Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(15):4604-4615. doi:10.7150/jca.58640 This issue Cite
Review
Circular RNAs as novel potential biomarkers for pancreatic cancer
1. Shanghai Public Health Clinical Center, Fudan University, 2901 Caolang Road, Jinshan District, Shanghai 201508, China.
2. King's College London, London, Nutritional Science Department, 150 Stamford street, waterloo, London, SE19NH, United Kingdom.
Received 2021-1-25; Accepted 2021-5-19; Published 2021-6-1
Abstract
Pancreatic cancer (PaCa) is the fourth leading cause of cancer-related deaths in the United States, and the vast majority of these malignancies are pancreatic ductal adenocarcinomas (PDAC), but there is still a lack of early detection biomarkers for PaCa. Unlike linear RNAs, circRNAs form covalently closed continuous loops and can act as mammalian gene regulators. They may be diagnostic or predictive biomarkers for some tumors, also be novel potential therapeutic targets in different diseases. This review focuses on (1) the biogenesis of circRNAs, RNA binding proteins (RBPs) and complementary sequences of circRNAs; (2) the characteristics of circRNAs which allow them to interact with miRNAs; (3) the roles of circRNAs playing in the regulation of gene expression, cell behavior and cancer, and their potential role as novel biomarkers and therapeutic targets in pancreatic cancer.
Keywords: circRNAs, pancreatic cancer, biomarkers, diagnostics, therapy
Introduction
Pancreatic cancer (PaCa) is an aggressive malignancy characterized by strong invasion. PaCa is also difficult to cure and has a poor prognosis, seriously deteriorating the patients' quality of life [1]. Due to the lack of effective biomarkers for the early diagnosis of this malignancy, patients often receive treatment when it is too late and the survival rate of patients diagnosed with PaCa after five years is < 6% [2]. Recently, circular RNA (circRNAs) is becoming a new research hotspot in the field of RNAs. circRNAs are widely dispersed in eukaryotic cells, and enriched and stabilized in many tissues, manipulating gene expressions [3-5]. circRNAs play a key role in the development and progression of human diseases and are involved in the proliferation, apoptosis, invasion and metastasis of various cancers [6, 7] including breast cancer (BCa) [8], colorectal cancer (CRC), gastrointestinal stromal tumor (GIST) [9], prostate cancer (PCa) [10], esophageal squamous cell carcinoma (ESCC) and pancreatic ductal adenocarcinoma (PDAC) [11-14]. circRNAs are promising diagnostic or predictive biomarkers for certain diseases, and in particular early stage of PaCa. In this review, we hypothesize that circRNAs may serve as targets for the development of early biomarkers of PaCa, which is significant for the early diagnosis of PaCa.
circRNAs biogenesis and regulatory mechanisms
Biogenesis of circRNAs, RNA binding proteins (RBPs) and complementary sequences
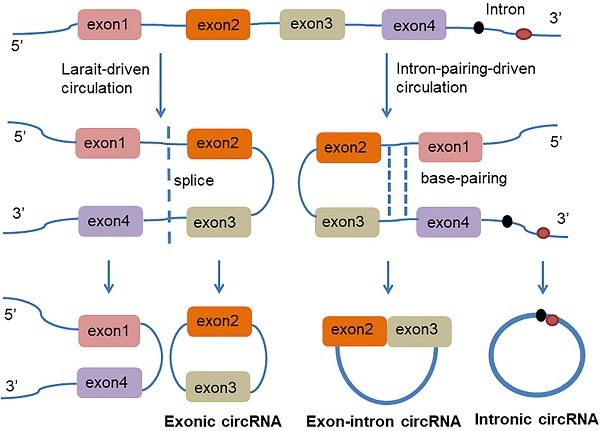
circRNA is a class of non-coding RNAs that are produced in the nucleus and are ubiquitous in eukaryotic cells. They are also characterized by a covalently closed continuous loop without 5' or 3' polarities structure. Because circRNAs are easily degraded by ribonuclease, they can be stably expressed in the cytoplasm. In some cases, the expression levels of circRNAs are tenfold greater than linear RNA and have a rich genetic diversity [15]. Most of them are transcribed from protein-coding genes by RNA polymerase II [16, 17]. Precursor messenger RNA (mRNA) containing exons and introns are produced in the nucleus, and then the pre-mRNA is transferred to the cytoplasm, which is cleaved into introns or exons. However, due to splicing diversity, circRNAs are mainly produced by rearrangement of exons. The recovery process involves RNA cyclization, which is facilitated by a covalent linkage between the downstream splice donor site (50 splice sites) and the upstream receptor splice site (30 splice sites). Therefore, circRNAs can be formed by lasso-driven circularization or exon jump model, intron-pair-driven circularization or direct back-stitching model [15], as shown in Figure 1. Intron pair-driven cyclization may be more frequent than lasso-driven cyclization and invert complement sequences [18] such as the inverted repeat Alu pair (IRAlus), which is an important sequence pair for circRNAs biogenesis [16, 19-25]. circRNA also has a high degree of conservation of reverse splicing, although circRNA is classified into intron circRNA and exon circRNA. The intron circRNA is composed of a 2'-5' chain, and the exon circRNA is composed of a 3'-5' chain without a 2'-5' chain [26].
