Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(16):4873-4882. doi:10.7150/jca.58859 This issue Cite
Review
Potential impact and mechanism of Long Non-coding RNAs on cancer and associated T cells
1. Department of Laboratory Medicine, the First Affiliated Hospital of Nanjing Medical University, Nanjing, China, 210029.
2. National Key Clinical Department of Laboratory Medicine, Nanjing, China, 210029.
Received 2021-1-30; Accepted 2021-5-30; Published 2021-6-11
Abstract
The discovery of many aberrant expressions of long non-coding RNAs (lncRNAs) in various cancers has focused attention on the effects of lncRNA on cancer cells themselves, including cell proliferation, growth inhibition, cell migration, cell immortality, vascular regeneration and cell viability. But with the increasing role of immunotherapy in cancer therapy, a large number of studies have revealed that the regulatory role of lncRNAs in immunity such as differentiation of immune cells can also influence the development and progression of cancer. In particular, recent publications have suggested that lncRNAs play critical roles in T-lymphocyte activation, proliferation, differentiation, function, apoptosis and metabolism. To elucidate the actual functions of lncRNAs at the molecular level of cancer pathogenesis, we summarize some of the current lncRNA regulatory mechanisms associated with T cell to discuss their effects in cancer in the hope of providing potential cancer therapeutic targets or cancer biomarkers. However, we all know that the differentiation and function of T cells is an extremely complex process that involves the expression and regulation of multiple lncRNAs. As a result, more regulatory mechanisms of lncRNAs need to be further studied.
Keywords: Long non-coding RNAs, cancer, T cell
1. Introduction
The regulatory role of non-coding RNAs (ncRNAs) in cancer has been favored by researchers in recent years. Among those, lncRNAs have gradually become an indispensable effective and regulatory component in different types of cancer, thus showing potential diagnostic and therapeutic value of cancer [1]. The involvement of lncRNAs in cancer not only interacts with the cell cycle and proliferation pathways of cancer cells, but also effects the immune system by regulating tumor microenvironment (TME), epithelial-mesenchymal transition (EMT), microbiota, metabolism, immune cell differentiation and function of immune cells [2].
The proper functioning of the immune system is the key to fighting off pathogens and cancers. Meanwhile, lncRNAs have been found to be involved in the regulation of gene expression in the immune system, affecting the differentiation and function of various cell types of innate immunity and adaptive immunity such as macrophage myeloid cell and T cell [3]. T cells, the most important participants in the process of innate and adaptive immunity, play a crucial role in regulating the immune response. The normal proliferation, differentiation and function of T cells is critical to maintaining the appropriate execution of the immune system. Many studies underscore the important role of lncRNAs in CD4+ T cell differentiation and have shown that the abnormal differentiation of CD4+ T-helper (Th) cells is influenced by lncRNAs and may lead to autoimmune diseases [4, 5]. Advances in analytical methods like real-time fluorescent quantitative PCR (qPCR) and microarrays have enabled more and more people to analyze the differential expression of lncRNAs in different T cell subtypes and at different stages of T cell growth and development. For example, Jinping Zhang et al. examined the expression profiles of lncRNAs and mRNAs in CD4-CD8- (DN), CD4+CD8+ (DP), CD4+CD8- and activated CD4+CD8- T cells in a microarray analysis and found specific expression profiles of lncRNAs at different stages of development [6]. Gangqing Hu et al. performed RNA-Seq of peripheral T cells at multiple time points during their differentiation and confirmed that the expression of lncRNA is cell-specific [7]. More importantly, one research found that immune-related lncRNAs in lung cancer patients have significantly increased expression in T cells [8]. Thus, in this review, we will provide an overview of current state of knowledge implicating lncRNAs and T cells, with main emphasis on the regulatory effects of lncRNAs on the activation, proliferation, differentiation, effector functions, apoptosis and metabolism of T cells and their potential implications in cancer.
2. Long non-coding RNAs
It is now well known that the human genome is almost completely transcribed [9]. However, numerous studies have shown that only a small fraction of the transcriptome is translated, and that most transcriptional output is ncRNA. LncRNAs are non-coding RNAs with lengths greater than 200 nucleotides (nt) and do not have an open reading frame [10, 11]. In this chapter, we will provide an overview of lncRNA biogenesis and function.
2.1. LncRNA biogenesis
LncRNA transcripts are longer than 200nt in length and they don't have the ability to code for proteins. Most known lncRNAs have been found to be transcribed by RNA polymerase II (Pol II). Therefore, they are structurally similar to messenger RNA (mRNA) and may have cap structures and poly A tails. The main difference between lncRNA and mRNA is that lncRNA contains transcriptional termination codons, so lncRNA has no protein-coding ability [12-16]. However, recent ribo-seq and mass spectrometry data suggested that lncRNAs may be translatable due to their small open reading frames encoding for short peptides [17, 18]. Liman Niu et al. even have verified a micropeptide encoded by lncRNA MIR155HG [19]. LncRNAs are often classified into five groups based on their genomic positions relative to protein-coding genes: sense lncRNAs, antisense lncRNAs, intronic lncRNAs, bidirectional lncRNAs and intergenic lncRNAs. Overlap with one or more exons of the encoding gene are sense lncRNAs, antisense lncRNAs are complementary to the transcriptional products on the opposite chain, intronic lncRNAs are produced in the introns of genes, bidirectional lncRNAs and protein-coding genes share the same promoter but the transcriptional direction is reversed and intergenic lncRNAs are transcribed independently by sequences located between protein-coding genes [20-22].
2.2. LncRNA function
More and more studies have proposed the regulation mechanism of lncRNA in diseases. Studies have shown that lncRNA mainly regulates gene expression from three aspects: epigenetics, transcriptional regulation and post-transcriptional regulation [23-25], and more specifically, lncRNA regulates gene expression, chromatin organization, cellular trafficking, RNA decay and translation. Furthermore, it is worth noting that lncRNA will also affect protein localization, function, decay and turnover [26]. LncRNA can play functions through RNA-DNA, RNA-RNA and RNA-protein interactions [27]. Previous studies have shown mechanisms of action of lncRNA: (I) interfere with the expression of adjacent protein-coding genes by transcription in the upstream promoter region of protein-coding genes; (II) affect gene expression by inhibiting RNA polymerase, or by mediating chromatin remodeling and histone modification; (III) Interfere with the splicing of mRNA by supplementing the double chain with transcripts of protein-coding genes; (IV) use the transcripts of protein-coding genes to complement the double chains and regulate gene expression through the production of endogenous small interference RNA by dicer enzymes; (V) regulate activity by binding to specific proteins; (VI) form nucleic acid protein complexes with protein as their structural components; (VII) change the cytoplasmic localization of proteins by binding to specific proteins; (VIII) as precursor molecules of small RNAs [15].
