Migration, Invasion, H2AFX
Introduction
Head and neck squamous cell carcinoma (HNSCC) accounted for more than 800,000 new cancers and 450,000 deaths in 2018 [1, 2]. Multidisciplinary treatment involving a combination of surgery, radiation, and chemotherapy are the effective therapies for most HNSCC patients. Unfortunately, once HNSCC has spread outside of the head and neck region, treatment is seldom curative. Thus, new targeted agents against migration and invasion can provide crucial countermeasures to improve the prognosis of patients with HNSCC.
MicroRNAs (miRNAs) serve as important regulators of gene expression at the post-transcriptional level through interacting with 3′-end and more rarely with 5′-end of mRNA transcribed from target genes [3]. There are over 60% human protein-coding genes paired with miRNAs[4]. Numerous studies over the past decade have been devoted to assess the role of miRNAs in various diseases, and have found that miRNAs serve as crucial onco-miRNAs or tumor suppressor miRNAs in cancer [5-9]. Thus, the role of miRNAs in cancer could not be ignored.
MiR-328-3p has been reported to be dysregulated in various diseases, such as pulmonary arterial hypertension, multiple sclerosis and epilepsy, as well as cancers [10-12]. Recent studies showed that miR-328-3p may prevent tumor progression in non-small cell lung cancer, colorectal cancer, hepatocellular carcinoma, bladder cancer and osteosarcoma [13-17], but served as an onco-miRNA in ovarian cancer [18], which demonstrated the complicated roles of miR-328-3p in different types of tumors. In regards to HNSCC, the effect of miR-328-3p on migration and invasion of HNSCC remains unclear. In this study, we showed that miR-328-3p could promote migration and invasion by targeting H2AFX in HNSCC and activate the mTOR pathway, which provide novel insights for elucidating the molecular mechanisms involving migration and invasion of HNSCC.
Materials and methods
Cell culture
The Tca8113, a cell line from human tongue squamous carcinoma, was bought from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. Tu686 cell line, established from a primary tongue tumor, was kindly provided by Dr. Zhuo Chen (Emory University Winship Cancer Institute, Atlanta, Georgia, USA). FaDu, the Human hypopharyngeal squamous cell carcinoma cell line, was purchased from American Type Culture Collection (ATCC, VA, USA). JHU011, the human laryngeal squamous cell carcinoma cell line, was obtained from Division of Head and Neck Cancer Research at the Johns Hopkins University. Dulbecco's modified Eagle's medium (DMEM; Gibco, USA) was the culture substrate of FaDu, while RPMI Medium 1640 basic (Gibco, USA) was used for Tca8113 and JHU011. Tu686 cells were cultured in DMEM/F12 medium (Gibco, USA). All these media were supplemented with 10% fetal bovine serum (FBS; Gibco, USA) and 1% Penicillin-Streptomycin. Cells were incubated at 37 °C in 5% CO2.
Transfection
Tca8113 and FaDu cells were transfected with miR-328-3p mimics (Genepharma, Suzhou, China) or negative control (NC), while JHU011 and Tu686 cells were transfected with miR-328-3p anti-sense molecules (also called inhibitor) or NC, using the si-MateTM (Genepharma) according to the manufacturer's protocol. Tca8113 and FaDu cells were transfected with H2AFX cDNA plasmid (EX-B0074-M98, GeneCopoeia Inc., MD, USA) using the FuGENE® 6 (Promega, Madison, WI, USA).
Transwell invasion assay
2×104 cells were starved in 200 μl serum-free medium, then seeded in the upper transwell chamber (Corning) which was covered with 200 μg/mL of Matrigel (Corning, NY), and 600 μl complete medium containing 10% FBS was added in the lower chamber at the same time. After 48 hours' incubation, cells invaded on the lower side of top chamber were fixed with 4% paraformaldehyde (Biosharp) for 20 min and stained with crystal violet (Beyotime) for 15 min. Cells on upper side of top chamber were softly removed by a cotton swab. Take pictures randomly under 50×, 100×, 200× Leica fluorescence microscope (Wetzlar, Germany). The invading cells were counted by Image J [19].
Wound healing assay
Wound healing assay was used to evaluate the ability of cell migration. Cells were seeded on a six-well plate with over 90% confluency. Then using a sterile micropipette tip to create scratch wound with the same width in each group. Cells were cultured in serum-free medium for at least 24 hours. Images were captured under an inverted microscope, and the wound closure areas were calculated by Image J [19].
Cell proliferation assay
Cell proliferation based on cell viability was assessed by using a Cell Counting Kit (Meilunbio). 3×103 cells were seeded on 96-well plates after transfection. After 0, 24, 48, 72, 96 hours, cells were co-incubated with 10 μl CCK8 and 100 μl serum-free medium per well at 37 °C in 5% CO2 incubator for 1 hour, while the medium's absorbance without a cell as blank control. The absorbance at 450 nm was detected using a Model 680 Microplate Reader (BioTek).
Colony formation assay
Colony formation assay was used to determine the multiplication capacity of HNSCC cells. 400 cells were seeded in each well of six-well plates. About 2 weeks later, cells were fixed with 4% paraformaldehyde (Biosharp) for 20 min and stained with crystal violet (Beyotime) for 15 min. Colonies with over 50 cells were counted by Image J [20].