Reverse complementary sequences or RNA binding proteins (RBPs) are also required for the formation of circRNAs [27]. circRNAs also may act as protein sponges, by binding RNA-binding proteins (RBPs) which can act as activators or inhibitors of circRNAs formation [16, 28]. Muscle blind protein (MBL) can strongly and specifically bind to the circRNAs which are generated from its own RNA. The RNA sequence between the MBL dimers forms a lariat structure, which allows the receptor and the donor to be spatially close to each other; thereby inducing RNA reverse splicing, hindering linear splicing, and stimulating circMBL production [16]. RNA pairing competition between or within a single flanking intron may significantly impact splicing selection and result in the processing of multiple circRNAs transcripts from a single gene by complementary sequences (repeats or non-repetitive sequences) which may be beneficial for exon cyclization. Exon circularization can also be mediated by complementary sequences in human introns [20].
circRNA interacts with miRNAs as miRNAs sponges
circRNAs are rich in miRNA binding sites and function as miRNA sponges, which can regulate the production of intracellular proteins. Competing endogenous RNAs (ceRNAs) regulate other RNA transcripts including mRNAs, long non-coding RNAs (lncRNAs) and pseudogenes by battling with shared microRNAs (miRNAs) [29]. circRNAs may have one or more miRNA response elements (MREs) [30, 31] which can bind to the corresponding miRNAs and induce the expression recovery of genes downstream of miRNAs, so that circRNA has the ability to bind to miRNAs, influencing miRNAs function and targets [31-33]. Thus, the presence or absence of ceRNA affects the ability of miRNAs to regulate gene expression, suggesting circRNAs could act as miRNAs sponges or potent ceRNA molecules, and could deplete polymorphisms at miRNAs binding sites [13, 31, 33-35].
The generation of Exon-intron circRNA and intronic circRNA. The mechanism by which circRNAs are formed is classified as model 1 "lasso-driven circularization" or "exon jump" (The generation of extronic circRNA); model 2 "intron-pair-driven circularization" or "direct back-stitching".
To date, circRNAs sponges are characterized by high expression levels and a large number of miRNAs binding sites. ciRS-7/CDR1as (a circular RNA sponge for miR-7 or CDR1 antisense) and SRY (the sex determining region Y) can be used as miRNAs sponges [12, 19, 33]. The brain degeneration-associated protein 1 (CDR1) gene can be translated into a natural circular antisense transcript, known as the antisense of the cerebellar degeneration-associated protein 1 transcript (CDR1as) [36]. SRY is carried out by long inverted repeats (IRs) of more than 15.5 kb in length. When one or two IRs are deleted, no cyclicization occurs [19]. The SRY gene can mediate the sex of a mammal and is highly expressed in the mouse testis and produces a circRNA containing 16 miR-138 binding sites, which acts as a miR-138 sponge [33, 37]. CDR1 is highly expressed in the brain and contains at least 60 miR-7 binding sites, overwhelming any known linear sponge [31, 33]. miR-7 has a wide range of functions and is involved in a variety of signaling pathways. ciRS-7 is also highly associated with Argonaute (AGO) protein in a miR-7-dependent manner and many tumors can be regulated by the ciRS-7-miR-7 axis [38]. Although circRNAs are fully resistant to miRNAs-mediated targeted destabilization, they strongly inhibit miR-7 activity, leading to elevated levels of miR-7 targets [33]. Thus, miR-7 can bind tightly to ciRS-7, and regulation of the miR-7/miR-671/ciRS-7 axis may play an important role in cancer-associated biological processes.
More importantly, the role of miR-7 and SRY in the development and progression of cancer has been demonstrated, suggesting that circRNAs may mediate physiological and pathological processes through binding to miRNAs [39]. Hence, circRNA sponges may have more research value than linear sponges, which not only can be involved in gene regulation but also play an important role in cancer. Our functional network analysis also showed that circ_0057558 and circ_0034467 regulated miR-6884, and circ_0062019 and circ_0060325 regulated miR-5008 as the key regulators of PCa [10].
circRNAs may regulate host gene expression
There are three mechanisms by which circRNA regulates parental gene expression. Firstly, circular intronic RNAs (ciRNAs) are produced by introns and bind to RNA polymerase II (Pol II) and promote transcription [40]. Secondly, exon-intron circRNAs (EIciRNA) bind to U1 snRNP (U1 small nuclear ribonucleoproteins) forming a EIciRNA-U1 snRNP complex that may interact with the RNA Pol II transcription complex to promote host gene expression [41]. Finally, circRNA acts as a miRNA sponge that increases transcript translation of its parent gene [13]. The formation of circRNAs depends on the critical flanking RNA elements that might be required for introns [42]. These circRNAs have little enrichment for miRNA target sites, indicating that they are functionally different [42]. Detailed studies have shown that some ciRNAs are prevalent in the nucleus and interact with Pol II mechanism and regulate the host transcriptional activity in a cis manner [26]. For example, EIciRNAs such as circEIF3J and circPAIP2 involved in RNA Pol II are mainly situated in the nucleus, interact with the U1-snRNPs and enhance their transcriptional parental genes in a cis-acting manner [43]. In short, circRNAs enhance the ability of transcription and translation by regulating parent genes.
circRNAs bind to proteins and regulate cell behavior
circRNA can also encode a variety of proteins with different functions. Synthetic circRNAs contain an inner chromosomal entry site (IRES) that can be efficiently translated into proteins [44]. circAmotl1 can bind to PDK1 and AKT1 in cardiomyocytes, resulting in AKT1 phosphorylation and transportation into the nucleus to protect the myocardium from damage [16]. It can also bind to STAT3 (signal transduction and transcriptional activator 3) and c-Myc which then transfer to the nucleus to promote cell proliferation, invasion and tumorigenesis [45], so that circAmotl1 may be used as a target for therapy for cancer.
