Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(21):6363-6371. doi:10.7150/jca.51338 This issue Cite
Research Paper
RUNX1 regulates the proliferation and chemoresistance of colorectal cancer through the Hedgehog signaling pathway
1. Guangdong Provincial Key Laboratory of Gastroenterology, Department of Gastroenterology, Nanfang Hospital, Southern Medical University, Guangzhou, Guangdong, China
2. The First School of Clinical Medicine, Southern Medical University, Guangzhou, Guangdong, China
* These authors contributed equally to this work.
Received 2020-7-31; Accepted 2021-8-17; Published 2021-9-3
Abstract
Background: Chemoresistance is one of the main causes of recurrence in colorectal cancer (CRC) patients and leads to a poor prognosis. To characterize RUNX1 expression in colorectal cancer (CRC) and elucidate its mechanistic involvement in the tumor biology of this disease.
Methods: The expression of RUNX1 in CRC and normal tissues was detected by bioinformatics analysis. Cell proliferation was measured by CCK-8 and clonogenic assays. In vivo tumor progression was assessed with a xenograft mouse model. Cell drug sensitivity tests and flow cytometry were performed to analyze CRC cell chemoresistance. RUNX1, key molecules of the Hedgehog signaling pathway, and ABCG2 were detected by qRT-PCR and Western blotting.
Results: RUNX1 expression is upregulated in CRC tissues. RUNX1 enhanced CRC cell resistance to 5-fluorouracil (5-FU), promoted proliferation, and inhibited 5-FU-induced apoptosis. Mechanistically, RUNX1 can activate the Hedgehog signaling pathway and promote the expression of ABCG2 in CRC cells.
Conclusions: Our study demonstrated that RUNX1 promotes CRC proliferation and chemoresistance by activating the Hedgehog signaling pathway and ABCG2 expression.
Keywords: colorectal cancer, RUNX1, Hedgehog signaling pathway, proliferation, chemoresistance
Background
Colorectal cancer (CRC) is one of the most common malignant tumors of the digestive tract and is a serious threat to human health, and its incidence is rising in young and middle-aged individuals[1]. 5-Fluorouracil (5-FU) is the backbone of therapy for patients with metastatic colorectal cancer (mCRC)[2]. Overexpression of the multidrug resistance (MDR) transporter ABCG2 in vitro has been shown to cause resistance to 5-FU, a component of the most commonly adopted regimens for treating CRC[3]. Abnormal proliferation and uncontrolled growth are the main characteristics of cancer, and chemotherapy failure is the major cause of recurrence and poor prognosis in CRC patients[4]. Thus, identifying the molecular mechanisms that influence CRC progression or chemotherapy is critical for improving the chemotherapy of the disease.
Runt-related transcription factor 1 (RUNX1) is a member of the RUNX family of transcription factors (RUNX1, RUNX2, RUNX3), and this family is highly conserved during evolution and plays a decisive role in encoding proteins associated with a variety of cell lineages[5,6]. Numerous studies have revealed that RUNX1 plays an oncogene or tumor suppressor role in a variety of solid tumors[6-11]. In particular, RUNX1 promotes the progression of CRC cells and plays a carcinogenic role[12,13].
In previous studies, the RUNX family and Hedgehog signaling pathway were shown to be closely related. We found that RUNX1 and the Hedgehog signaling pathway are interdependent in hemogenic endothelium (HE) and hematopoietic stem and progenitor cells (HSPCs)[14,15], and the Hedgehog signaling pathway can be induced directly by RUNX2 and RUNX3 in osteoblastic cells[16,17]. In CRC, RUNX1 has been reported to promote metastasis by activating the Wnt/β-catenin signaling pathway[12]. At the same time, more than one of the pathways is active in tumors, and these pathways are unbalanced; moreover, impaired crosstalk contributes to tumor development[18,19].
In the present study, we report that RUNX1 acts as an oncogene in CRC by activating the Hedgehog signaling pathway to promote cancer cell proliferation and regulating ABCG2 expression to reduce the sensitivity of cancer chemotherapy.
Materials and Methods
Bioinformatics analysis
UALCAN (http://ualcan.path.uab.edu/) is an effective online analysis and mining website for in-depth analyses of gene expression data using The Cancer Genome Atlas (TCGA) RNA-sequencing (RNA-seq) and clinical data from 31 cancer types[20]. The UCSC XENA (https://xenabrowser.net/) website provides information on the 31 cancer types and clinical data from the TCGA database[21]. The biological functions of these differentially expressed RUNX1 genes were comprehensively assessed by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. All enrichment analyses were carried out by WebGestalt (http://www.webgestalt.org/)[22]. Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/about.html) is a newly developed interactive web server for analyzing the RNA-seq expression data of 9,736 tumors and 8,587 normal samples from the TCGA and Genotype-Tissue Expression (GTEx) projects using a standard processing pipeline[23].
Cell culture, plasmid construction, lentiviral construction, and cell transfection
Three human CRC cell lines (HCT116, SW480, and RKO) were purchased from the Cell Bank of Type Culture Collection (CBTCC, Chinese Academy of Sciences, Shanghai, China) and were cultured in Dulbecco's modified Eagle medium (DMEM) (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA). Cells were maintained at 37℃ in a humidified 5% CO2 atmosphere.