Data source and EMT score
Publicly available dataset of HNSCC cohort, including gene expression profile of 522 HNSCC patients, was downloaded from TCGA data portal (http://cancergenome.nih.gov/). Based on the expression level of 76-gene EMT signature obtained from the study of Lauren Averett Byers et al. [21], every HNSCC sample got an EMT score, which was calculated as the average expression level of “mesenchymal” genes minus the average expression level of “epithelial” genes. The 522 tumor samples were ranked by EMT score from lowest to highest, then the first 174 samples were defined as “epithelial” HNSCC while the last 174 samples were defined as “mesenchymal” HNSCC.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cell lines using the AG RNAex Pro reagent (Accurate Biology). 2 μg total RNA was used to synthesized single-stranded cDNA by All-in-OneTM miRNA or mRNA cDNA synthesis kit (GeneCopoeia Inc., MD, USA). qRT-PCR was carried out on the ABI QuantStudio 7 Flex Real-Time PCR System using All-in-One Kit (GeneCopoeia Inc., MD, USA) and sets of gene-specific primers. The reaction parameters included an initial step at 95 °C for 10 min, 40 cycles of 95 °C for 10 seconds, 62.5 °C for 20 seconds, and 72 °C for 15 seconds. U6 and GAPDH were used as endogenous control for miRNA and mRNA, respectively, and data were gauged through the classical 2-ΔΔCt method. Sequences of primers used in this study were listed in Table S1.
Western blotting
Total protein was extracted by using RIPA lysis buffer (NCM Biotech) with protease inhibitor cocktail and phosphatase inhibitor after transfection for 72 hours. 16-30 μg protein was loaded and electrophoresed on 8-12% SDS-PAGE gels, then transferred to a PVDF membranes (Millipore, Bedford, MA, USA). After sealing in NcmBlot blocking buffer (NCM Biotech) for 15 min, the membrane was incubated within primary antibody overnight at 4 °C. Then a horseradish peroxidase-conjugated secondary antibody (1:2000, Cell Signaling Technology) was used. Target protein bands were visualized using super sensitive ECL luminescence reagent (Dalian Meilun Biotechnology Co, Ltd.) in a Chemiluminescence and Fluorescence Imaging System (Bio-Rad). The antibodies used in this study were listed in Table S2.
miRNA target prediction
The potential targets of miR-328-3p were predicted with the Encyclopedia of RNA Interactomes (ENCORI, http://starbase.sysu.edu.cn/) database, which include six algorithms (PITA, miRmap, microT, miRanda, Pictar and TargetScan). And ENCORI was also used to acquire the predicted binding sites between miR-328-3p and the target gene.
Dual-luciferase activity assay
Cells were seeded into 24-well plate, and co-transfected with 50 nM of either miR-328-3p mimics or NC oligos in combination with 150 ng of wild-type or mutated 3'-UTR H2AFX plasmids (GeneCopoeia Inc., MD, USA) via si-MateTM (Genepharma) and the FuGENE® 6 (Promega, Madison, WI, USA), respectively. Then, the luciferase activity was appraised through utilizing the dual-luciferase assay system (Promega Corporation, WI) on the basis of the manufacturer's information.
Statistical analyses
GraphPad Prism (version 5.01, GraphPad Software, Version X; La Jolla, CA) was used for statistical analysis of the data. Unpaired t-test and Mann-Whitney U test were performed to analyze the significance of differences between groups. P-values less than 0.05 were considered as statistically significant.
Results
Overexpression of miR-328-3p promotes HNSCC migration and invasion
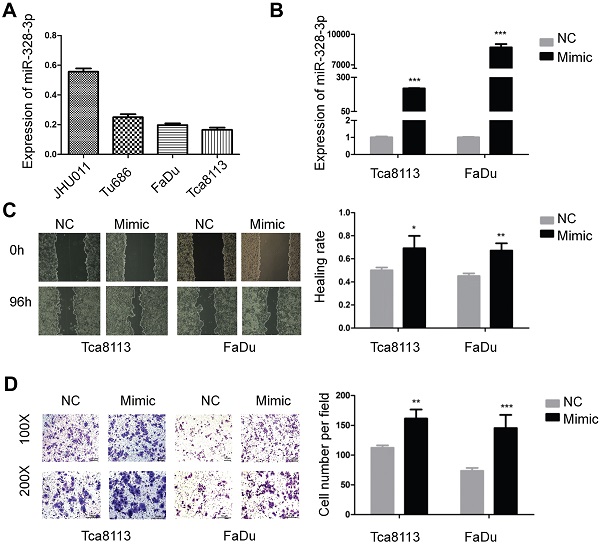
Firstly, we investigated the basic expression level of miR-328-3p in four HNSCC cell lines and found that the expression of miR-328-3p was higher in JHU011 and Tu686, and lower in Tca8113 and FaDu (Fig. 1A), which were selected for loss and gain of function experiments, respectively.
For the gain of function experiments, miR-328-3p mimic was transfected in the Tca8113 and FaDu cells and a significant overexpression of miR-328-3p was confirmed by qRT-PCR (Fig. 1B). The wound healing and transwell invasion assays showed that overexpression of miR-328-3p significantly increased the ability of migration and invasion in HNSCC cells (Fig. 1C, D). Furthermore, we found that the elevated miR-328-3p had no obvious effect on the proliferation of HNSCC cells via cell proliferation assay and colony formation assay (Fig. S1A, B).