Deregulated circRNAs are associated with cancer
A large number of circRNAs have been identified to play key regulating roles in major tumor diseases, which have enlarged the regulatory networks of ceRNAs and provided a new direction for our in-depth study of the pathogenesis and human malignancies which are involved in the regulation of tumor cell production and growth in a variety of cancers. For example, in hypoxia-induced human umbilical vein endothelial cells (HUVEC) circ_0010729 was co-expressed with hypoxia-inducible factor 1α (HIF-1α) and negatively correlated with miR-186 [46], revealing that the key regulation of circ_0010729 on vascular endothelial cell proliferation and apoptosis was determined by targeting the miR-186/HIF-1α axis. Esophageal cancer is the 8th most common cancer, one of which is ESCC. circITCH expression is usually down-regulated in ESCC, and circITCH might impose anti-tumor function in ESCC [13]. Several studies have shown that miR-7 is lowly expressed in many types of tumors and negatively correlated with tumor growth and invasion [47-49]. circITCH acts by interaction with miRNAs such as miR-7, with increased levels of miR-17 and miR-214 and ITCH, which promotes protein-mediated degradation of Dvl2 and reduces expression of the oncogene c-Myc. Thus, this process inhibits classical Wnt signaling and can prevent esophageal tumorigenesis [13]. Based on the latest research, it was also found that the expression of circ_0067934 in ESCC was significantly higher than that in normal tissues. circ_0067934 is located in the chromosomal region 3q26.2, which consists of two exons joined by reverse splicing [50]. Notably, circ_0067934 expression was also found to be involved in ESCC differentiation, T stage and TNM stage, as well as tumors with a lower degree of differentiation, higher expression levels of circ_0067934. In addition, circ_0067934 can promote ESCC cell proliferation and migration in vitro, and si-circ_0067934 can block the cell cycle of G2 phase.
We have analyzed the profiles of differentially expressed circRNAs in BCa [8], PCa [10] and GIST [9] by competing in endogenous RNAs microarray. A total of 4,370, 1,021, and 5,770 differentially expressed circRNAs were found in BCa, PCa and GISTs tumor tissue. Eight circRNAs, such as circ_0069094, circ_0062558, circ_0074026, circ_0079876, circ_0017536, circ_0023302, circ_0017650, and circ_0017545 were verified differentially expressed in BCa tissue and associated with TNM stage, lymph node infiltration, and Ki67 [8]. The expression levels of circ_0057558 and circ_0062019 in PCa tumor tissues were validated by qRT-PCR analysis [10]. circ_0069765, circ_0084097, and circ_0079471 were also verified differentially expressed in 68 pairs of GISTs, comparing with the adjacent normal gastrointestinal tissues by qRT-PCR [9]. We further demonstrated that the specific KIT-related regulation networks involved these three circRNAs, their host genes and miR-142-5p, miR-144-3p and miR-485-3p, which may be key regulators of GISTs [9].
circRNAs are also associated with hepatocellular carcinoma (HCC). circMTO1 acts as a sponge of miR-9, inhibits the progression of liver cancer, reduces the expression of circMTO1, and implies poor survival prognosis [3]. The expression of circ_0001649 in HCC was lower than that in adjacent tissues, and its expression was related to tumor size and tumor embolus. It is worth noting that the larger the tumor size, the lower the expression of circ_0001649. Therefore, circ_0001649 might be involved in tumor growth and metastasis [5]. High-throughput circRNA microarrays showed circ_0000520, circ_0005075 and circ_0066444 were significantly and abnormally expressed in HCC tissues [51]. At the same time, ciRS-7 expression was significantly associated with liver MVI in serum AFP and HCC patients. Furthermore, ciRS-7 expression together with MVI expression was negatively correlated with miR-7 and synergistically associated with PIK3CD and p70S6K, which are targets of miR-7. Therefore, it means that ciRS-7 has become a new biomarker of liver MVI [52]. circ_0007534 can bind to miR-761 and increase ZIC5 expression to promote the progression of glioma cells [53]. In colorectal cancer, small interfering RNA (siRNA) knockdown of circ_0007534 significantly attenuated SW-620 and LoVo cell proliferation and promoted apoptosis. Tumor stage and lymph node metastasis are two diagnostic factors closely related to the expression of circ_0007534 [54]. Coincidentally, circ_0014130 (circPIP5K1A) is also involved in tumorigenesis in non-small cell lung cancer (NSCLC) [55]. circ_0014130 can act as a miR-600 sponge to promote NSCLC proliferation and metastasis by promoting HIF-1α, suggesting circ_0014130 could be a novel candidate therapeutic target for NSCLC through the circ_0014130/miR-600/HIF-1 alpha axis. Through the circRNA-miRNA-mRNA axis, circRNA can upregulate or downregulate gene expression and affect tumor progression. Emerging evidence showed that circ_103809 was involved in the development of colorectal, lung and liver cancer through the circRNA-miRNA axis by participating in the proliferation and migration of cancer cells, and could thus provide new ideas for clinical treatment of cancer [56-58].