Plasmid construction, lentiviral construction, and cell transfection
RUNX1 overexpression and knockdown were performed using a lentiviral packaging system. To construct overexpressing exogenous and RNA-interfered endogenous RUNX1 cell lines, full-length RUNX1 (NM_001754) was cloned into the expression vector pLenti-EF1a-EGFP-P2A-Puro-CMV (Obio Technology, Shanghai, China) and transfected into HCT116 and RKO cell lines according to the manufacturer's instructions. Knockdown of endogenous RUNX1 was mediated by designed shRNAs (Cyagen, Guangzhou, China) that were transfected into SW480 and RKO cell lines according to the manufacturer's instructions. The RUNX1 shRNA sequences were sense 5'-CCAGGTTGCAAGATTTAAT-3' and 5'-GGCAGAAACTAGATGATCA-3', and the scramble sequence was sense 5'-CCTAAGGTTAAGTCGCCCTCG-3'. Transduced cells were selected in a medium containing puromycin (#EZ2811D376, BioFroxx, Germany) (2 μg/ml) and maintained in a medium containing puromycin (1 μg/ml).
RNA isolation and qRT-PCR
Total RNA was extracted from cells or tissues with TRIzol reagent (TaKaRa, China). qRT-PCR was performed using the PrimeScript RT Reagent Kit (#RR035A, TaKaRa, China) and SYBR Premix Ex Taq (#RR820A, TaKaRa, Dalian, China) following the manufacturer's instructions. Our results were normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6. The specific primers used are listed in Table S1. The qRT-PCR results were analyzed to obtain the Ct values of the amplified products, and the data were analyzed by the 2-ΔΔCt method.
Western blotting and immunohistochemistry (IHC) analysis
We performed Western blotting according to the methods of a previous study. Protein lysates were prepared, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto polyvinylidene difluoride (PVDF) membranes, and blotted according to standard methods using antibodies to the following: RUNX1 (#4336, CST), PTCH1 (#2468, CST), PTCH2 (#2470, CST), GLI1 (#3538, CST), Shh (#2207, CST), ABCG2 (#42078, CST) and GAPDH (60004-1-Ig, Proteintech).
IHC was performed following the manufacturer's instructions (PV-6001, ZSGB-BIO, Beijing, China) using Ki-67 (27309-1-AP, Proteintech). One independent pathologist used ImageJ software to calculate gray values for pathological scoring.
Cell counting kit assay
Cells transfected with the appropriate plasmids were seeded into a 96-well plate at 1,000 cells per well and cultured at 37 °C and 5% CO2 for 6 h. For the cell counting kit-8 (CCK-8) assay, the cells were incubated with 10 μL of CCK-8 (Dojindo, Japan) for 2 h at 37°C and their density was measured at a wavelength of 450 nm using the Paradigm Detection Platform (Beckman, CA, USA). The cells were further incubated for 5 days following the CCK-8 assay.
Colony formation assays
Colony formation assays were performed in 6-well culture plates. Cells were seeded at a density of 5×102 per well. The cells were incubated at 37°C in a humidified atmosphere with 5% CO2 and the medium was replaced every 3-4 days. The colonies were counted and analyzed in about 2 weeks. The experiment was performed with at least three replicates for each cell line.
Apoptosis assays
The cells were analyzed by FACS according to the standard protocol provided by the manufacturer (BD FACSAria II). Apoptosis was measured by using an Annexin V-PE/7-Amino-Actinomycin (7-AAD) Apoptosis Detection Kit (#559763, BD Biosciences Pharmingen, US). After treatment with 5-FU (#9648, TargetMol, China) at the indicated concentrations for 48 hours, the cells were harvested by trypsinization, washed with cold phosphate-buffered saline (PBS), and then resuspended in 1X binding buffer. Then, 5 µL of PE Annexin V and 5 µL of 7-AAD were added to each tube. The suspension was then mixed well and incubated for 15 min in the dark at room temperature (RT) (25°C). After resuspension, the samples were analyzed by flow cytometry.
In vivo experiments
Female athymic 4- to 5-week-old Balb/c (nu/nu) mice were purchased from the Laboratory Animal Services Centre of Guangdong Province and were maintained in a specific pathogen-free facility. For the tumor growth assay, 5 × 106 cells were subcutaneously injected into the right and left sides of the back of the nude mice (n =5/group). The tumor volume was calculated using the following formula: V = 0.5 × D × d2 (where V represents volume, D represents the longitudinal diameter, and d represents the latitudinal diameter).
Statistical analysis
GraphPad Prism 7.0, SPSS 22.0, and Microsoft Excel 2016 were used for statistical analysis. The data are expressed as the mean ± standard error of the mean unless otherwise stated. The student's t-test was used to detect significance between the groups, and the χ2 test was used for measuring the data. Analysis of variance (ANOVA) was used to determine differences among three or more groups followed by post hoc analysis. Spearman's correlation analysis was performed to detect the expression correlation.