Knockdown of miR-328-3p suppresses HNSCC migration and invasion
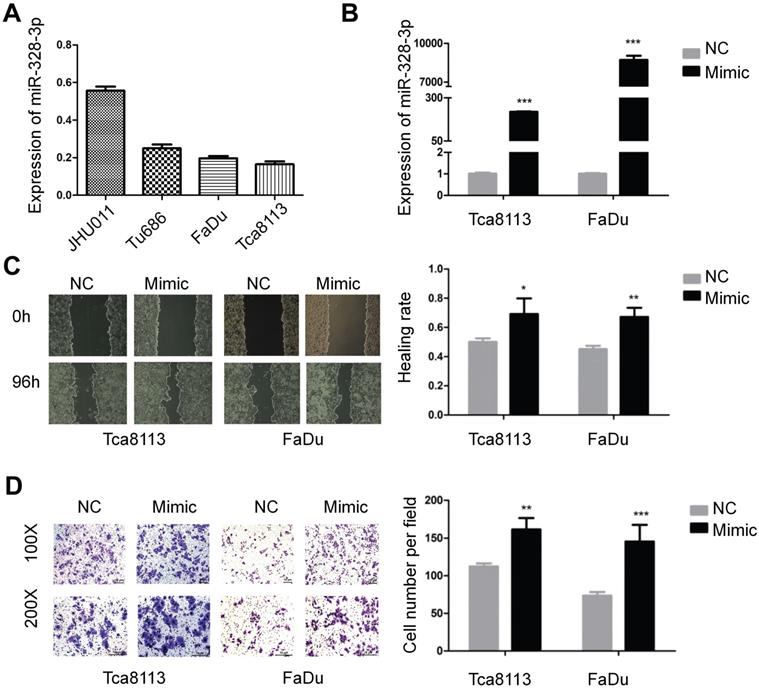
To further explore the function of miR-328-3p in HNSCC, miR-328-3p anti-sense molecules (inhibitor) was adopted to silence the expression of miR-328-3p. qRT-PCR showed that following transfection of inhibitor for 48 hours, the expression level of miR-328-3p was down-regulated by at least 50% in JHU011 and Tu686 cells (Fig. 2A). Then the wound healing and transwell invasion assays demonstrated that silencing miR-328-3p suppressed cell migration and invasion of JHU011 and Tu686 cells (Fig. 2B, C).
Upregulation of miR-328-3p promotes EMT in HNSCC
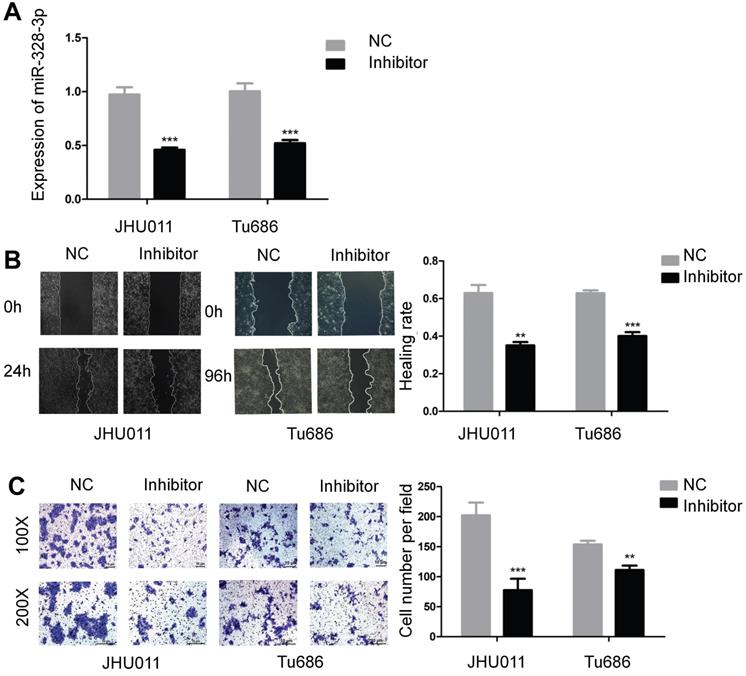
Epithelial-mesenchymal transition (EMT) plays a key role during carcinoma pathogenesis by imparting the mesenchymal phenotypes associated with the cells of highly aggressive tumors [22]. According to the scoring method of tumor EMT status [23], the expression of miR-328-3p in “Mesenchymal” HNSCC was higher than that in “Epithelial” HNSCC (Fig. 3A). To further explore the correlation between miR-328-3p and EMT in HNSCC, we detected the mRNA and protein level of hallmarks of EMT, such as epithelial cadherin (E-cadherin), Vimentin, neural cadherin (N-cadherin) and transcription factors that repress the epithelial phenotype and activate the mesenchymal phenotype. The results of qRT-PCR showed that miR-328-3p overexpression in Tca8113 cells resulted in the upregulation of mesenchymal marker Vimentin, N-cadherin and Fibronectin, with the increase of transcription factors Snail1, Zeb2 (Fig. 3B). Meanwhile, in FaDu cells, upregulated miR-328-3p decreased the expression of E-cadherin and promoted the expression of Vimentin and Twist1 (Fig. 3C). Additionally, Western blotting was used to confirm that miR-328-3p overexpression suppressed the levels of E-cadherin and elevated the levels of Vimentin, Snail1 and Twist1 in both Tca8113 and FaDu cells (Fig. 3D, Fig. S2A-B). In addition, immunofluorescent analysis confirmed that miR-328-3p overexpression elevated the levels of Vimentin in Tca8113 cells (Fig. 3E). These results indicated that upregulation of miR-328-3p promoted EMT in HNSCC.
Overexpression of miR-328-3p promoted migration and invasion in HNSCC. A. Different expression levels of miR-328-3p in HNSCC cell lines. B. miR-328-3p expression was significant upregulated in Tca8113 and FaDu cells after transfection with miR-328-3p mimic. C and D. Wound healing assays (C) and transwell invasion assays (D) showed overexpression of miR-328-3p could promote the ability of migration and invasion in Tca8113 and FaDu cells. NC: negative control. Each experiment was repeated in triplicate. Data are presented as the mean ± SD. Student's unpaired t-test, *, P <0.05; **, P < 0.01; ***, P < 0.001.
Overexpression of miR-328-3p activates the mTOR pathway
Previous studies have demonstrated that the mTOR pathway may be associated with EMT of carcinomas [24]. To further investigate whether miR-328-3p could exert its effect through mTOR signaling pathway, the mRNAs level and the phosphorylated protein level of mTOR, p70S6K, S6RP and 4E-BP1 were determined by qRT-PCR (Fig. 4A) and Western blotting (Fig. 4B) in Tca8113 and FaDu cells transfected with miR-328-3p mimic. The phosphorylated protein level was upregulated in miR-328-3p-mimic group while the non-phosphorylated proteins showed no obvious differences, suggesting that miR-328-3p could activate the mTOR pathway in HNSCC.