3. LncRNAs and regulation of T cell activation, proliferation, differentiation, effector functions, apoptosis and metabolism
A better understanding of the role of lncRNAs in T cells with extensively investigated disease such as autoimmune disease may provide new lncRNA-T cell-based therapeutic target for cancer. Many lncRNAs have been explored for their role in regulating T cell activation, proliferation, differentiation, effector functions, apoptosis and metabolism. In this chapter, we will show some typical lncRNAs with fully-elucidated role in regulating T cell.
3.1. Activation
The activation of T cells depends on the double signal of antigen and costimulatory molecules. Antigen-presenting cell (APC) is a kind of immune cell that captures, processes and presents antigens to antigen-specific lymphocytes. APC provides at least two independent signals to stimulate T cell activation. Major histocompatibility complexes (MHC) and costimulatory molecules expressed by APC are the sites where many lncRNAs regulate. Dendritic cells (DC), the most important APC, has been found in many studies to be extensively regulated by lncRNA in the activation of T cells. Yiling Ding et al. have identified a lncRNA specifically expressed in DC can promote the over-maturation of DC and induce a strong immune response [28]. On the contrary, many researchers have noticed that some lncRNAs can induce DC to become tolerogenic dendritic cells (tDC). For example, Jian Wu et al. demonstrated that lncRNA MALAT1 induces DC towards tolerant type and MALAT1 promoted dendritic cell-specific intercellular adhesion molecule-3 grabbing nonintegrin (DC-SIGN) expression by functioning as an miR155 sponge. This maintains tolerance of DC by secreting interleukin-10 (IL-10), leading to low level of CD80, CD86 and MHC expressed by DC [29]. In addition to MALAT1, NEAT1 has been demonstrated to induce DC to develop into tolerogenic phenotype by regulating the NACHT, LRR, and PYD domains-containing protein3 (NLRP3) inflammasome as a competitor for miR-3076-3p [30].
LncRNA not only affects the double signal provided by APC cells for T cell activation, but also specifically affects the expression of MHC molecules and costimulatory molecules. Previous studies showed that lncRNA IFNG-AS1 is related to the production of IFN-γ, but in a study by Mengchuan Luo et al., they found IFNG-AS1 affects the expression of HLA-DRB1 to control the immune response in myasthenia gravis (MG) [31]. Additionally, according to Liu Qian et al's research, lncRNA GAS5 upregulates adenovirus E4 binding protein 4 (E4BP4) which targets CD40L to inhibit activation of CD4+ T cell by inhibiting miR-92a-3p in systemic lupus erythematosus (SLE) [32]. In addition to positive costimulatory molecules, negative costimulatory molecules represented by PD-1/PD-L1 have also attracted extensive attention in recent years. Multiple lncRNAs have been shown to be involved in the process. For instance, Bo Hu et al. revealed that lncRNA XLOC_003810 decreased the proportion of CD4+ PD-1+ T cells in patients with MG [33].
3.2. Proliferation and differentiation
Naive T cells are activated by double signals, proliferate and differentiate into different functional subsets under the action of microenvironmental cytokines. It has been well established that lncRNAs are also widely involved in the differentiation of helper T cells (Th) and regulatory T cells (Treg). For example, Martina Gast et al. used NEAT1-/- mice as a model of lncRNA NEAT1 deficiency to show anomalous Treg and Th cell differentiation. They finally found a shift of CD4+ T cell balance towards Th cell proliferation both in the spleens and in the circulating blood [34]. Furthermore, the differentiation of Th is also orchestrated by numerous lncRNAs. Many studies have shown that the differentiation of CD4+ T cells into Th17 cells is a major cause of the occurrence and development of autoimmune diseases. Xiaolong Shui et al. determined the involvement of lncRNA NEAT1 in the differentiation of Th17 cells by targeting STAT3 protein level in rheumatoid arthritis (RA) [35]. Fang Zhang et al. determined lncDDIT4 inhibits DDIT4/mTOR signaling by targeting DNA-damage-inducible transcript 4 (DDIT4) to suppress Th17 cells differentiation in multiple sclerosis (MS) [36]. RORγt is the main transcription factor for Th17 cell differentiation, and Manuel B. Braga-Neto et al. described a novel lincRNA XLOC_000261 which appears to negatively regulate RORγt protein expression in Th17 cells in Crohn's disease (CD) [37]. Besides, Th1/Th2 lineage differentiation requires the involvement of many lncRNAs. Valeria Ranzani et al. found that down-regulation of linc-MAF-4 by mediating transcription of MAF through epigenetics enables T cells to differentiate toward the Th2 subtype instead of Th1 subtype [38]. The signal transducer and activator of transcription (STAT) 6 is an important transcription factor for the differentiation of Th2 cells and Shuman Huang et al. demonstrated that lncRNA NEAT1 promotes Th2 differentiation through EZH2/ITCH/STAT6 axis [39]. GATA3 is also considered the master regulator of Th2 and Hunter R. Gibbons et al. demonstrated that divergent lncRNA GATA3-AS1 is necessary for the transcription of GATA3, thus promoting the differentiation of Th2, but it cannot induce total Th2 polarization independent of other factors such as c-Maf or Stat6 [40]. Moreover, lncRNAs in exosomes have also been found to regulate T cell differentiation. Xiaoyuan Zhu et al. detected the expression of lncRNA GAS5 in exosomes which were isolated from allergic rhinitis (AR) patients and found GAS5 suppresses Th1 differentiation and promotes Th2 differentiation via downregulating EZH2 and T-bet [41]. Similarly, lncRNAs play vital role in Treg differentiation. TGF-β signaling is an important pathway to induce Treg cells, and this process has also been shown to be actively regulated by lncRNA. For instance, Meng Xia et al. identified lnc-smad3 inhibits the H3K4 methyltransferase Ash1l-mediated Smad2/3-TGF-β signaling pathway and thus inhibits induced Tregs (iTreg) polarization [42]. Treg-specific transcription factor Foxp3 plays a key role in the differentiation of Treg cells and it has been well documented that lncRNA regulates the expression of Foxp3 to promote Treg cell differentiation. Aleksandra Brajic et al. identified a novel lncRNA Flatr which anticipates Foxp3 expression in Tregs [43] and David Zemmour et al. found that lncRNA Flicr modulates Foxp3 expression through modification of chromatin accessibility [44].