Results
Higher RUNX1 expression was found in colon adenocarcinoma (COAD) tissues
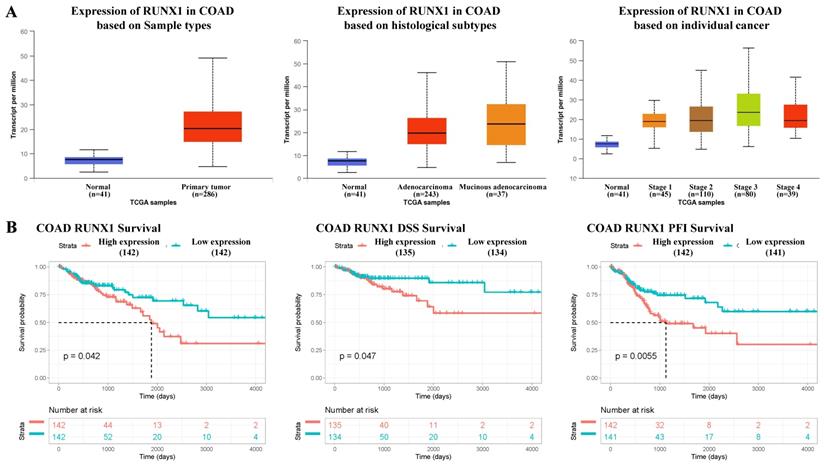
According to the mRNA-seq results of the TCGA COAD dataset, RUNX1 was upregulated in COAD tissues compared with normal tissues, and the expression of RUNX1 increased to varying degrees in different histological types and cancer stages (Fig. 1A). Further, the survival analysis results suggest that high RUNX1 expression is associated with worse overall survival (OS) and disease-specific survival (DSS) and a shorter platinum-free interval (PFI) (Fig. 1B).
RUNX1 promoted the proliferation of CRC cells in vitro
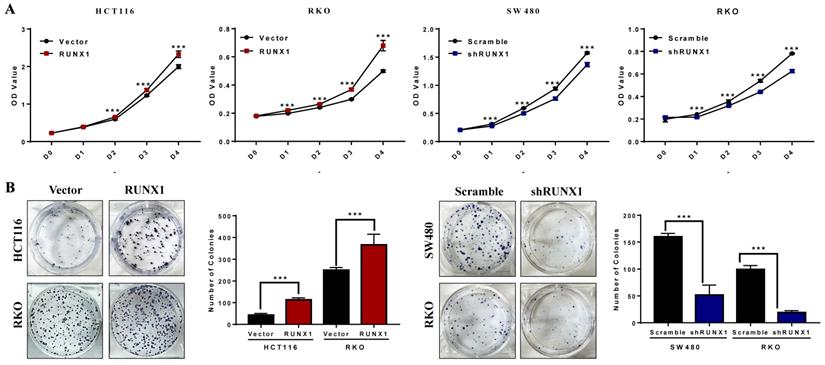
The CCK-8 assays demonstrated that the proliferation rates of HCT116 and RKO cells were increased when RUNX1 was overexpressed, and the proliferation rates of SW480 and RKO cells were decreased when RUNX1 was downregulated (Fig. 2A). The number of cell colonies was increased when RUNX1 was overexpressed, and the number of cell colonies was decreased when RUNX1 was downregulated (Fig. 2B).
RUNX1 promoted the proliferation of CRC cells in vivo
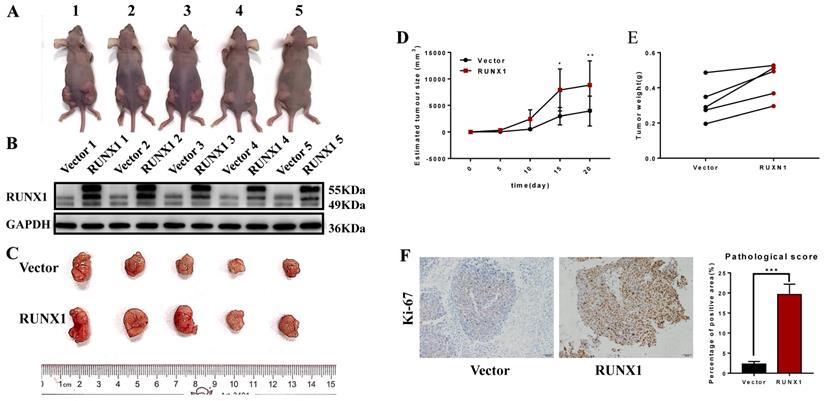
Furthermore, we also achieved the same results in the in vivo proliferation assays. HCT116 cells stably transfected with RUNX1 and vector were injected into the right and left hips of female nude mice. We found that after 20 days, HCT116-RUNX1 promoted tumor growth compared to the control group (Fig. 3A-E). Consistent with these findings, higher Ki-67 expression levels were shown by IHC in the RUNX1-overexpressing subcutaneous tumors of the nude mice (Fig. 3F). These results strongly suggest that RUNX1 is involved in enhancing the proliferative capacity of CRC.
RUNX1 expression in colorectal cancer tissues and its relationship with prognosis. A. RUNX1 is highly expressed in the colorectal cancer and the expression of RUNX1 is increased in different histological subtypes and cancer stages (UALCAN). B. Percent of overall survival (OS), disease-specific survival (DSS) and platinum-free interval (PFI) survival with low RUNX1 expression was higher than that with high RUNX1 expression of colon adenocarcinoma (COAD), given by TCGA database.