Downregulation of miR-328-3p suppressed migration and invasion in HNSCC. A. miR-328-3p expression was significantly inhibited in JHU011 and Tu686 cells after transfection with miR-328-3p anti-sense molecules (inhibitor). B and C. Wound healing assays (B) and transwell invasion assays (C) showed down-regulation of miR-328-3p could suppress the ability of migration and invasion in JHU011 and Tu686 cells. Each experiment was repeated in triplicate. All data are represented as the mean ± SD. Statistical analysis was performed using Student's t- test. *, P <0.05; **, P < 0.01, ***, P < 0.001.
H2AFX is a direct target of miR-328-3p
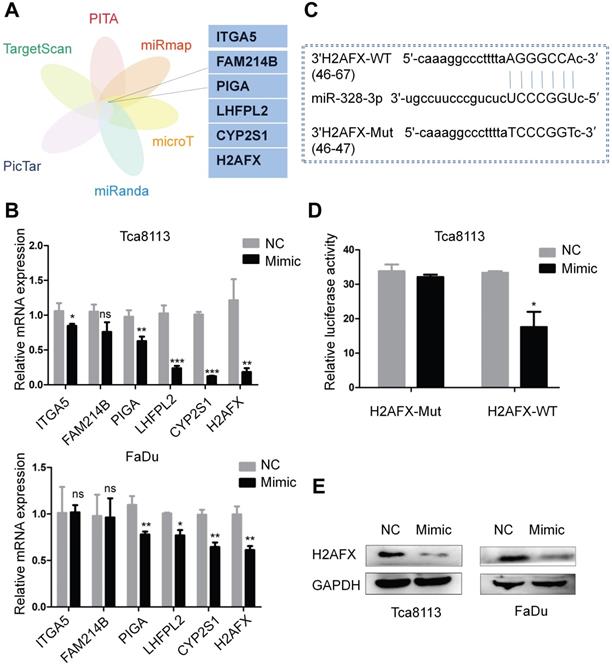
To explore the target genes of miR-328-3p, we selected candidate genes at ENCORI website, which included data from six algorithms (PITA, miRmap, microT, miRanda, Pictar and TargetScan) (Fig. 5A). H2AFX was selected as the target gene owing to its most obvious downregulation of mRNA expression compared with the other candidate genes after transfecting with miR-328-3p mimic in Tca8113 and FaDu cells (Fig. 5B). Then the luciferase reporter assay further confirmed the direct binding of miR-328-3p to the 3'UTR of H2AFX mRNA (Fig. 5C, D). Western blotting validated the dysregulation of H2AFX protein by miR-328-3p in Tca8113 and FaDu cells (Fig. 5E). These data showed that H2AFX is a direct target of miR-328-3p in HNSCC.
H2AFX is partially involved in the migration and invasion of HNSCC mediated by miR-328-3p
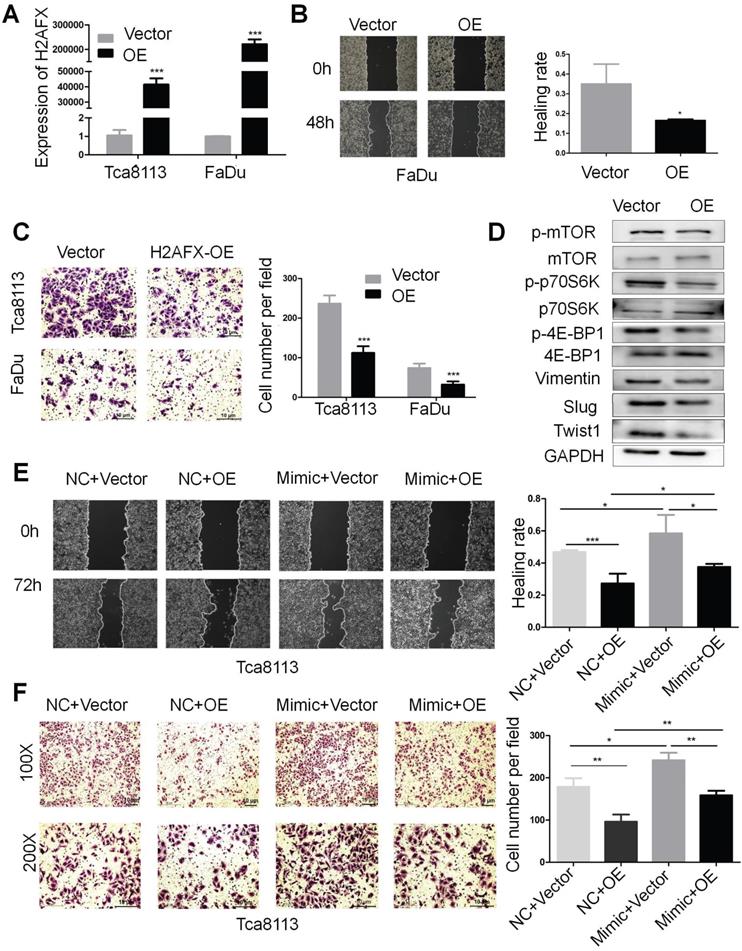
In order to investigate the role of H2AFX in HNSCC, the H2AFX cDNA plasmid was transfected in Tca8113 and FaDu cells and the expression of H2AFX mRNA was successfully upregulated (Fig. 6A). Then, the wound healing and transwell invasion assays showed that upregulation of H2AFX suppressed the migration and invasion of HNSCC (Fig. 6B, C). Additionally, H2AFX overexpression significantly reduced the protein levels of Vimentin, Slug and Twist1 in Tca8113 cells, and the biomarkers of mTOR pathway including p-mTOR, p-p70S6K and p-4E-BP1 were also decreased (Fig. 6D, Fig. S2C).