3.3. Effector functions
Distinct effector T cell subsets have their own characteristics and effects. CD4+ T cells are divided into a series of multifunctional immune cell subsets, including Th1, Th2, Th17, and Treg cells as well as unconventional T helper subsets. Cytotoxic CD8+ T cells are the main component that clears virus infection and resists tumor in adaptive immunity. Different subtypes of Th cells secrete different cytokines to function, and the possibility that lncRNAs participate in this process has been investigated. IFN-γ production is considered to be the most representative marker of Th1 cells and is often associated with inflammation and autoimmune diseases. Studies have shown that lncRNA IFNG-AS1, also known as Tmevpg1, regulates the production of IFN-γ [45]. For example, Carl Robert Rankin et al. verified IFNG-AS1 promotes the production of IFN-γ, thus exacerbating inflammatory bowel disease (IBD) [46] and Juan Wang et al. found TMEVPG1 enhances the immune response of Th1 cells in patients with Sjogren syndrome [47]. In addition, a large number of other studies have shown that IFNG-AS1 enhances the immune response of Th1 and is associated with autoimmune diseases such as Multiple sclerosis (MS) and Hashimoto's Thyroiditis (HT) [48, 49].
Compared with Th1 cells, Th2 cells mainly mediate humoral immunity by secreting cytokines such as IL-4, IL-5, IL-6 and IL-10. Weijie Yin et al. have identified a noncoding interleukin 4 (IL-4) RNA that promotes IL-4 mRNA translation in Th2 [50]. James P. Hewitson et al. demonstrated lncRNA Malat1 suppresses immunity to infection through promoting expression of Maf, a key transcriptional regulator of IL-10 in Th cells [51]. However, this finding is contrary to the conclusion of another study conducted by Yingpeng Yao and his colleagues that Malat1 is not essential for response to lymphocytic choriomeningitis virus (LCMV) infection [52]. Of course, they used different models of mouse infection but actually further research has yet to be confirmed.
Th17 cells mainly secrete IL-17 to participate in the inflammatory response. Yuying Qiu et al. demonstrated lncRNA-MEG3 acts as a microRNA-17 sponge to promote the production of Th17-related cytokines such as IL-17 and IL-22 and disrupt the balance of Th17 and Treg cells, thus contributing to the pathogenesis of asthma [53]. Likewise, Jianqin Li et al. also found that MEG3 induces the imbalance of Th17 and Treg cells by inhibiting the expression of miR-125A-5P in immune thrombocytopenic purpura (ITP) [54].
Treg cells are a group of T cells with immunosuppressive function, mainly including natural Tregs (nTregs) and induced Tregs (iTregs). Treg cells can reduce tissue damage caused by autoimmunity in chronic immune responses. In parallel, they also create opportunities for tumor immune escape. Many findings reveal the mechanisms by which Treg cells inhibit immune response. Xinhong Pei et al. demonstrated that lncRNA SNHG1 regulated IDO by targeting miR-448 to promote Treg cell differentiation and IL-10 secretion to mediate the immune escape of breast cancer (BC) [55]. Jing Wang et al. demonstrated that lncRNA DQ786243 upregulates Foxp3+ Treg cells in oral lichen planus (OLP) and suppresses the secretion of IFN-γ and IL-17 by other CD4+ T cells such as Th1 and Th17 through Foxp3-miR-146a-NF-κB axis [56]. In comparison to αβT cells, γδT cells have been less studied. But recently Chao Ni et al. showed that breast tumour cell-derived exosomes (TDEs) could transmit lncRNA SNHG16, which induces CD73+ γδ1 Treg cells through TGF-β1/lncRNA SNHG16/miR-16-5p/SMAD5 pathway. Notably, CD73+ γδ1 Treg cells inhibit CD4+ T cells from producing IFN-γ and CD8+ T cells from producing perforin and granzyme, and secreting IL-10 and TNF-β depending on the adenosine pathway [57].
Cytotoxic CD8+ T cell whose effector function is also regulated by lncRNA plays an irreplaceable role in the body's resistance to pathogen infection and tumor cells by secreting IFN-γ and TNF-α. Yang Wang et al. showed that lncRNA-CD244 which is induced by CD244+ CD8+ T cell in active tuberculosis (TB) infection inhibits IFN-γ/TNF-α expression in CD8+ T cells through recruitment of EZH2 to IFNG and TNFA loci for repressive chromatin states [58]. Moreover, Jiansong Wu et al. have found that lncRNA‑CD160 bind to histone‑modification enzyme gene histone deacetylases 11 (HDAC11) to form a complex and inhibit the function of HDAC11, which further inhibits the secretion of IFN‑γ and TNF-α in in hepatitis B virus infection [59].
3.4. Apoptosis
LncRNA regulates extrinsic and intrinsic apoptosis of T cells. Extrinsic apoptotic pathway is mediated by death receptors such as tumor necrosis factor related apoptosis-inducing ligand (TRAIL) and Fas-ligand (Fas-L). Intrinsic apoptotic pathway refers to increased permeability of mitochondrial outer membrane. Di Huang et al. examined that the JAK1-STAT1 signaling pathway in activated T cells upregulates the expression of NKILA lncRNA and inhibits NF-κB activity, thus leading to sensitivity of tumor-specific cytotoxic T lymphocytes (CTLs) and type 1 helper T (Th1) cells to activation-induced cell death (AICD) in the tumor microenvironment of breast cancer and lung cancer [60]. Similarly, a research found lncRNA TANCR regulates the cytotoxic function of γδ T cells by affecting the expression of TRAIL [61].
In addition, lncRNAs are widely involved in intrinsic apoptosis pathway. S. M. Ali Hosseini Rad et al. showed that a divergently transcribed lncRNA LOC107985203 negatively modulated Mcl-1 expression, a member of the Bcl-2 anti-apoptotic protein family to affect the lifespan of T lymphocytes [62]. Besides, Wiam Saadi et al. have found a lncRNA XLOC_000895 (Robnr), located downstream of the anti-apoptotic gene Bcl2, affected the activation of Bcl2 in the development of T lymphocytes [63]. In addition to influencing Bcl2 anti-apoptotic protein family, lncRNA was also found to be involved in the regulation of apoptotic factors such as Par-4. Lin Zhang et al. demonstrated that knockdown of a novel lncRNA, which is characterized from the Jurkat leukemic T-cell line can induce apoptosis of T-cell acute lymphoblastic leukemia (T-ALL) cells by promoting the formation of par-4 /THAP1 protein complex, thus increasing the activation of caspase-3 and the expression of pro-apoptotic smac protein [64]. Apoptosis-inducing factor, mitochondrion-associated 2 (AIFM2) has also been found to be upregulated by lncRNA MEG3 through targeting miR-214 in T-cell lymphoblastic lymphoma (T-LBL) [65] Actually, considerable studies have explored specific lncRNAs involved in the regulation of the apoptosis of T lymphocytes, but the specific mechanism of apoptosis remains unclear. For example, Hongbing Liu et al. found that knockout of lncRNA NEAT1 increases sensitivity of Jurkat CD4+ T cell lines to apoptosis during HIV-1 replication, but no specific mechanism has been found [66].