RUNX1 promotes the proliferation of CRC in vitro. A. Overexpressed RUNX1 stimulates cell proliferation as determined by CCK8 assay, while CRC cells with RUNX1 silencing inhibited cell proliferation. B. Overexpression of RUNX1 in HCT116 and RKO cells promoted CRC cells to form more colonies comparing to control cells. SW480 and RKO cells with RUNX1 silencing inhibited cells to form colonies; error bars, SD.
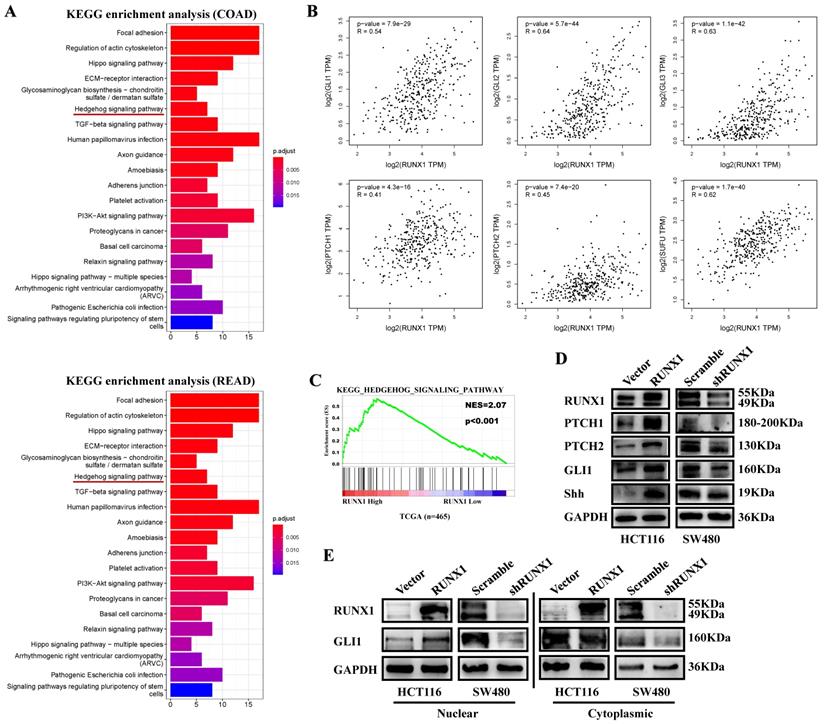
RUNX1 enhances Hedgehog pathway activation in CRC
To determine the potential function of RUNX1 in CRC, we analyzed the top 300 genes most similar to RUNX1 in COAD and rectum adenocarcinoma (READ) tissue samples from the TCGA database by KEGG enrichment, and the results showed that there was a correlation between RUNX and the Hedgehog signaling pathway (Fig. 4A). Through mRNA expression correlation analysis on the GEPIA website, we found that the expression of RUNX1 is positively correlated with multiple molecules of the Hedgehog pathway, such as GLI1, GLI2, GLI3, PTHC1, and PTCH2, and is statistically significant (Fig. 4B). Gene set enrichment analysis (GSEA) of the TCGA database showed that RUNX1 expression was associated with the Hedgehog signaling pathway (Fig. 4C). We used Western blotting to detect the expression of molecules in the Hedgehog pathway, and the results showed that RUNX1 increased the expression of proteins, including PTCH1, PTCH2, GLI1, and Shh, that are associated with the Hedgehog pathway (Fig. 4D). Moreover, a nuclear and cytoplasmic separation assay suggested that the expression changes of GLI1 protein were primarily shown in the nucleus (Fig. 4E).