circRNAs play key roles in pancreatic cancer
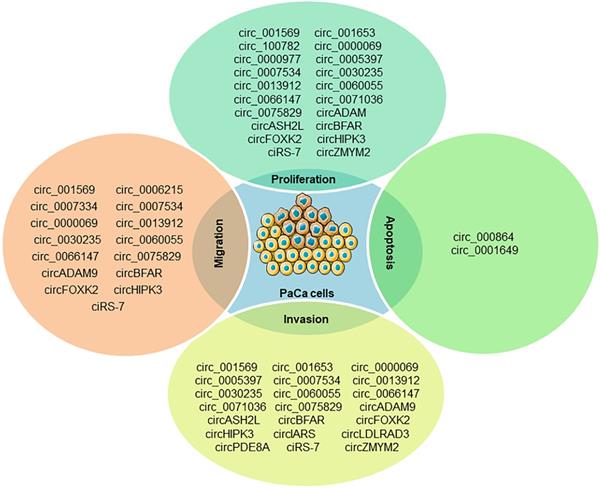
PaCa is one of the most fatal malignancies in the digestive system. Understanding the molecular mechanisms underlying the initiation and progression of pancreatic cancer may promote the development of diagnostic and therapeutic strategies. miRNAs and circRNAs have been identified as important regulators of human cancer development [59, 60]. A comprehensive literature search was executed in PubMed using the medical subject headings (MeSH) terms “pancreatic cancer” and “circRNA”, there are a total of 129 relevant studies for PaCa involved in circRNAs and only five review articles for pancreatic cancer found in our initial study, which revealed that circRNAs could serve as diagnostic, therapeutic or prognostic biomarkers for PaCa [61-65]. Several oncogenic and antioncogenic circRNAs (circ_000864, circ_001587 and circNFIB1) have been discovered to regulate the proliferation, migration, invasion and angiogenesis of PaCa cells (Figure 2). Most of them could regulate PaCa-related signaling pathways including through miRNA sponges as shown in Figure 3.
The known roles of circRNAs in PaCa progression.
circRNAs involved in miRNA-associated gene regulatory pathway in PaCa.
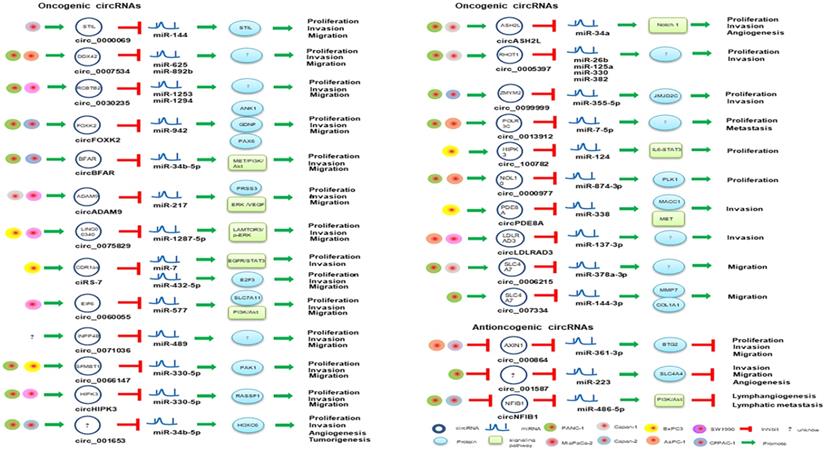
Oncogenic circRNAs in PaCa
circRHOT1 (circ_0005397) is produced by the sequence located at chr17: 30500849-30503232, and the splicing sequence is 233 nt in length. circRHOT1 might bind to miR-26b, miR-125a, miR-330 and miR-382 and regulate a variety of tumor-associated pathways [66]. circRHOT1 was upregulated in PaCa tissues and cell lines, and silencing circRHOT1 could inhibit pancreatic cell proliferation, invasion and migration [66], which demonstrated that circRHOT1 might play an important role in pancreatic cancer through spongiform tumor-associated miRNAs as a novel potential therapeutic target for PaCa. The expression of circLDLRAD3 was significantly upregulated in PaCa tissue and PaCa plasma samples. circLDLRAD3 expression was associated with lymphatic invasion, venous invasion and metastasis [67]. Functional studies of circLDLRAD3 may also improve our understanding of the pathogenesis of PaCa [67]. Therefore, circLDLRAD3 has the potential to be a novel indicative biomarker for tumor invasion in the diagnosis of PaCa.
IL6-JAK2-STAT3 signaling pathway is activated in pancreatic cell lines and PDAC tissue samples [68]. Activation of STAT3 is closely related to tumorigenesis, and STAT3 can cause tumor cell proliferation and migration [69]. circ_100782 plays a vital role in PDAC. Chen et al. revealed that circ_100782 was significantly upregulated in pancreatic cancer. Silencing circ_100782 through the sponge effect of circRNAs regulates the STAT3 pathway by targeting miR-124, which subsequently inhibits the proliferation of pancreatic cancer cell BxPC-3. Thus, circ_100782 plays an important role in the pathogenesis of PaCa.