3.5. Metabolism
The transition from naive T cells to activated T cells mediated by TCR and co-stimulation signals involves a metabolic transition from oxidative phosphorylation (OXPHOS) to glycolysis [67] although some studies have shown that Tregs and memory CD8+ T cells depend on OXPHOS [68]. Many signaling pathways of TCR in the activation of T cells are associated with the metabolic process. Among those, PI3K/Akt/mTOR signaling pathway is demonstrated to affect transcription of several glycolytic genes [69]. Jiayao Fu et al. have found that lncRNA PVT1 regulates the expression of Myc through the PI3K/Akt signaling pathway, thus affecting the activation and proliferation of CD4+ T cells by reprogramming glycolysis [70]. Specific metabolic programs regulated by lncRNAs are adopted when T cells run through the activation, proliferation, differentiation and effector functions. LncRNA regulatory mechanisms responsible for key metabolic enzymes or transporters of energetic nutrients in the process may provide an explanation for metabolic reprogramming.
4. Role of lncRNAs in cancer associated with T cell
Considering the role of T cell in immunotherapy of cancer, lncRNAs involved in T cell activation, proliferation, differentiation, effector function, apoptosis and metabolism can impact cancer progression. Thus, lncRNAs can play an oncogenic or a cancer suppressive role through regulation of T cells, helping explore immunotherapy targets and overcome immunotherapy resistance in cancer. In this chapter, T cell associated lncRNAs in cancer and the possible pathways involved as well as potential clinic applications will be discussed.
4.1. T cell-associated lncRNAs in cancer
One of the central roles of T cell in immunity is the recognition and elimination of malignant transformations. Tumor cells inhibit T cell activation by reducing their antigenicity and upregulating immunosuppressive molecules. Qingsong Hu et al. reported that the transformed mammary gland epithelial cells downregulate antigen presentation machinery upon expression of LINK-A which facilitated crosstalk between phosphatidylinositol-(3,4,5)-trisphosphate and inhibitory G-protein-coupled receptor (GPCR) pathways, damping protein kinase A-mediated phosphorylation of the E3 ubiquitin ligase TRIM71, enhancing K48-polyubiquitination-mediated degradation of the antigen peptide-loading complex (PLC) in human triple-negative breast cancer (TNBC) [71] and Hao Li et al. demonstrated that long intergenic non-protein coding RNA 2195 (LINC02195) was closely related to antigen processing and presentation by affecting genes encoding MHC I molecules in head and neck squamous cell carcinoma (HNSCC) [72]. Besides, the possibility that lncRNAs are involved in regulating the expression of PD1/PD-L1 in cancer and affecting the activation of T cells has been raised. For example, PD-L1 is upregulated in pancreatic cancer (PC) by lncRNA LINC00473 as a sponge of microRNA-195-5p [73] and lncRNA HOTTIP promotes IL-6 transcription by regulating transcription factor c-jun to activate the STAT3/PD-L1 pathway in neutrophils in ovarian cancer (OC) [74]. Moreover, Hasmeena Kathuria et al. have found that genes regulated by NKX2-1-AS1 are related to PD-L1/PD-1 checkpoint pathways in human lung carcinoma [75] and Qing-Ming Wang et al. revealed that MALAT1, PD-L1 were upregulated in diffuse large B cell lymphoma (DLBCL) tissues by targeting miR-195 [76]. Compared with PD-L1, PD1 expression on lymphocytes was also found to be regulated by lncRNAs. For instance, AFAP1-AS1 and PD-1 are co-expressed in infiltrating lymphocytes in Nasopharyngeal carcinoma (NPC) [77].
Another central role of T cell immunity is the differentiation of distinct effector T cell subsets. Lnc-SGK1 induced by Helicobacter pylori infection and high salt diet suggests a possibility of promoting Th2 and Th17 differentiation in human gastric cancer (GC) by SGK1/Jun B signaling [78] and lnc-sox5 was found to promote the expression of indoleamine 2,3-dioxygenase 1 (IDO1) to modulate the infiltration and cytotoxicity of CD3+CD8+ T cells in colorectal cancer (CRC) [79]. Additionally, differentiation of Tregs is associated with the progression of cancer. For instance, linc-POU3F3 activates TGF- β signal pathway to increase Treg distribution and thereby promote gastric cancer (GC) cell proliferation [80] and the transcription factor FOXC1 mediates LINC00301 increases Treg through targeting TGF-β in non-small cell lung cancer (NSCLC) is verified by Cheng-Cao Sun et al. [81].