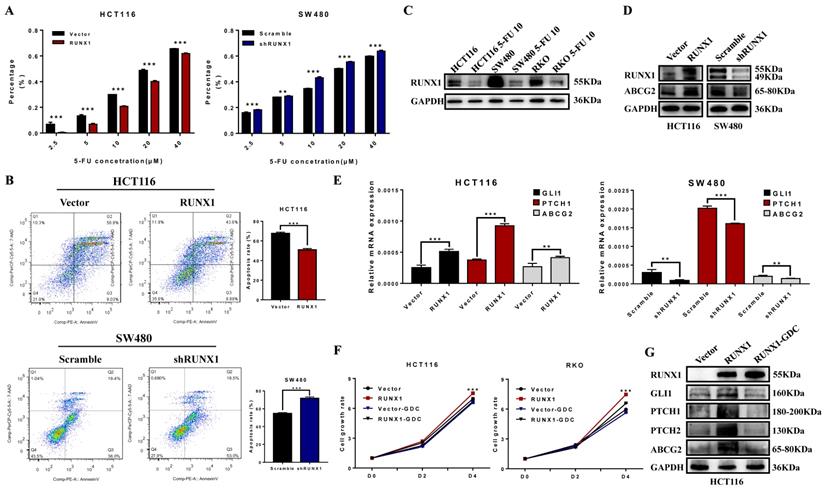
RUNX1 reduced the chemosensitivity of CRC cells to 5-FU
To explore the possible role of RUNX1 in chemotherapy resistance, the cell growth inhibition rate was detected after treatment with a concentration gradient of 5-FU, and the results demonstrated that RUNX1 weakened the cytostatic action of 5-FU (Fig. 5A). The cell apoptosis rate was determined by an apoptosis kit and flow cytometry. The apoptosis rate was increased in the RUNX1 knockdown group, while it was dramatically decreased in the RUNX1-overexpressing group during treatment with 5-FU (Fig. 5B). In addition, we investigated the protein expression level of RUNX1 and found that RUNX1 was downregulated after treatment with 5-FU (10 µM) in the HCT116, SW480, and RKO cell lines (Fig. 5C). Furthermore, there was a positive correlation between the expression of RUNX1 and ABCG2 (Fig. 5D), and ABCG2 is a part of the superfamily of ATP-binding cassette (ABC) transporters. The mRNA expression level of GLI1, PTCH1, ABCG2 were significantly altered by RUNX1 upregulation. Similar results were observed in HCT116 cells with RUNX1 overexpression and SW480 cells with RUNX1 silencing (Fig. 5E). GDC-0449, a Hedgehog signaling pathway-specific inhibitor. Using CCK8 assays, we found that when cells were treated with GDC-0449, the proliferation ability of HCT116 and RKO RUNX1-overexpressing cell lines was decreased (Fig. 5F). Moreover, the results of Western blot can suggest that GDC-0449 can inhibit the expression of ABCG2 and targeted genes activated by the Hedgehog signaling pathway induced by RUNX1 (Fig. 5G).
Discussion
It is generally believed that RUNX1 dosage is important during normal development and in the homeostasis of hematopoietic tissues, and growing experimental evidence implicates RUNX1 in crucial hallmarks of cancer progression, such as cell proliferation, epithelial-mesenchymal transition (EMT), or DNA repair[24]. In studies of CRC, RUNX1 regulates tumor metastasis by activating the Wnt/β-catenin signaling pathway and EMT. The lncRNA RNCR3 can be activated by RUNX1 and exerts its oncogenic role by regulating the miR-1301-3p/AKT1 axis[25]. However, no report has shown that RUNX1 has a role in chemoresistance.
RUNX1 promotes CRC proliferation in vivo. A. HCT116/Vector and HCT116/RUNX1 cells were injected in the backside of nude mice. B. RUNX1 expression in the subcutaneous tumors of mice were detected by western blotting. C. External whole-tumors images by subcutaneous injection of HCT116/Vector and HCT116/RUNX1 cells were obtained. D. Tumor sizes were measured on the indicated days to assess the effect of RUNX1 on subcutaneous tumor growth. E. Tumor weight of subcutaneous tumors were obtained. F. IHC of Ki-67 in subcutaneous tumors were assessed. Relevant pathological scores of each were also obtained; error bars, SD.
RUNX1 promotes CRC proliferation via Hedgehog signaling pathway. A. Enrichment of Hedgehog signaling pathway with RUNX1 expression in CRC was shown by enrichment analysis of KEGG. B. Relationship of RUNX1 and multiple molecules of the Hedgehog signaling pathway C. Enrichment of Hedgehog signaling pathway with RUNX1 expression in CRC was shown by enrichment analysis of GSEA. D. Expression of targeted genes activated by Hedgehog signaling pathway in HCT116/Vector, HCT116/RUNX1, SW480/scramble and SW480/shRUNX1 groups. E. GLI1 of the Hedgehog signaling pathway expression in HCT116/Vector and HCT116/RUNX1, SW480/scramble and SW480/shRUNX1 groups (nuclear and cytoplasmic separation assay).
An unresolved question is whether RUNX1 functions to promote tumor proliferation. First, in this report, we found that RUNX1 expression was upregulated in CRC tissue by analysis of the TCGA database. Next, we identified RUNX1 as an oncogene that promotes CRC cell proliferation both in vitro and in vivo, and the downregulation of RUNX1 had the opposite effect. Moreover, we found that RUNX1 promotes 5-FU chemoresistance in HCT116 and SW480 cells. Mechanistically, RUNX1 promotes CRC proliferation by activating the Hedgehog signaling pathway and regulating ABCG2 expression to promote chemoresistance.
Aberrant activation of the Hh signaling pathway is associated with tumorigenesis in various tissues, and most studies specify that the Hedgehog signaling pathway participates in the oncogenesis of CRC[26]. Some oncogenes promote the stemness and drug resistance of CRC by aberrant activation of the Hedgehog signaling pathway[27-30].
In the ABC protein superfamily, human breast cancer resistance protein (BCRP, ABCG2) is the main efflux transporter, and its overexpression in cancer-resistant cell lines leads to MDR[31]. In human multiple myeloma, the expression of ABCG2 is regulated by the PI3K/Akt signaling pathway, which affects the drug resistance of cancer cells[32,33]. The expression of ABCG2 in non-small cell lung cancer affected the efficacy of some antitumor drugs[34]. Upregulation of ABCG2 was reported to play an important role in the chemoresistance of breast cancer cells[35], and similar conclusions have been drawn from the study of gastric cancer[36]. ABCG2 was reported to be an essential factor for MDR in CRC[37]; however, whether RUNX1 can lead to MDR by regulating ABCG2 expression has not been confirmed. It has been suggested that by upregulating the expression of ABC transporter family members, the transcription factors inducing EMT can participate in the production of MDR[38]. Our previous studies have shown that RUNX1 is associated with EMT transcription factors in CRC[12].