circCDR1as not only promotes the migration, invasion and proliferation of PaCa cells via regulating E2F3 expression by sponging miR-432-5p [70], but mediates EGFR/STAT3 signaling pathway as a sponge of miR-7 [71]. circ_0000977 was abnormally upregulated in pancreatic cancer, and silencing circ_0000977 inhibited pancreatic cancer cell proliferation and induced cell cycle arrest by stimulating miR-874-3p and inhibiting PLK1 expression [72]. circ_0007534 participates in the development of colorectal cancer and glioma, and the formation of PDAC [73]. circ_0007534 was upregulated in PDAC tissues and PANC-1, SW1990 and BxPC-3 cells, accelerated cell proliferation, migration and invasion, inhibited cell apoptosis, and was positive correlation with poor PDAC phenotype [73]. Previously, miR-625 and miR-892b were identified as tumor suppressor molecules in cancer. Hao et al. confirmed that circ_0007534 can directly interact with miR-625 and miR-892b [73], which implied that circ_0007534/miR-625/miR-892b regulation axis might contribute to the development of PDAC. circ_0013912, circ_0000069, circ_0071036, circ_0075829, circ_0099999 could also facilitate the proliferative, migratory and invasive rates of PaCa cells through sponging miR-7-5p, miR-144, miR-489, miR-1287-5p and miR-335-5p, respectively [74-78]. circSFMBT1 serves as a miR-330-5p sponge and also promotes PaCa growth and metastasis via regulating miR-330-5p/ PAK1 axis [79]. Silencing of circ_001653 in PDAC cells can inhibit cell proliferation, cell-cycle progression, angiogenesis, and invasive properties by binding to miR-377 [80]. Besides, circBFAR, circASH2L, circFOXK2, circADAM9, circEIF6 and circRNA chr7:154954255-154998784+ were significantly up-regulated in PDAC by sponging miR-34b-5p, miR-34a, miR-942, miR-217, miR-557 and miR-4459, respectively [81-86], which play key roles in tumor invasion and might therefore be useful diagnosis biomarkers of PDAC.
Antioncogenic circRNAs in PaCa
Several antioncogenic circRNAs such as circ_000864, circ_001587, circ_0001649 and circNFIB1 were repressed in PaCa tumor or cancer cells [87-90]. circ_000864 was downregulated in AsPC-1 and MiaPaCa-2 cells, and it could upregulate BTG2 expression and inhibited the proliferation, invasion and migration of PaCa cells as miR-361-3p sponges [87]. circ_001587 mediated miR-223/SLC4A4 axis and also could inhibit PaCa cell migration, invasion and angiogenesis [88]. Besides, circNFIB1 was suppressed in PANC-1 and Capan-2 cells and regulated PI3K/Akt signaling pathway by attenuating the oncogenic effect of miR-486-5p, which was negatively associated with lymph node metastasis in PaCa patients [89]. circ_0001649 was also downregulated in PaCa tissues and cell lines, and its low expression has been associated with advanced tumor stage and poor tissue grade. Exogenous administration of circ_0001649 can inhibit the proliferative ability of PaCa cell lines and induce apoptosis, suggesting it could be used as an exogenous anticancer agent [90].
circRNAs were associated with gemcitabine (GEM) resistance in PaCa
CircHIPK3 as a sponge for miR-330-5p which directly bounds to the 3' UTR of RASSF1 could promote cell proliferation, invasive, migration and EMT, which was associated with GEM resistance in PaCa cells [91]. Ding et al. showed the expression profiles of circRNAs in gemcitabine-resistant PaCa cell lines were clearly different to normal PaCa cell lines. They also observed that the differentially expressed circRNAs played a role in chemoresistance in PaCa by acting as a miRNA sponge that affected MRPK and mTOR signaling pathways [92]. Huang et al. analyzed circRNA expression profiles in PANC-1-GR (gemcitabine resistant cell line) and PANC-1, and found two circRNAs (chr14:101402109-101464448+, chr4:52729603-52780244+) to be significantly different. They also found that silencing the expression of the genes restored the sensitivity of PaCa resistant cell lines to gemcitabine while their overexpression weakened this sensitivity [93].
Clinical significance of circRNAs for PaCa
| circRNA ID | Host gene | Patients (n) | Expression (High/Low) | Differentiation (Well/Poorly) | Clinicopathological association | Biomarker type | Ref. |
|---|---|---|---|---|---|---|---|
| circ_0001649 | SHPRH | 58 | 25 /33 | 31 /27 | T stage | Prognostic biomarker | [90] |
| circ_001569 | IRF4 | 26 | 13 /13 | 19 /7 | T stage | Diagnostic and prognostic marker | [107] |
| circ_0007534 | DDX42 | 60 | 30 /30 | 34 /26 | T stage, lymph node invasion | Prognostic biomarker | [73] |
| circ_0030235 | RCBTB2 | 62 | 32 /30 | 34 /28 | T stage, lymph node invasion | Prognostic biomarker | [106] |
| circASH2L | ASH2L | 90 | 45 /45 | 9 /66 | Lymphatic invasion, TNM stage | Diagnostic marker | [82] |
| circIARS | IARS | 85 | 42 /43 | 65 /20 | Liver metastasis, vascular invasion, TNM stage | Diagnostic and prognostic marker | [104] |
| circLDLRAD3 | LDLRAD3 | 30 | 18 /12 | - | Venous invasion, lymph node invasion | Diagnostic marker | [67] |
| circPDE8A | PDE8A | 93 | 46 /47 | 72 /21 | Lymphatic invasion, TNM stage | Diagnostic and prognostic marker | [105] |
| circZMYM2 | ZMYM2 | 106 | 73 /33 | 88 /18 | - | - | [74] |
| ciRS-7 | CDR1as | 41 | - | - | Lymph node metastasis, venous invasion | - | [71] |
circRNAs were involved in protein translation
circRNAs not only play a role in gene transcription, but they can be involved in protein translation. Studies have shown that N6-methyladenosine (m6A) is the most ubiquitous base modification of RNA, and a single m6A site is sufficient to drive translation initiation, which can promote the efficient initiation of protein translation of circRNA in human cells. The methylation of circRNAs synergizes with IRES to improve the efficiency of circRNAs in translating proteins. The p16 gene is involved in cell cycle regulation and tumor suppressor processes, and its inactivation mechanisms include deletions, mutations, and aberrant methylation of 5'-CpG islands. Abnormal methylation of 5'-CpG islands has been found in a variety of tumors as the primary mechanism for their inactivation [94]. Translation of circRNAs may be more prevalent in cancer cells because circRNAs contain large amounts of m6A modifications sufficient to drive protein translation in a cap-independent manner to promote cancer cell development, apoptosis and cell cycle regulation [95]. circZNF609 contains an open reading frame and is translated into protein in a cap-free manner, which can specifically control the proliferation of myoblast [96]. Moreover, circ-ZNF609 acts as a competitive endogenous RNA, regulating AKT3 expression by a sponge miR-150-5p in Hirschsprung disease [97]. circZNF609 can also induce the expression of cancer-related proteins, regulate cell proliferation, and participate in the tumor suppressor process in PaCa.