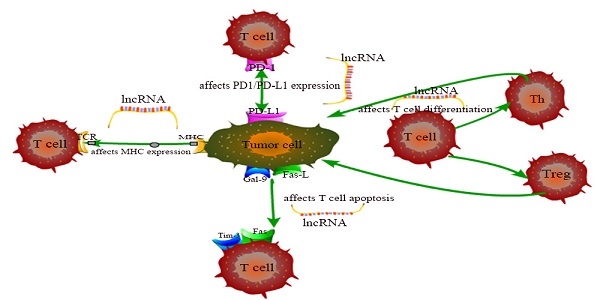
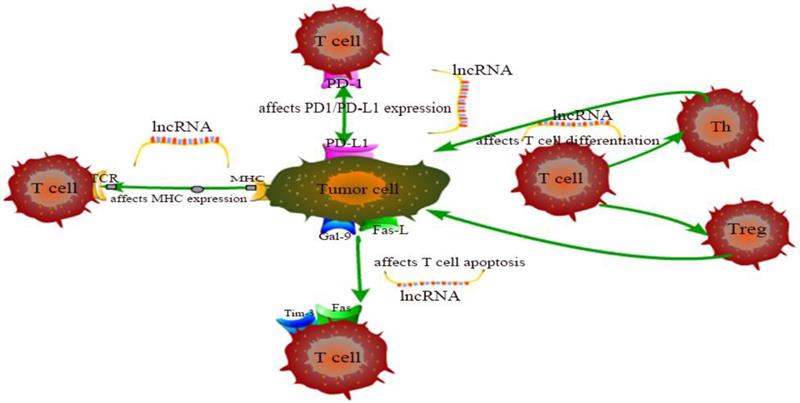
The exhaustion and apoptosis of T cells affect central role of T cell immunity, and more and more studies have proved that lncRNAs are involved in the regulation of this process. For example, T cell immunoglobulin mucin 3 (Tim-3), an inhibitory receptor, is regulated by lncRNA to alter the immune response of CD8+ T cells. Jie Ji et al. demonstrated that lnc-Tim3 promotes T cell exhaustion via suppressing Tim-3-Bat3 signaling and downstream signaling pathway NFAT1 and AP-1 in hepatocellular carcinoma (HCC) [82]. Similarly, Kai Yan et al. showed that down-regulation of lncRNA NEAT1 can limit the apoptosis of CD8+ T cells through miR-155/Tim-3 pathway in HCC [83]. In the tumor microenvironment of breast cancer and lung cancer, Di Huang et al. examined that the JAK1-STAT1 signaling pathway in activated T cells upregulates the expression of NKILA lncRNA and inhibits NF-κB activity, thus leading to sensitivity of tumor-specific cytotoxic T lymphocytes (CTLs) and type 1 helper T (Th1) cells to activation-induced cell death (AICD) [60]. Aside from that, Lalit Sehgal et al. found that a lncRNA corresponding to an antisense transcript of Fas (FAS-AS1) regulates alternative splicing of Fas in lymphomas and therefore impairing Fas-mediated apoptosis in B-cell lymphoma [84]. These are just a few of lncRNA examples representative to carcinogenesis. LncRNAs involved in cancer regulation associated with T cells in this chapter are shown in Table 1 and the specific regulation of lncRNAs on T cells is shown in Figure 1. Although the regulation of lncRNAs in the pathogenesis of cancer is not fully understood, lncRNA-mediated T cell is an important direction.
The specific regulation of lncRNAs on T cells.
Summary of some representative lncRNAs involved in cancer regulation.
| LncRNA | Function | Pathway | Cancer | Ref |
|---|---|---|---|---|
| LINK-A | Downregulates antigen presentation | GPCR | TNBC | [71] |
| LINC02195 | Promotes MHC I molecules | Unknown | HNSCC | [72] |
| LINC00473 | Upregulates PD-L1 | miR-195-5p | PC | [73] |
| HOTTIP | Upregulates PD-L1 | STAT3/PD-L1 | OC | [74] |
| NKX2-1-AS1 | Downregulates PD-L1 | NKX2-1/CD274 | LC | [75] |
| MALAT1 | Upregulates PD-L1 | miR-195 | DLBCL | [76] |
| AFAP1-AS1 | Upregulates PD-1 | Unknown | NPC | [77] |
| Lnc-SGK1 | Promotes Th2 and Th17 differentiation | SGK1/Jun B | GC | [78] |
| Lnc-sox5 | Downregulates CD3+CD8+ T cells | IDO1 | CRC | [79] |
| Linc-POU3F3 | Increases Treg distribution | TGF-β | GC | [80] |
| LINC00301 | Increases Treg distribution | TGF-β | NSCLC | [81] |
| Lnc-Tim3 | Promotes CD8+ T cell exhaustion | Bat3/NFAT1/AP | HCC | [82] |
| NEAT1 | Promotes the apoptosis of CD8+ T cells | miR-155/Tim-3 | HCC | [83] |
| NKILA | Sensitizes CTL, Th1 to AICD | JAK1-STAT1 | BC, LC | [60] |
| FAS-AS1 | Upregulates Fas | Unknown | BCL | [84] |
TNBC: triple-negative breast cancer; HNSCC: head and neck squamous cell carcinoma; PC: pancreatic cancer; OC: ovarian cancer; LC: lung cancer; DCBLC: diffuse large B cell lymphoma; NPC: nasopharyngeal carcinoma; GC: gastric cancer; CRC: colorectal cancer; NSCLC: non-small cell lung cancer; HCC: hepatocellular carcinoma; BC: breast cancer; BCL: B-cell lymphoma; GPCR: G-protein-coupled receptor.
4.2. Possible pathways of lncRNAs on cancer and T cell
LncRNAs are emerging as key regulators of gene expression and play a key role in the immunity related to progression of cancer. However, only a small number of T cell-associated lncRNAs as well as possible pathways have been found to play a role in cancer regulation. Yongsheng Li et al. demonstrated that not all immune-related pathways are equally associated with lncRNAs; cytokine and cytokine receptor pathways are likely to be correlated with more lncRNAs [8]. In fact, chemokines and chemokine receptors have long been reported to become new targets for cancer immunotherapy [85] and Mu Xu et al. found that SATB2-AS1 inhibits colorectal cancer (CRC) cell metastasis and regulates TH1-type chemokines expression [86]. Moreover, in the research conducted by Yongsheng Li et al., several immune-related lncRNA are associated with TCR signaling pathway, hippo pathway and cell cycle pathway in lung cancer [8]. Based on this, we hypothesized that these signaling pathways are might also associated with T cells in cancer. Identification of the targets of lncRNAs remains to be further investigated.
4.3. Potential clinic applications of T cell-associated lncRNAs
Cancer-associated lncRNAs are being favored by researchers and widely studied. Adam M. Schmitt et al. summarized the mechanisms of cancer-related lncRNAs in the diagnosis, treatment and prognosis of various types of cancer [87]. However, with the key role of immunotherapy in cancer, some researchers have not only focused on the regulation of lncRNAs on cancer cells, but also tried to find immune-related lncRNAs to explore new targets in cancer, and simultaneously make contributions to overcoming the resistance of immunotherapy. Yuwen Zhou et al. discussed the mechanisms of lncRNA participation in immunotherapy resistance from the aspects of antigen presentation, PD-L1 expression regulation, modulation of CD8+ T cells and control of Tregs and myeloid-derived suppressor cells (MDSCs) [88]. In our review, we discussed the mechanisms by which lncRNAs affect T cell activation, differentiation, effector function and apoptosis in cancer. Upregulation or downregulation of related lncRNAs are also expected to help overcome immunotherapy resistance.