Overexpression of miR-328-3p promoted EMT in HNSCC. A. The expression of miR-328-3p in “Epithelial” or “Mesenchymal” HNSCC in TCGA dataset. B-D. qRT-PCR (B and C) and Western blotting (D) were used to quantify the expression of the EMT markers of E-cadherin, Vimentin, Fibronectin and the related transcription factors Snail1, Slug, Zeb2 and Twist1 in Tca8113 and FaDu cells. GAPDH was used as a loading control. E. Representative immunofluorescence staining images showed the expression level of Vimentin in Tca8113 cells. ns, not significant. Each experiment was repeated in triplicate and all data are presented as mean ± SD. Mann-Whitney U test was used for Figure 3A, while Student's unpaired t-test was used for the others. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To finally ascertain whether upregulation of H2AFX is responsible for the inhibition of miR-328-3p-mediated migration and invasion of HNSCC, Tca8113 cells were treated with H2AFX overexpressed plasmid and empty vector following the transfection with miR-328-3p-mimic and NC. As expected, the migration and invasion capacities of HNSCC cells co-transfected with miR-328-3p mimic and H2AFX plasmid was weaker than cells co-transfected with miR-328-3p mimic and empty vector, suggesting that H2AFX partially reversed the promotion effect on cell migration and invasion caused by the miR-328-3p in Tca8113 cells. In addition, HNSCC cells co-transfected with miR-328-3p mimic and H2AFX plasmid exhibited stronger migration and invasion capacities than those co-transfected with NC and H2AFX plasmid, which again proved that miR-328-3p can promote the migration and invasion of HNSCC cells (Fig. 6E, F). Moreover, H2AFX partially reversed the protein change of EMT biomarkers caused by miR-328-3p (Fig. S3A). These findings demonstrated that H2AFX was partially involved in miR-328-3p mediated migration and invasion in HNSCC.
Discussion
MicroRNA plays a crucial role in cancer migration and invasion by incorporating within an RNA-induced silencing complex (RISC) and inducing post-transcriptional gene silencing by base pairing with a target mRNA and interfering with its translation to protein [6, 25]. Several studies have revealed the function and mechanism of miR-328-3p on cancer migration and invasion. For examples, in osteosarcoma, colorectal cancer, liver cancer and breast cancer, miR-328-3p overexpression inhibited their migration, invasion, and EMT by directly inhibiting MMP-16, Girdin, Endoplasmic Reticulum Metallo Protease 1 (ERMP1) and CD44, respectively [17, 26-29]. Intriguingly, there was an opposing effect of miR-328-3p in ovarian cancer, in which miR-328-3p overexpression could maintain the CSC population and promote tumor metastasis [18]. Then a recent study revealed that miR-328-3p may promote bone metastasis by activating Wnt/β-catenin pathway in non-small cell lung cancer [30]. Another study showed that hypoxic bone marrow mesenchymal cell (BMSC)-derived extracellular vesicles could deliver miR-328-3p to lung cancer cells to target the NF2 gene, which ultimately promoted the invasion, migration and EMT of lung cancer by inhibiting the Hippo pathway [31]. These studies indicated that the function of miR-328-3p could be tissue- and tumor-specific, which was further confirmed by our study. In the current study, miR-328-3p served as an onco-miRNA and promoted the migration and invasion of HNSCC.
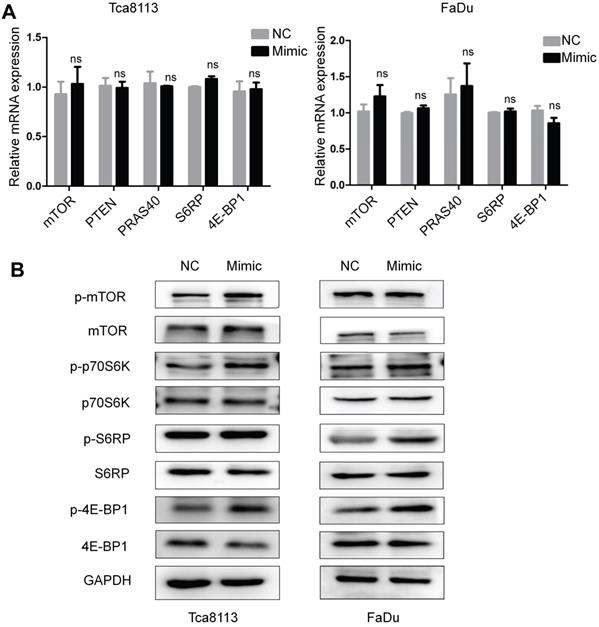
Overexpression of miR-328-3p activated the mTOR pathway in HNSCC. A and B. Following miR-328-3p overexpression in Tca8113 and FaDu cells, qRT-PCR (A) showed the mRNA expression of the markers of mTOR pathway, including mTOR, PTEN, PRAS40, S6RP and 4E-BP1 did not change significantly. Western blotting (B) showed the phosphorylated proteins level of mTOR, p70S6K, S6RP and 4E-BP1 were elevated. GAPDH served as an internal control.