circRNAs and exosomal circRNAs as diagnostic and prognostic biomarkers for pancreatic cancer
Wu et al. found that three circRNAs (circ_004183, circ_079265 and circ_105039) were downregulated in plasma from children with CHD (congenital heart diseases), suggesting they may be crucial in the development of CHD and serve as novel non-invasive biomarkers for the diagnosis of CHD in children [98]. circRNAs are frequently upregulated in gastric cancer (GC) tissues and promote cell growth through members of the spongy miR-125 family, and as such have been described as potential disease biomarkers in human saliva [99]. circ_002059 was downregulated in the GC and could represent a potential novel biomarker for GC diagnosis [100-102]. Combination of circ_0057558, circ_0062019 and PSA level showed significantly increased AUC, sensitivity and specificity of PCa than PSA alone [10]. We also found that circ_0069094, circ_0079876, circ_0017650, and circ_0017526 were upregulated in the plasma of those patients with BCa, in contrast to normal controls [8], which suggested that plasma circRNAs might be potential biomarkers for cancer. In addition, circ_0007334 promoted the expression levels of matrix metallopeptidase 7 (MMP7) and collagen type I alpha1 chain (COL1A1) by blocking the functions of miR-144-3p and miR-577 in PDAC [103] and may be used as a potential biomarker of diagnosis and therapy for PDAC. Furthermore, the therapeutic value of circRNAs as biomarkers has been explored in many studies. circIARS, ciRS-7 and circLDLRAD3 were involved in vascular or lymph node invasion [67, 71, 104]. circIARS, circPDE8A, circ_0001649, circ_0007534, circ_001569 and circ_0030235 could be as a classifier with TNM stages to evaluate the risk of recurrence [73, 90, 104-107]. Therefore, these circRNAs may be correlated with cancer progression and could be a novel diagnostic and prognostic markers for PaCa (Table 1).
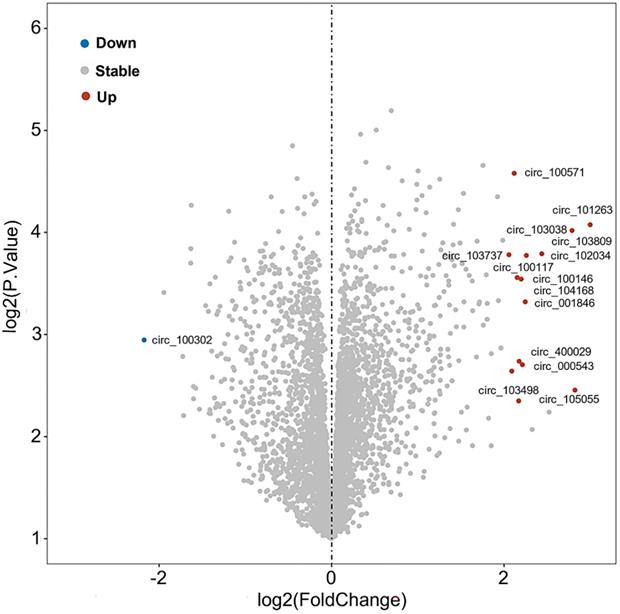
Further, the expression profile of circRNAs in PDAC has been analyzed in adjacent tissues of 6 pairs of PDAC patients using microarray technology [108]. Comparing normal tissues with pancreatic tumor tissues, it was shown that sixteen circRNAs were significantly deregulated, specifically fifteen circRNAs were upregulated and one circRNA (circ_100302) was repressed, which were recorded in Gene Expression Omnibus (GEO; No. GSE69362) [108] (Figure 4, Table 2). Zhao et al also analyzed the circRNA expression profiles of pancreatic cancer from GSE79634 and GSE69362 datasets and constructed a ceRNA network which was involved in the NF-kappa B, PI3K-Akt, and Wnt signaling pathways [109]. Thus, we speculated that these differentially expressed circRNAs were critical to PaCa progression and could be developed as a novel biomarker, therapeutic targets for PaCa need further exploration.