5. Discussion
The activation, proliferation, differentiation, effector functions, apoptosis and metabolism of T cell have been studied a long time ago and the regulation of lncRNAs in these processes has also attracted attention over the years. Although lncRNAs do not seem to be as critical as T cells themselves in cancer immunotherapy to some extent, it is obvious that lncRNAs are vital to this regulatory network by providing a background support to enable T cells to play a more effective role in cancer immunity.
The most important characteristic of lncRNA-mediated regulation on T cells is that lncRNAs can bind to signal molecules such as immune receptors and transcriptional regulators. Given the complexity of T cell differentiation and function, it is very difficult to try to elucidate carcinogenesis with the function of a single lncRNA. In the above review of lncRNAs in T cells, we found that in every disease model, multiple lncRNAs are involved in a complex regulatory network and the same lncRNA also plays different roles in different disease models. More importantly, some newly discovered lncRNAs have no clear molecular target and no signaling pathway has been found, leaving many difficulties for lncRNAs to be used as cancer therapeutic targets or cancer biomarkers in the future. The uncertainty of molecular targets also explains the functional diversity of lncRNAs. It is important to keep in mind that lncRNAs alone may not be sufficient to affect cancer immunity; likewise, T cells may not take effect on their own or they alone may not work efficiently in cancer immunity. Since the regulation of lncRNAs on T cells is still in the early stage of research, we hope to find more lncRNAs in regulation of T cells and their specific signaling pathways in the future. Further characterization of these lncRNAs will provide new insight into how lncRNAs and signaling molecules work together to impact T cell in cancer.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81772779), “Professionals from Six-Pronged Top-Talent Program” of Jiangsu Province (No. LGY2017068), “The Six Top Talent Project” of Jiangsu Province (No. 2015-WSN-034), Medical Talent of Empowering Medicine through Science and Education Program of Jiangsu Province (No. ZDRCA2016003) and Key Laboratory for Medicine of Jiangsu Province of China (No. ZDXKB2016005).
Author Contributions
Wenxiu Chen performed the literature search and data analysis and wrote the manuscript. Fang Wang designed and supervised the study. Shuna Liu polished the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer research. 2017;77:3965-81
2. Denaro N, Merlano MC, Lo Nigro C. Long noncoding RNAs as regulators of cancer immunity. Molecular oncology. 2019;13:61-73
3. Chen Y, Satpathy A, Chang H. Gene regulation in the immune system by long noncoding RNAs. Nature immunology. 2017;18:962-72
4. Kanduri K, Tripathi S, Larjo A, Mannerstrom H, Ullah U, Lund R. et al. Identification of global regulators of T-helper cell lineage specification. Genome medicine. 2015;7:122
5. Ranzani V, Arrigoni A, Rossetti G, Panzeri I, Abrignani S, Bonnal RJ. et al. Next-Generation Sequencing Analysis of Long Noncoding RNAs in CD4+ T Cell Differentiation. Methods in molecular biology (Clifton, NJ). 2017;1514:173-85
6. Xia F, Dong F, Yang Y, Huang A, Chen S, Sun D. et al. Dynamic transcription of long non-coding RNA genes during CD4+ T cell development and activation. PloS one. 2014;9:e101588
7. Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA. et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nature immunology. 2013;14:1190-8
8. Li Y, Jiang T, Zhou W, Li J, Li X, Wang Q. et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nature communications. 2020;11:1000
9. Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799-816
10. Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science (New York, NY). 2007;316:1484-8
11. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer discovery. 2011;1:391-407
12. Vishnubalaji R, Shaath H, Elango R, Alajez NM. Noncoding RNAs as potential mediators of resistance to cancer immunotherapy. Seminars in cancer biology. 2020;65:65-79
13. Zhang X, Wang W. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int J Mol Sci. 2019;20:5573
14. Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nature reviews Genetics. 2014;15:394-408
15. Chen J, Ao L, Yang J. Long non-coding RNAs in diseases related to inflammation and immunity. Annals of translational medicine. 2019;7:494
16. Euskirchen GM, Rozowsky JS, Wei CL, Lee WH, Zhang ZD, Hartman S. et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome research. 2007;17:898-909
17. Ruiz-Orera J, Messeguer X, Subirana J, Alba M. Long non-coding RNAs as a source of new peptides. eLife. 2014;3:e03523
18. Ingolia N, Brar G, Stern-Ginossar N, Harris M, Talhouarne G, Jackson S. et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell reports. 2014;8:1365-79
19. Niu L, Lou F, Sun Y, Sun L, Cai X, Liu Z. et al. A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation. Science advances. 2020;6:eaaz2059
20. Fernandes J, Acuña S, Aoki J, Floeter-Winter L, Muxel S. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Non-coding RNA. 2019;5:17
21. Hadjicharalambous MR, Lindsay MA. Long Non-Coding RNAs and the Innate Immune Response. Non-coding RNA. 2019;5:34
22. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775-89
23. Heo JB, Lee YS, Sung S. Epigenetic regulation by long noncoding RNAs in plants. Chromosome research: an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2013;21:685-93
24. Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends in genetics: TIG. 2014;30:348-55
25. Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics, proteomics & bioinformatics. 2017;15:177-86
26. Kulkarni S, Lied A, Kulkarni V, Rucevic M, Martin M, Walker-Sperling V. et al. CCR5AS lncRNA variation differentially regulates CCR5, influencing HIV disease outcome. Nature immunology. 2019;20:824-34
27. Satpathy AT, Chang HY. Long noncoding RNA in hematopoiesis and immunity. Immunity. 2015;42:792-804
28. Zhang W, Zhou Y, Ding Y. Lnc-DC mediates the over-maturation of decidual dendritic cells and induces the increase in Th1 cells in preeclampsia. Am J Reprod Immunol. 2017;77:e12647
29. Wu J, Zhang H, Zheng Y, Jin X, Liu M, Li S. et al. The Long Noncoding RNA MALAT1 Induces Tolerogenic Dendritic Cells and Regulatory T Cells via miR155/Dendritic Cell-Specific Intercellular Adhesion Molecule-3 Grabbing Nonintegrin/IL10 Axis. Frontiers in immunology. 2018;9:1847
30. Zhang M, Zheng Y, Sun Y, Li S, Chen L, Jin X. et al. Knockdown of NEAT1 induces tolerogenic phenotype in dendritic cells by inhibiting activation of NLRP3 inflammasome. Theranostics. 2019;9:3425-42
31. Luo M, Liu X, Meng H, Xu L, Li Y, Li Z. et al. IFNA-AS1 regulates CD4(+) T cell activation in myasthenia gravis though HLA-DRB1. Clinical immunology (Orlando, Fla). 2017;183:121-31
32. Liu Q, Deng Y, Li C, Xie H, Liu Q, Ming S. et al. LncRNA GAS5 suppresses CD4(+) T cell activation by upregulating E4BP4 via inhibiting miR-92a-3p in systemic lupus erythematosus. Immunology letters. 2020;227:41-7
33. Hu B, Niu L, Jiang Z, Xu S, Hu Y. LncRNA XLOC_003810 promotes T cell activation and inhibits PD-1/PD-L1 expression in patients with myasthenia gravis-related thymoma. 2020; 92: e12886.