H2AFX was a direct target of miR-328-3p. A. Potential target genes were obtained by integrating six algorithms (PITA, miRmap, microT, miRanda, Pictar and TargetScan) in ENCORI. B. The relative mRNA expression of potential target genes after transfecting with miR-328-3p mimic was examined by qRT-PCR in Tca8113 and FaDu cells. C. Predicted binding site between miR-328-3p and the wild-type 3'-UTR of H2AFX was obtained by ENCORI. D. Dual luciferase reporter assay conducted in Tca8113 cells confirmed that H2AFX was a direct target of miR-328-3p. E. Western blotting showed that H2AFX protein expression was down-regulated in Tca8113 and FaDu cells after transfecting with the miR-328-3p mimic. Each experiment was repeated in triplicate. Data are presented as the mean ± SD. Student's unpaired t-test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
H2AFX is a variant of the H2A protein family, a component of the histone octamer in nucleosomes [32]. H2AFX would be phosphorylated on residue serine 139 in cells triggered by double-stranded breaks (DSB) [33]. γ-H2AFX can recruit DNA repair proteins and other DNA double strand breaks signaling to the damaged site. Once DNA is repaired, γ-H2AFX is dephosphorylated, thereby γ-H2AFX is widely used as a DNA damage marker in vitro [34, 35]. For the role of H2AFX in cancer, researchers found that H2AFX can help prevent aberrant repair of both programmed and general DNA breakage and serves as a suppressor of genomic instability and tumors in mice, which is dosage-dependent [36]. Moreover, loss of a single H2AFX allele compromises genomic integrity and enhances the susceptibility to cancer in the absence of p53 [37], and H2AFX variants are associated with an increased risk of breast cancer [38, 39]. Based on these results, H2AFX may serve as a tumor suppressor. However, in lung adenocarcinoma, patients with low expression of H2AFX have a significantly better overall survival [40], and H2AFX can promote radioresistance in non-small cell lung cancer and osteosarcoma [13, 41], which reveal that H2AFX may also be a tumor promoter. Despite these previous findings, studies involved the role of H2AFX in cancer migration and invasion and EMT remained scarce, and our study was the first to demonstrate that H2AFX functioned as a tumor suppressor to inhibit HNSCC migration and invasion in vitro. Additionally, upregulation of H2AFX partially abrogated increased migration and invasion induced by miR-328-3p, suggesting that H2AFX was a major target of miR-328-3p in HNSCC.
H2AFX was partially involved in the migration and invasion of HNSCC mediated by miR-328-3p. A. H2AFX mRNA expression was significant upregulated in Tca8113 and FaDu cells after transfection with H2AFX plasmid. B and C. Following H2AFX overexpression in Tca8113 and FaDu cells, wound healing (B) and transwell invasion assays (C) were performed. D. Western blotting showed the alternation of marker proteins for EMT and mTOR pathway. E and F. Cells transfected with miR-328-3p mimics or NC were subsequently treated with H2AFX plasmid or vector. Wound healing assays (E) and transwell invasion assays (F) showed overexpression of H2AFX could rescue the inhibitory effect on cell migration and invasion caused by the miR-328-3p-mimic in Tca8113 cells. OE, H2AFX overexpression. Each experiment was repeated in triplicate. Data are presented as the mean ± SD. P-values were calculated using the Student's t-test. *, P <0.05; **, P < 0.01.
The mTOR signaling pathway encompasses two functionally distinct protein complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) [42], and only the former one is sensitive to the macrolide fungicide rapamycin. The mTORC1 contains two positive regulatory subunits, raptor and mLST8, and two negative regulators, PRAS40 and DEPTOR [43]. The p70 ribosomal S6 kinase/S6 ribosomal protein (P70S6K/S6RP) and eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) are two downstream targets of mTORC1 [44]. The mobilized P70S6K will induce the S6 activation, which is essential for protein synthesis [45, 46]. In quiescent cells, 4E-BP1 competes with eIF-4G for binding to eIF-4E and represses translation by displacing the initiation factor 4F from the mRNA. Once 4E-BP1 is phosphorylated, its affinity for eIF-4E will decrease and release the block on cap-dependent translation [47]. The mTORC2 activates the mTORC1 by activating Akt [48]. Previous studies had showed that mTOR was not only a central regulator of cell growth, proliferation, differentiation and survival, but also played a critical role in the regulation of tumor cell motility, invasion and metastasis [49, 50]. Berven et al. reported that Fibronectin, a mesenchymal marker, stimulated stress fiber formation in the absence of growth factors and caused an inactivation of p70S6K to regulate cell migration [51]. Lamouille et al. found that the mTORC2 pathway was an essential downstream branch of TGF-β signaling. TGF-β can rapidly induce mTORC2 kinase activity in cells undergoing EMT, and control epithelial cell progression through EMT [52]. Liu et al. found low expression of the apelin receptor promoted EMT in nasopharyngeal carcinoma cells by activating the PI3K-mTOR signaling pathway [53]. These studies revealed that mTOR pathway was associated with EMT and tumor metastasis [54, 55]. Additionally, several groups demonstrated anti-cancer effect of mTOR inhibitors in HNSCC xenografts either as a single agent or when combined with chemotherapy/radiotherapy [56-58]. Clinical trials were also conducted to investigate the function of several mTOR inhibitors used either alone or in combination with chemotherapy or radiotherapy in HNSCC [59, 60]. These results indicated that it was significant to explore the mechanism of mTOR pathway in HNSCC. In the current study, miR-328-3p can increase the phosphorylation protein level of mTOR, P70S6K, S6RP and 4E-BP1, and overexpression of the target gene H2AFX reduced the protein levels of p-mTOR, p-p70S6K and p-4E-BP1, suggesting that miR-328-3p/H2AFX axis could activate the mTOR pathway in HNSCC.
Taken together, our results demonstrated that miR-328-3p enhanced migration and invasion of HNSCC through targeting H2AFX and activated the mTOR pathway, which may be a potential therapeutic target in the treatment of HNSCC patients.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
Funding
Grants were provided by the National Natural Science Foundation of China (Nos. 81773243, 81974424, 81874133, 81772903 and 82073009), the Natural Science Foundation of Hunan Province (Nos. 2019JJ40481 and 2019JJ50944), the Huxiang Young Talent Project (No. 2018RS3024) and the Youth Science Foundation of Xiangya Hospital (No. 2020Q03).