Deregulated circRNAs in pancreatic tumor tissue
| circRNAs ID | Alias in circBase | Host gene | Length | Strand | Position | Expression | Ref. |
|---|---|---|---|---|---|---|---|
| circ_100571 | circ_0018004 | PDSS1 | 195 | + | chr10:27024168-27024508 | Upregulated in PaCa tumor tissue | [108] |
| circ_101263 | circ_0030235 | RCBTB2 | 318 | - | chr13:49075877-49077050 | Upregulated in PaCa tumor tissue | [108] |
| circ_103038 | circ_0060055 | EIF6 | 906 | - | chr20:33866724-33872064 | Upregulated in PaCa tumor tissue | [108] |
| circ_103809 | circ_0072088 | ZFR | 693 | - | chr5:32379220-32388780 | Upregulated in PaCa tumor tissue | [108] |
| circ_102034 | circ_0005397 | RHOT1 | 233 | + | chr17:30500849-30503232 | Upregulated in PaCa tumor tissue | [108] |
| circ_103737 | circ_0070934 | LARP1B | 745 | + | chr4:128995614-129012667 | Upregulated in PaCa tumor tissue | [108] |
| circ_100117 | circ_0007895 | EYA3 | 429 | - | chr1:28362054-28384605 | Upregulated in PaCa tumor tissue | [108] |
| circ_100146 | circ_0011385 | EIF3I | 278 | + | chr1:32691771-32692131 | Upregulated in PaCa tumor tissue | [108] |
| circ_104168 | circ_0008514 | RTN4IP1 | 463 | - | chr6:107031202-107050797 | Upregulated in PaCa tumor tissue | [108] |
| circ_001846 | circ_0000520 | RPPH1 | 123 | - | chr14:20811436-20811559 | Upregulated in PaCa tumor tissue | [108] |
| circ_400029 | circ_0092337 | RPL13 | 360 | + | chr16:89628179-89628539 | Upregulated in PaCa tumor tissue | [108] |
| circ_000543 | circ_0000326 | TCONS_l2_00004572 | 96 | + | chr11:65272490-65272586 | Upregulated in PaCa tumor tissue | [108] |
| circ_400068 | circ_0092314 | RANBP1 | 340 | + | chr22:20113099-20113439 | Upregulated in PaCa tumor tissue | [108] |
| circ_105055 | circ_0001946 | CDR1 | 1485 | + | chrX:139865339-139866824 | Upregulated in PaCa tumor tissue | [108] |
| circ_103468 | circ_0067260 | COPG | 169 | + | chr3:128973510-128973920 | Upregulated in PaCa tumor tissue | [108] |
| circ_100302 | circ_0013587 | LRIG2 | 291 | + | chr1:113661854-113662145 | Downregulated in PaCa tumor tissue | [108] |
Volcano analysis of differentially expressed circRNAs in pancreatic tumor from GEO (No. GSE69362).
Exosomes are critical mediators of intercellular communication that can regulate a diverse range of biological processes between cells [110]. Exosomal circRNAs can originate from tumor cells, or other cells, such as activated human platelets and adipose cells, and enriched in the circulation and urine [111], can transfer biological information to specific cells, and might stimulate cancer [112]. Exosomal circRNAs have also been found in platelet-derived extracellular vesicles [113], PaCa [105], and cholangiocarcinoma [114]. Recent RNA sequencing data indicated that many circRNAs are stable and enriched in exosomes and could be a promising biomarker for cancer diagnosis. Exosomal circPDE8A can act as a ceRNA for miR-338 to regulate MACC1 and promote invasive growth via the miR-338/MACC1/MET pathway in PaCa [105] (Figure 3). Also, exosomal circPDE8A could be detected in blood circulation and correlated with progression and prognosis in PaCa patients. Thus, exosomal circRNAs would be potential therapeutic targets. Exosomal circRNAs are expected to become remarkable biomarkers and therapy tools for PaCa.
Summary and prospect for circular RNAs
In recent years, as the number of circRNAs has increased, the functions of circRNAs have extended enormously. As a large class of RNA with an extensive ability to regulate genes [31], circRNAs are more stable and highly conserved than linear mRNAs. As a sponge for miRNAs, circRNAs can bind miRNAs to regulate transcription or influence parental gene expression, and mediate the entire process of physiology and pathology. circRNAs can also regulate the production of intracellular proteins, regulate cell behavior and participate in defense mechanisms in vivo. In terms of tumor formation, circRNAs can further regulate tumorigenesis, participate in tumor formation, metastasis, invasion, and cancer-related pathways through the circRNA-miRNA axis. The roles of circRNAs in tumor immunity, extracellular transfer, involvement in protein regulation and their transformation into functional proteins can also affect tumor progression. Considered as novel biomarkers in cancer development and progression, circRNAs have been used in tumor targeted therapies.
Since the pathological and biological characteristics of PDAC lead to the lack of specificity of early symptoms [115], the heterogeneity of PaCa makes it difficult to cure. Although the value of circRNAs in PDAC has been gradually identified, there are still some challenges in early diagnosis of PDAC. Based on the current research of circRNAs, it is believed that circRNAs can be used as novel biomarkers for the diagnosis of pancreatic cancer, solving the problem of early diagnosis of pancreatic cancer, and providing a new therapeutic target for the treatment of this malignancy.