ABCG2 is the target of the Hedgehog signaling pathway. The abnormal expression of the transcription factors Gli1 and Gli2 in the Hh signaling pathway regulates the expression of ABCG2 in many cancers. The activation of the Hh signaling pathway regulates drug sensitivity by directly upregulating ABCG2 expression in diffuse large B-cell lymphoma (DLBCL)[39], ovarian cancer[40,41], and gastric cancer[42]. Combined with the results of this study, we consider that RUNX1 regulates ABCG2 by activating the Hedgehog signaling pathway.
Conclusions
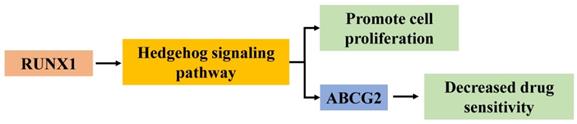
In summary, the current study illustrates that RUNX1 functions as an oncogene to facilitate the progression and chemotherapy resistance of CRC by enhancing the Hedgehog signaling pathway and promoting the expression of ABCG2 (Fig. 6). These findings enhance our understanding of CRC progression and chemotherapy. RUNX1 might be regarded as a potential prognostic marker and as an effective therapeutic target for CRC.
RUNX1 decreased 5-FU sensitivity of CRC cells via Hedgehog signaling pathway. A. 5-FU sensitivity measured by CCK8 assays in HCT116/RUNX1, SW480/shRUNX1, HCT116/Vector and SW480/Scramble cells with the different concentrations of 5-FU treated; error bars, SD. B. Apoptosis rate measured by flow cytometer in HCT116/RUNX1, SW480/shRUNX1, HCT116/Vector and SW480/Scramble cells with 5-FU treated; error bars, SD. C. Expression of RUNX1 in HCT116, SW480 and RKO cell lines with 5-FU treated. D. Expression of ABCG2 in HCT116/RUNX1, SW480/shRUNX1, HCT116/Vector and SW480/Scramble groups. E. Expression levels of GLI1 PTCH1 ABCG2 mRNA in HCT116/Vector and HCT116/RUNX1, SW480/scramble and SW480/shRUNX1 group; error bars, SD. F. HCT116 and RKO cell proliferation determined by CCK8 assay in Vector and RUNX1, vector with GDC and RUNX1 with GDC groups G. Expression of ABCG2 and targeted genes activated by Hedgehog signaling pathway in HCT116/Vector, HCT116/RUNX1, HCT116/RUNX1 with GDC groups.
Hypothesized molecular mechanism of RUNX1 in proliferation and 5-FU sensitivity decrease of CRC via Hedgehog signaling pathway.
Abbreviations
CRC: colorectal cancer; mCRC: metastatic colorectal cancer; RUNX1: Runt-related transcription factor 1; 5Fu: 5-fluorouracil; MDR: multidrug resistance; ABCG2: ATP binding cassette subfamily G member 2; HE: hemogenic endothelium; HSPCS: Hematopoietic stem and progenitor cells; KEGG: Kyoto encyclopedia of genes and genomes; TCGA: the cancer genome atlas; SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis; PVDF: polyvinylidene difluoride; CCK-8: cell count kit-8; COAD: colon adenocarcinoma; READ: rectum adenocarcinoma; OS: overall survival; DSS: disease-specific survival; PFI: platinum-free interval; EMT: epithelial to mesenchymal transition.
Supplementary Material
Supplementary table.
Acknowledgements
This study was supported by Guangdong Provincial Bio-engineering Research Center for Gastroenterology Diseases.
Author Contributions
Aimin Li, Side Liu, and Qingyuan Li conceived and designed the study. Qingyuan Li, Qiuhua Lai, Chengcheng He, and Qun Yan performed the experiments. Qingyuan Li, Qiuhua Lai, Chengcheng He, Yuxin Fang, and Haonan Zhang performed qPCR, cell culture, H&E, and IHC. Qingyuan Li, Qiuhua Lai, Chengcheng He, Qun Yan, and Haolin Li conducted WB and animal experiments. Qingyuan Li, Qiuhua Lai, Xingzhu Pan performed Immunofluorescence assays. Qingyuan Li, Qiuhua Lai, and Qun Yan interpreted and analyzed the data. Aimin Li, Side Liu, Qingyuan Li, and Xingzhu Pan wrote and reviewed the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Funds of China (82072758, 12026605), the Guangdong Basic and Applied Basic Research Fund (2020A1515110916, 2021A1515010992), the Guangdong Medical Science and Technology Research Fund Project (A2020143), and the College Students' Innovative Entrepreneurial Training Plan Program (X202012121322, S202012121089, S202012121090), The China's Postdoctoral Science Fund (2019M663005) and the Foundation of Nanfang Hospital, Southern Medical University (2018C027, 2019B014), and the Guangdong Science and Technology Plan Project (2017B020209003), the National Program on Key Research Project (2016YFC0901402).