34. Gast M, Rauch BH, Haghikia A, Nakagawa S, Haas J, Stroux A. et al. Long noncoding RNA NEAT1 modulates immune cell functions and is suppressed in early onset myocardial infarction patients. Cardiovascular research. 2019;115:1886-906
35. Shui X, Chen S, Lin J, Kong J, Zhou C, Wu J. Knockdown of lncRNA NEAT1 inhibits Th17/CD4(+) T cell differentiation through reducing the STAT3 protein level. 2019; 234: 22477-84.
36. Zhang F, Liu G, Li D, Wei C, Hao J. DDIT4 and Associated lncDDIT4 Modulate Th17 Differentiation through the DDIT4/TSC/mTOR Pathway. Journal of immunology (Baltimore, Md: 1950). 2018;200:1618-26
37. Braga-Neto MB, Gaballa JM, Bamidele AO, Sarmento OF, Svingen P, Gonzalez M. et al. Deregulation of Long Intergenic Non-coding RNAs in CD4+ T Cells of Lamina Propria in Crohn's Disease Through Transcriptome Profiling. Journal of Crohn's & colitis. 2020;14:96-109
38. Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJ, Curti S. et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nature immunology. 2015;16:318-25
39. Huang S, Dong D, Zhang Y, Chen Z, Geng J, Zhao Y. NEAT1 regulates Th2 cell development by targeting STAT6 for degradation. Cell cycle (Georgetown, Tex). 2019;18:312-9
40. Gibbons HR, Shaginurova G, Kim LC, Chapman N, Spurlock CF 3rd, Aune TM. Divergent lncRNA GATA3-AS1 Regulates GATA3 Transcription in T-Helper 2 Cells. Frontiers in immunology. 2018;9:2512
41. Zhu X, Wang X, Wang Y, Zhao Y. Exosomal long non-coding RNA GAS5 suppresses Th1 differentiation and promotes Th2 differentiation via downregulating EZH2 and T-bet in allergic rhinitis. Molecular immunology. 2020;118:30-9
42. Xia M, Liu J, Liu S, Chen K, Lin H, Jiang M. et al. Ash1l and lnc-Smad3 coordinate Smad3 locus accessibility to modulate iTreg polarization and T cell autoimmunity. 2017; 8: 15818.
43. Brajic A, Franckaert D, Burton O, Bornschein S, Calvanese AL, Demeyer S. et al. The Long Non-coding RNA Flatr Anticipates Foxp3 Expression in Regulatory T Cells. Frontiers in immunology. 2018;9:1989
44. Zemmour D, Pratama A, Loughhead SM, Mathis D, Benoist C. Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E3472-e80
45. Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. Journal of immunology (Baltimore, Md: 1950). 2012;189:2084-8
46. Rankin CR, Shao L, Elliott J, Rowe L, Patel A, Videlock E. The IBD-associated long noncoding RNA IFNG-AS1 regulates the balance between inflammatory and anti-inflammatory cytokine production after T-cell stimulation. 2020; 318: G34-g40.
47. Wang J, Peng H, Tian J, Ma J, Tang X, Rui K. et al. Upregulation of long noncoding RNA TMEVPG1 enhances T helper type 1 cell response in patients with Sjögren syndrome. Immunologic research. 2016;64:489-96
48. Hosseini A, Teimuri S, Ehsani M, Rasa SMM, Etemadifar M, Nasr Esfahani MH. et al. LncRNAs associated with multiple sclerosis expressed in the Th1 cell lineage. Journal of cellular physiology. 2019;234:22153-62
49. Peng H, Liu Y, Tian J, Ma J, Tang X, Rui K. et al. The Long Noncoding RNA IFNG-AS1 Promotes T Helper Type 1 Cells Response in Patients with Hashimoto's Thyroiditis. Scientific reports. 2015;5:17702
50. Yin W, Song Y, Chang X. Single-cell RNA-Seq analysis identifies a noncoding interleukin 4 (IL-4) RNA that post-transcriptionally up-regulates IL-4 production in T helper cells. 2019; 294: 290-8.
51. Hewitson JP, West KA. Malat1 Suppresses Immunity to Infection through Promoting Expression of Maf and IL-10 in Th Cells. 2020; 204: 2949-60.
52. Yao Y, Guo W, Chen J, Guo P, Yu G, Liu J. et al. Long noncoding RNA Malat1 is not essential for T cell development and response to LCMV infection. 2018; 15: 1477-86.
53. Qiu YY, Wu Y, Lin MJ, Bian T, Xiao YL, Qin C. LncRNA-MEG3 functions as a competing endogenous RNA to regulate Treg/Th17 balance in patients with asthma by targeting microRNA-17/ RORγt. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;111:386-94
54. Li JQ, Hu SY, Wang ZY, Lin J, Jian S, Dong YC. et al. Long non-coding RNA MEG3 inhibits microRNA-125a-5p expression and induces immune imbalance of Treg/Th17 in immune thrombocytopenic purpura. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2016;83:905-11
55. Pei X, Wang X, Li H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. International journal of biological macromolecules. 2018;118:24-30
56. Wang J, Zhai X, Guo J, Li Y, Yang Y, Wang L. et al. Long non-coding RNA DQ786243 modulates the induction and function of CD4(+) Treg cells through Foxp3-miR-146a-NF-κB axis: Implications for alleviating oral lichen planus. International immunopharmacology. 2019;75:105761
57. Ni C, Fang QQ, Chen WZ, Jiang JX, Jiang Z, Ye J. et al. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+γδ1 Treg cells. Signal transduction and targeted therapy. 2020;5:41
58. Wang Y, Zhong H, Xie X, Chen CY, Huang D, Shen L. et al. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E3883-92
59. Wu J, Niu Q, Yuan J, Xu X, Cao L. lncRNA-CD160 decreases the immunity of CD8(+) T cells through epigenetic mechanisms in hepatitis B virus infection. Oncology letters. 2020;20:235-47
60. Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L. et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat Immunol. 2018;19:1112-25
61. Yang C, Feng T, Lin F, Gong T, Yang S, Tao Y. et al. Long noncoding RNA TANCR promotes γδ T cells activation by regulating TRAIL expression in cis. Cell & bioscience. 2020;10:15
62. Ali Hosseini Rad SM, Min Yi Tan G, Poudel A, He K, McLellan AD. Regulation of human Mcl-1 by a divergently-expressed antisense transcript. Gene. 2020;762:145016
63. Saadi W, Kermezli Y. A critical regulator of Bcl2 revealed by systematic transcript discovery of lncRNAs associated with T-cell differentiation. 2019; 9: 4707.