Author Contributions
HM, YL, and YQ were responsible for the study design.HM was responsible for drafting and editing of the original article, data acquisition and data analysis. HM, WY, and JH performed the experiments. HM, SZ, and DH were responsible for data interpretation and methodology. XZ, YL, and YQ were responsible for supervision. CL and YL revised the manuscript. All authors have read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68:394-424
2. Chai AWY, Lim KP, Cheong SC. Translational genomics and recent advances in oral squamous cell carcinoma. Seminars in cancer biology. 2020;61:71-83
3. Gulyaeva LF, Kushlinskiy NE. Regulatory mechanisms of microRNA expression. Journal of translational medicine. 2016;14:143
4. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19:92-105
5. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nature reviews Cancer. 2015;15:321-33
6. Yang CX, Sedhom W, Song J, Lu SL. The Role of MicroRNAs in Recurrence and Metastasis of Head and Neck Squamous Cell Carcinoma. Cancers. 2019;11:395
7. Zhang S, He Y, Liu C, Li G, Lu S, Jing Q. et al. miR-93-5p enhances migration and invasion by targeting RGMB in squamous cell carcinoma of the head and neck. Journal of Cancer. 2020;11:3871-81
8. Zhang S, Li G, Liu C, Lu S, Jing Q, Chen X. et al. miR-30e-5p represses angiogenesis and metastasis by directly targeting AEG-1 in squamous cell carcinoma of the head and neck. Cancer science. 2020;111:356-68
9. Liu C, Li G, Yang N, Su Z, Zhang S, Deng T. et al. miR-324-3p suppresses migration and invasion by targeting WNT2B in nasopharyngeal carcinoma. Cancer cell international. 2017;17:2
10. Xing Y, Zheng X, Fu Y, Qi J, Li M, Ma M. et al. Long Noncoding RNA-Maternally Expressed Gene 3 Contributes to Hypoxic Pulmonary Hypertension. Molecular therapy: the journal of the American Society of Gene Therapy. 2019;27:2166-81
11. Ioriatti ES, Cirino MLA, Lizarte Neto FS, Velasco TR, Sakamoto AC, Freitas-Lima P. et al. Expression of circulating microRNAs as predictors of diagnosis and surgical outcome in patients with mesial temporal lobe epilepsy with hippocampal sclerosis. Epilepsy research. 2020;166:106373
12. Yang Q, Pan W, Qian L. Identification of the miRNA-mRNA regulatory network in multiple sclerosis. Neurological research. 2017;39:142-51
13. Ma W, Ma CN, Zhou NN, Li XD, Zhang YJ. Up- regulation of miR-328-3p sensitizes non-small cell lung cancer to radiotherapy. Scientific reports. 2016;6:31651
14. Ji D, Qiao M, Yao Y, Li M, Chen H, Dong Q. et al. Serum-based microRNA signature predicts relapse and therapeutic outcome of adjuvant chemotherapy in colorectal cancer patients. EBioMedicine. 2018;35:189-97
15. Li JZ, Li J, Liu BZ. MicroRNA-328-3p inhibits malignant progression of hepatocellular carcinoma by regulating MMP-9 level. European review for medical and pharmacological sciences. 2019;23:9331-40
16. Yan T, Ye XX. MicroRNA-328-3p inhibits the tumorigenesis of bladder cancer through targeting ITGA5 and inactivating PI3K/AKT pathway. European review for medical and pharmacological sciences. 2019;23:5139-48
17. Zhang M, Zhang J, Zhou Q. Elevated expression of microRNA-328-3p suppresses aggressive malignant behaviors via targeting matrix metalloprotease 16 in osteosarcoma. OncoTargets and therapy. 2019;12:2063-70
18. Srivastava AK, Banerjee A, Cui T, Han C, Cai S, Liu L. et al. Inhibition of miR-328-3p Impairs Cancer Stem Cell Function and Prevents Metastasis in Ovarian Cancer. Cancer research. 2019;79:2314-26
19. Venter C, Niesler CU. Rapid quantification of cellular proliferation and migration using ImageJ. BioTechniques. 2019;66:99-102
20. Guzmán C, Bagga M, Kaur A, Westermarck J, Abankwa D. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PloS one. 2014;9:e92444
21. Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M. et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:279-90
22. Derynck R, Weinberg RA. EMT and Cancer: More Than Meets the Eye. Developmental cell. 2019;49:313-6
23. Lou Y, Diao L, Cuentas ER, Denning WL, Chen L, Fan YH. et al. Epithelial-Mesenchymal Transition Is Associated with a Distinct Tumor Microenvironment Including Elevation of Inflammatory Signals and Multiple Immune Checkpoints in Lung Adenocarcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016;22:3630-42
24. Pearlman RL, Montes de Oca MK, Pal HC, Afaq F. Potential therapeutic targets of epithelial-mesenchymal transition in melanoma. Cancer letters. 2017;391:125-40
25. Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nature structural & molecular biology. 2012;19:145-51
26. Shi J, An G, Guan Y, Wei T, Peng Z, Liang M. et al. miR-328-3p mediates the anti-tumor effect in osteosarcoma via directly targeting MMP-16. Cancer cell international. 2019;19:104
27. Pan S, Ren F, Li L, Liu D, Li Y, Wang A. et al. MiR-328-3p inhibits cell proliferation and metastasis in colorectal cancer by targeting Girdin and inhibiting the PI3K/Akt signaling pathway. Experimental cell research. 2020;390:111939
28. Lu H, Hu J, Li J, Lu W, Deng X, Wang X. miR-328-3p overexpression attenuates the malignant proliferation and invasion of liver cancer via targeting Endoplasmic Reticulum Metallo Protease 1 to inhibit AKT phosphorylation. Annals of translational medicine. 2020;8:754
29. Al-Othman N, Hammad H, Ahram M. Dihydrotestosterone regulates expression of CD44 via miR-328-3p in triple-negative breast cancer cells. Gene. 2018;675:128-35
30. Yang XR, Pi C, Yu R, Fan XJ, Peng XX, Zhang XC. et al. Correlation of exosomal microRNA clusters with bone metastasis in non-small cell lung cancer. Clinical & experimental metastasis. 2021;38(1):109-117