Availability of data and materials
All data analyzed during this study are included in this manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel R, Miller K, Goding Sauer A, Fedewa S, Butterly L, Anderson J. et al. Colorectal cancer statistics, 2020. CA: a cancer journal for clinicians. 2020;70:145-64
2. Byrne M, Saif M. Selecting treatment options in refractory metastatic colorectal cancer. OncoTargets and therapy. 2019;12:2271-8
3. To K, Leung W, Ng S. Exploiting a novel miR-519c-HuR-ABCG2 regulatory pathway to overcome chemoresistance in colorectal cancer. Experimental cell research. 2015;338:222-31
4. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J. et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170:548-63.e16
5. de Bruijn M, Dzierzak E. Runx transcription factors in the development and function of the definitive hematopoietic system. Blood. 2017;129:2061-9
6. Ito Y, Bae S, Chuang L. The RUNX family: developmental regulators in cancer. Nature reviews Cancer. 2015;15:81-95
7. Han R, Wei J, Zhang H, Su X, Chu X, Chen Y. et al. Influence of TS (rs34743033) and RUNX1 (rs2014300) gene polymorphisms on survival outcomes of fluorouracil-based chemotherapy in Chinese advanced gastric cancer patients. Cancer management and research. 2018;10:1429-37
8. Hong D, Fritz A, Finstad K, Fitzgerald M, Weinheimer A, Viens A. et al. Suppression of Breast Cancer Stem Cells and Tumor Growth by the RUNX1 Transcription Factor. Molecular cancer research: MCR. 2018;16:1952-64
9. Deltcheva E, Nimmo R. RUNX transcription factors at the interface of stem cells and cancer. The Biochemical journal. 2017;474:1755-68
10. Chimge N, Ahmed-Alnassar S, Frenkel B. Relationship between RUNX1 and AXIN1 in ER-negative versus ER-positive Breast Cancer. Cell cycle (Georgetown, Tex). 2017;16:312-8
11. Chimge N, Little G, Baniwal S, Adisetiyo H, Xie Y, Zhang T. et al. RUNX1 prevents oestrogen-mediated AXIN1 suppression and β-catenin activation in ER-positive breast cancer. Nature communications. 2016;7:10751
12. Li Q, Lai Q, He C, Fang Y, Yan Q, Zhang Y. et al. RUNX1 promotes tumour metastasis by activating the Wnt/β-catenin signalling pathway and EMT in colorectal cancer. Journal of experimental & clinical cancer research: CR. 2019;38:334
13. Rooney N, Mason S, McDonald L, Däbritz J, Campbell K, Hedley A. et al. RUNX1 Is a Driver of Renal Cell Carcinoma Correlating with Clinical Outcome. Cancer research. 2020;80:2325-39
14. Gao L, Tober J, Gao P, Chen C, Tan K, Speck N. RUNX1 and the endothelial origin of blood. Experimental hematology. 2018;68:2-9
15. Cortes M, Liu S, Kwan W, Alexa K, Goessling W, North T. Accumulation of the Vitamin D Precursor Cholecalciferol Antagonizes Hedgehog Signaling to Impair Hemogenic Endothelium Formation. Stem cell reports. 2015;5:471-9
16. Li C, Zheng Z, Zhang X, Asatrian G, Chen E, Song R. et al. Nfatc1 Is a Functional Transcriptional Factor Mediating Nell-1-Induced Runx3 Upregulation in Chondrocytes. International journal of molecular sciences. 2018 19
17. Komori T. Regulation of skeletal development by the Runx family of transcription factors. Journal of cellular biochemistry. 2005;95:445-53
18. Pelullo M, Zema S, Nardozza F, Checquolo S, Screpanti I, Bellavia D. Wnt, Notch, and TGF-β Pathways Impinge on Hedgehog Signaling Complexity: An Open Window on Cancer. Frontiers in genetics. 2019;10:711
19. Chatterjee S, Sil P. Targeting the crosstalks of Wnt pathway with Hedgehog and Notch for cancer therapy. Pharmacological research. 2019;142:251-61
20. Chandrashekar D, Bashel B, Balasubramanya S, Creighton C, Ponce-Rodriguez I, Chakravarthi B. et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia (New York, NY). 2017;19:649-58
21. Haeussler M, Zweig A, Tyner C, Speir M, Rosenbloom K, Raney B. et al. The UCSC Genome Browser database: 2019 update. Nucleic acids research. 2019;47:D853-D8
22. Liao Y, Wang J, Jaehnig E, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic acids research. 2019;47:W199-W205
23. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic acids research. 2017;45:W98-W102
24. Lie-A-Ling M, Mevel R, Patel R, Blyth K, Baena E, Kouskoff V. et al. RUNX1 Dosage in Development and Cancer. Molecules and cells. 2020;43:126-38
25. Xu G, Wang H, Yuan D, Yao J, Meng L, Li K. et al. RUNX1-activated upregulation of lncRNA RNCR3 promotes cell proliferation, invasion, and suppresses apoptosis in colorectal cancer via miR-1301-3p/AKT1 axis in vitro and in vivo. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2020;22:1762-77