64. Zhang L, Xu HG, Lu C. A novel long non-coding RNA T-ALL-R-LncR1 knockdown and Par-4 cooperate to induce cellular apoptosis in T-cell acute lymphoblastic leukemia cells. Leukemia & lymphoma. 2014;55:1373-82
65. Fan FY, Deng R, Yi H, Sun HP, Zeng Y, He GC. et al. The inhibitory effect of MEG3/miR-214/AIFM2 axis on the growth of T-cell lymphoblastic lymphoma. International journal of oncology. 2017;51:316-26
66. Liu H, Hu PW, Couturier J, Lewis DE, Rice AP. HIV-1 replication in CD4(+) T cells exploits the down-regulation of antiviral NEAT1 long non-coding RNAs following T cell activation. Virology. 2018;522:193-8
67. von Meyenn L, Bertschi NL, Schlapbach C. Targeting T Cell Metabolism in Inflammatory Skin Disease. Frontiers in immunology. 2019;10:2285
68. Ho PC, Liu PS. Metabolic communication in tumors: a new layer of immunoregulation for immune evasion. Journal for immunotherapy of cancer. 2016;4:4
69. Yin Z, Bai L, Li W, Zeng T, Tian H, Cui J. Targeting T cell metabolism in the tumor microenvironment: an anti-cancer therapeutic strategy. Journal of experimental & clinical cancer research: CR. 2019;38:403
70. Fu J, Shi H, Wang B, Zhan T, Shao Y, Ye L. et al. LncRNA PVT1 links Myc to glycolytic metabolism upon CD4(+) T cell activation and Sjögren's syndrome-like autoimmune response. Journal of autoimmunity. 2020;107:102358
71. Hu Q, Ye Y, Chan LC, Li Y, Liang K, Lin A. et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nature immunology. 2019;20:835-51
72. Li H, Xiong HG, Xiao Y, Yang QC, Yang SC, Tang HC. et al. Long Non-coding RNA LINC02195 as a Regulator of MHC I Molecules and Favorable Prognostic Marker for Head and Neck Squamous Cell Carcinoma. Frontiers in oncology. 2020;10:615
73. Zhou WY, Zhang MM, Liu C, Kang Y, Wang JO, Yang XH. Long noncoding RNA LINC00473 drives the progression of pancreatic cancer via upregulating programmed death-ligand 1 by sponging microRNA-195-5p. J Cell Physiol. 2019;234:23176-89
74. Shang A, Wang W, Gu C, Chen C, Zeng B, Yang Y. et al. Long non-coding RNA HOTTIP enhances IL-6 expression to potentiate immune escape of ovarian cancer cells by upregulating the expression of PD-L1 in neutrophils. Journal of experimental & clinical cancer research: CR. 2019;38:411
75. Kathuria H, Millien G, McNally L, Gower AC, Tagne JB, Cao Y. et al. NKX2-1-AS1 negatively regulates CD274/PD-L1, cell-cell interaction genes, and limits human lung carcinoma cell migration. Scientific reports. 2018;8:14418
76. Wang QM, Lian GY, Song Y, Huang YF, Gong Y. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life sciences. 2019;231:116335
77. Tang Y, He Y, Shi L, Yang L, Wang J, Lian Y. et al. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2017;8:39001-11
78. Yao Y, Jiang Q, Jiang L, Wu J, Zhang Q, Wang J. et al. Lnc-SGK1 induced by Helicobacter pylori infection and highsalt diet promote Th2 and Th17 differentiation in human gastric cancer by SGK1/Jun B signaling. Oncotarget. 2016;7:20549-60
79. Wu K, Zhao Z, Liu K, Zhang J, Li G, Wang L. Long noncoding RNA lnc-sox5 modulates CRC tumorigenesis by unbalancing tumor microenvironment. Cell cycle (Georgetown, Tex). 2017;16:1295-301
80. Xiong G, Yang L, Chen Y, Fan Z. Linc-POU3F3 promotes cell proliferation in gastric cancer via increasing T-reg distribution. American journal of translational research. 2015;7:2262-9
81. Sun CC, Zhu W, Li SJ, Hu W, Zhang J, Zhuo Y. et al. FOXC1-mediated LINC00301 facilitates tumor progression and triggers an immune-suppressing microenvironment in non-small cell lung cancer by regulating the HIF1α pathway. Genome medicine. 2020;12:77
82. Ji J, Yin Y, Ju H, Xu X, Liu W, Fu Q. et al. Long non-coding RNA Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell death & disease. 2018;9:478
83. Yan K, Fu Y, Zhu N, Wang Z, Hong JL, Li Y. et al. Repression of lncRNA NEAT1 enhances the antitumor activity of CD8(+)T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. The international journal of biochemistry & cell biology. 2019;110:1-8
84. Sehgal L, Mathur R, Braun FK, Wise JF, Berkova Z, Neelapu S. et al. FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia. 2014;28:2376-87
85. Mollica Poeta V, Massara M, Capucetti A, Bonecchi R. Chemokines and Chemokine Receptors: New Targets for Cancer Immunotherapy. Frontiers in immunology. 2019;10:379
86. Xu M, Xu X, Pan B, Chen X, Lin K, Zeng K. et al. LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Molecular cancer. 2019;18:135
87. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer cell. 2016;29:452-63
88. Zhou Y, Zhu Y, Xie Y, Ma X. The Role of Long Non-coding RNAs in Immunotherapy Resistance. Frontiers in oncology. 2019;9:1292
Author contact
![]() Corresponding author: Fang Wang, E-mail: wangfangedu.cn, ORCID: 0000-0002-6631-2045.
Corresponding author: Fang Wang, E-mail: wangfangedu.cn, ORCID: 0000-0002-6631-2045.

 Global reach, higher impact
Global reach, higher impact