31. Liu X, Jiang F, Wang Z, Tang L, Zou B, Xu P. et al. Hypoxic bone marrow mesenchymal cell-extracellular vesicles containing miR-328-3p promote lung cancer progression via the NF2-mediated Hippo axis. Journal of cellular and molecular medicine. 2021;25:96-109
32. Foster ER, Downs JA. Histone H2A phosphorylation in DNA double-strand break repair. The FEBS journal. 2005;272:3231-40
33. Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. The Journal of biological chemistry. 1998;273:5858-68
34. Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell research. 2008;18:134-47
35. Kuo LJ, Yang LX. Gamma-H2AX - a novel biomarker for DNA double-strand breaks. In vivo (Athens, Greece). 2008;22:305-9
36. Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M. et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359-70
37. Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA. et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371-83
38. Podralska M, Ziółkowska-Suchanek I, Żurawek M, Dzikiewicz-Krawczyk A, Słomski R, Nowak J. et al. Genetic variants in ATM, H2AFX and MRE11 genes and susceptibility to breast cancer in the polish population. BMC cancer. 2018;18:452
39. Lu J, Wei Q, Bondy ML, Brewster AM, Bevers TB, Yu TK. et al. Genetic variants in the H2AFX promoter region are associated with risk of sporadic breast cancer in non-Hispanic white women aged <or=55 years. Breast cancer research and treatment. 2008;110:357-66
40. Zhao M, Chen Z, Zheng Y, Liang J, Hu Z, Bian Y. et al. Identification of cancer stem cell-related biomarkers in lung adenocarcinoma by stemness index and weighted correlation network analysis. Journal of cancer research and clinical oncology. 2020;146:1463-72
41. Yang Z, Wa QD, Lu C, Pan W, Lu Z, Ao J. miR-328-3p enhances the radiosensitivity of osteosarcoma and regulates apoptosis and cell viability via H2AX. Oncology reports. 2018;39:545-53
42. Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D. et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Molecular cell. 2002;10:457-68
43. Pópulo H, Lopes JM, Soares P. The mTOR signalling pathway in human cancer. International journal of molecular sciences. 2012;13:1886-918
44. Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:1432-7
45. Chauvin C, Koka V, Nouschi A, Mieulet V, Hoareau-Aveilla C, Dreazen A. et al. Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene. 2014;33:474-83
46. Tavares MR, Pavan IC, Amaral CL, Meneguello L, Luchessi AD, Simabuco FM. The S6K protein family in health and disease. Life sciences. 2015;131:1-10
47. Martineau Y, Azar R, Bousquet C, Pyronnet S. Anti-oncogenic potential of the eIF4E-binding proteins. Oncogene. 2013;32:671-7
48. Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science (New York, NY). 2005;307:1098-101
49. Zhou H, Huang S. Role of mTOR signaling in tumor cell motility, invasion and metastasis. Current protein & peptide science. 2011;12:30-42
50. Tian Y, Shen L, Li F, Yang J, Wan X, Ouyang M. Silencing of RHEB inhibits cell proliferation and promotes apoptosis in colorectal cancer cells via inhibition of the mTOR signaling pathway. Journal of cellular physiology. 2020;235:442-53
51. Berven LA, Willard FS, Crouch MF. Role of the p70(S6K) pathway in regulating the actin cytoskeleton and cell migration. Experimental cell research. 2004;296:183-95
52. Lamouille S, Connolly E, Smyth JW, Akhurst RJ, Derynck R. TGF-β-induced activation of mTOR complex 2 drives epithelial-mesenchymal transition and cell invasion. Journal of cell science. 2012;125:1259-73
53. Liu Y, Liu Q, Chen S, Liu Y, Huang Y, Chen P. et al. APLNR is involved in ATRA-induced growth inhibition of nasopharyngeal carcinoma and may suppress EMT through PI3K-Akt-mTOR signaling. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2019;33:11959-72
54. Murugan AK. mTOR: Role in cancer, metastasis and drug resistance. Seminars in cancer biology. 2019;59:92-111
55. Zhou C, Liu C, Liu W, Chen W, Yin Y, Li CW. et al. SLFN11 inhibits hepatocellular carcinoma tumorigenesis and metastasis by targeting RPS4X via mTOR pathway. Theranostics. 2020;10:4627-43
56. Ekshyyan O, Rong Y, Rong X, Pattani KM, Abreo F, Caldito G. et al. Comparison of radiosensitizing effects of the mammalian target of rapamycin inhibitor CCI-779 to cisplatin in experimental models of head and neck squamous cell carcinoma. Molecular cancer therapeutics. 2009;8:2255-65
57. Cassell A, Freilino ML, Lee J, Barr S, Wang L, Panahandeh MC. et al. Targeting TORC1/2 enhances sensitivity to EGFR inhibitors in head and neck cancer preclinical models. Neoplasia (New York, NY). 2012;14:1005-14
58. D'Amato V, Rosa R, D'Amato C, Formisano L, Marciano R, Nappi L. et al. The dual PI3K/mTOR inhibitor PKI-587 enhances sensitivity to cetuximab in EGFR-resistant human head and neck cancer models. British journal of cancer. 2014;110:2887-95
59. Day TA, Shirai K, O'Brien PE, Matheus MG, Godwin K, Sood AJ. et al. Inhibition of mTOR Signaling and Clinical Activity of Rapamycin in Head and Neck Cancer in a Window of Opportunity Trial. Clinical cancer research: an official journal of the American Association for Cancer Research. 2019;25:1156-64
60. Wang Z, Valera JC, Zhao X, Chen Q, Gutkind JS. mTOR co-targeting strategies for head and neck cancer therapy. Cancer metastasis reviews. 2017;36:491-502
Author contact
![]() Corresponding authors: Yong Liu and Yuanzheng Qiu, Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, 87 Xiangya Road, Changsha, Hunan 410008, China. Tel.: +86073184327469; Fax: +86073184327469; E-mail: liuyongentedu.cn (Y.L); xyqyzcom (Y.Q).
Corresponding authors: Yong Liu and Yuanzheng Qiu, Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, 87 Xiangya Road, Changsha, Hunan 410008, China. Tel.: +86073184327469; Fax: +86073184327469; E-mail: liuyongentedu.cn (Y.L); xyqyzcom (Y.Q).

 Global reach, higher impact
Global reach, higher impact