26. Wu C, Zhu X, Liu W, Ruan T, Tao K. Hedgehog signaling pathway in colorectal cancer: function, mechanism, and therapy. OncoTargets and therapy. 2017;10:3249-59
27. Ren Y, Deng R, Cai R, Lu X, Luo Y, Wang Z. et al. TUSC3 induces drug resistance and cellular stemness via Hedgehog signaling pathway in colorectal cancer. Carcinogenesis. 2020;41:1755-66
28. Yang Z, Zhang C, Feng Y, Quan M, Cui Y, Xuan Y. Tenascin-C predicts poor outcomes for patients with colorectal cancer and drives cancer stemness via Hedgehog signaling pathway. Cancer cell international. 2020;20:122
29. Zhou H, Xiong Y, Peng L, Wang R, Zhang H, Fu Z. LncRNA-cCSC1 modulates cancer stem cell properties in colorectal cancer via activation of the Hedgehog signaling pathway. Journal of cellular biochemistry. 2020;121:2510-24
30. Lai Q, Li Q, He C, Fang Y, Lin S, Cai J. et al. CTCF promotes colorectal cancer cell proliferation and chemotherapy resistance to 5-FU via the P53-Hedgehog axis. Aging. 2020;12:16270-93
31. Mao Q, Unadkat J. Role of the breast cancer resistance protein (BCRP/ABCG2) in drug transport-an update. The AAPS journal. 2015;17:65-82
32. Wang L, Lin N, Li Y. The PI3K/AKT signaling pathway regulates ABCG2 expression and confers resistance to chemotherapy in human multiple myeloma. Oncology reports. 2019;41:1678-90
33. Porcelli L, Giovannetti E, Assaraf Y, Jansen G, Scheffer G, Kathman I. et al. The EGFR pathway regulates BCRP expression in NSCLC cells: role of erlotinib. Current drug targets. 2014;15:1322-30
34. Zhang G, Zhang Y, Wang Y, Gupta P, Ashby C, Alqahtani S. et al. Epidermal growth factor receptor (EGFR) inhibitor PD153035 reverses ABCG2-mediated multidrug resistance in non-small cell lung cancer: In vitro and in vivo. Cancer letters. 2018;424:19-29
35. Zhou S, Liao L, Chen C, Zeng W, Liu S, Su J. et al. CD147 mediates chemoresistance in breast cancer via ABCG2 by affecting its cellular localization and dimerization. Cancer letters. 2013;337:285-92
36. Zhang H, Jiang H, Zhang H, Liu J, Hu X, Chen L. Ribophorin II potentiates P-glycoprotein- and ABCG2-mediated multidrug resistance via activating ERK pathway in gastric cancer. International journal of biological macromolecules. 2019;128:574-82
37. Zhang Y, Wang Y, Lei Z, Zhang G, Zhang X, Wang D. et al. Regorafenib antagonizes BCRP-mediated multidrug resistance in colon cancer. Cancer letters. 2019;442:104-12
38. Saxena M, Stephens M, Pathak H, Rangarajan A. Transcription factors that mediate epithelial-mesenchymal transition lead to multidrug resistance by upregulating ABC transporters. Cell death & disease. 2011;2:e179
39. Singh R, Kunkalla K, Qu C, Schlette E, Neelapu S, Samaniego F. et al. ABCG2 is a direct transcriptional target of hedgehog signaling and involved in stroma-induced drug tolerance in diffuse large B-cell lymphoma. Oncogene. 2011;30:4874-86
40. Chen Y, Bieber M, Teng N. Hedgehog signaling regulates drug sensitivity by targeting ABC transporters ABCB1 and ABCG2 in epithelial ovarian cancer. Molecular carcinogenesis. 2014;53:625-34
41. Zhang W, Yu F, Wang Y, Zhang Y, Meng L, Chi Y. Rab23 promotes the cisplatin resistance of ovarian cancer via the Shh-Gli-ABCG2 signaling pathway. Oncology letters. 2018;15:5155-60
42. Yu B, Gu D, Zhang X, Liu B, Xie J. The role of GLI2-ABCG2 signaling axis for 5Fu resistance in gastric cancer. Journal of genetics and genomics = Yi chuan xue bao. 2017;44:375-83
Author contact
![]() Corresponding authors: Aimin Li, Department of Gastroenterology, Nanfang Hospital, Southern Medical University, No. 1838, Guangzhou Avenue North, Guangzhou, People's Republic of China. E-mail: lam0725com. Side Liu, Department of Gastroenterology, Nanfang Hospital, Southern Medical University, No. 1838, Guangzhou Avenue North, Guangzhou, People's Republic of China. E-mail: liuside2011com
Corresponding authors: Aimin Li, Department of Gastroenterology, Nanfang Hospital, Southern Medical University, No. 1838, Guangzhou Avenue North, Guangzhou, People's Republic of China. E-mail: lam0725com. Side Liu, Department of Gastroenterology, Nanfang Hospital, Southern Medical University, No. 1838, Guangzhou Avenue North, Guangzhou, People's Republic of China. E-mail: liuside2011com

 Global reach, higher impact
Global reach, higher impact