Abbreviations
AGO: Argonaute; BC: breast cancer; CDR1as: antisense of the cerebellar degeneration-associated protein 1 transcript; CHD: congenital heart diseases; ceRNAs: Competing endogenous RNAs; ciRNAs: circular intronic RNAs; circRNAs: circular RNAs; CRC: colorectal cancer; COL1A1: collagen type I alpha1 chain; ESCC: esophageal squamous cell carcinoma; EIciRNA: exon-intron circRNAs; GC: gastric cancer; GEO: Gene Expression Omnibus; GIST: gastrointestinal stromal tumor; HCC: hepatocellular carcinoma; HIF-1α: hypoxia-inducible factor 1α; HUVEC: human umbilical vein endothelial cells; lncRNAs: long non-coding RNAs; IRAlus: inverted repeat Alu pair; IRES: inner chromosomal entry site; IRs: inverted repeats; m6A: N6-methyladenosine; MBL: Muscleblind protein; microRNAs: miRNAs; MREs: miRNA response elements; MMP7: matrix metallopeptidase 7; mRNA: messenger RNA; PaCa: Pancreatic cancer; NSCLC: non-small cell lung cancer; PDAC: pancreatic ductal adenocarcinomas; PCa: prostate cancer; RBPs: RNA binding proteins; Pol II: RNA polymerase II; siRNA: small interfering RNA; SRY: sex determining region Y; STAT3: signal transduction and transcriptional activator 3; U1 snRNP: U1 small nuclear ribonucleoproteins.
Acknowledgements
Funding
This research was supported by a grant from Science and Technology Commission of Shanghai (20Y11900700), a grant from Special Research Fund of Youan Medical Alliance for the Liver and Infectious Diseases (LM202020), a grant (2018ZX10302103-003) from the National Special Research Program of China for Important Infectious Diseases and a grant from the National Natural Science Foundation of China (81672383).
Author Contributions
Conceived and designed the study: J. W. and S. L. Collected the literature: S. L. and Y. M. Wrote the manuscript: S. L. Revised the manuscript: J.W., Q.L. and C.C. provided funding support: J. W. All the authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sun H, Ma H, Hong G. et al. Survival improvement in patients with pancreatic cancer by decade: a period analysis of the SEER database, 1981-2010. Sci Rep. 2014;4:6747
2. Wang J, Paris PL, Chen J. et al. Next generation sequencing of pancreatic cyst fluid microRNAs from low grade-benign and high grade-invasive lesions. Cancer Lett. 2015;356:404-9
3. Han D, Li J, Wang H. et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151-64
4. Li Y, Zheng Q, Bao C. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981-4
5. Qin M, Liu G, Huo X. et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161-9
6. Zhang HD, Jiang LH, Sun DW. et al. MiR-139-5p: promising biomarker for cancer. Tumor Biol. 2015;36:1355-65
7. Liu M, Huang F, Zhang D. et al. Heterochromatin Protein HP1γ Promotes Colorectal Cancer Progression and Is Regulated by miR-30a. Cancer Res. 2015;75:4593-604
8. Li Z, Chen Z, Hu G. et al. Profiling and integrated analysis of differentially expressed circRNAs as novel biomarkers for breast cancer. J Cell Physiol. 2020;235:7945-59
9. Jia N, Tong H, Zhang Y. et al. CeRNA Expression Profiling Identifies KIT-Related circRNA-miRNA-mRNA Networks in Gastrointestinal Stromal Tumour. Front Genet. 2019;10:825
10. Xia Q, Ding T, Zhang G. et al. Circular RNA Expression Profiling Identifies Prostate Cancer- Specific circRNAs in Prostate Cancer. Cell Physiol Biochem. 2018;50:1903-15
11. Burd CE, Jeck WR, Yan L. et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. Plos Genet. 2010;6:e1001233
12. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609-12
13. Fang L, Liyuan Z, Wei L. et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001-13
14. Chen YY, Jiang MJ, Tian L. Analysis of exosomal circRNAs upon irradiation in pancreatic cancer cell repopulation. BMC Med Genomics. 2020;13(1):107
15. Jeck WR, Sorrentino JA, Wang K. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-57
16. Ashwalfluss R, Meyer M, Pamudurti NR. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55-66
17. Li P, Qing YX, Cheng LG. The emerging landscape of circular RNA ciRS-7 in cancer. Oncol Rep. 2015;33:2669-74
18. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453-61
19. Dubin RA, Kazmi MA, Ostrer H. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene. 1995;167:245-8
20. Zhang XO, Wang HB, Zhang Y. et al. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-47
21. Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233-47
22. Andranik I, Sebastian M, Emanuel W. et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170-7
23. Starke S, Jost I, Rossbach O. et al. Exon Circularization Requires Canonical Splice Signals. Cell Rep. 2015;10:103-11
24. Vicens Q, Westhof E. Biogenesis of Circular RNAs. Cell. 2014;159:13-4
25. Petkovic S, Müller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454-65
26. Zhang Y, Zhang XO, Chen T. et al. Circular Intronic Long Noncoding RNAs. Mol Cell. 2013;51:792-806
27. Shibin Q, Xisheng Y, Xiaolei L. et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141-8
28. Hentze MW, Thomas P. Circular RNAs: splicing's enigma variations. Embo J. 2014;32:923-5
29. Xuefei S, Ming S, Hongbing L. et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159-66
30. Julia S, Chen RE, Olsen MN. et al. Cell-type specific features of circular RNA expression. Plos Genet. 2013;9:e1003777
31. Memczak S, Jens M, Elefsinioti A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
32. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J of Biotechnol. 2016;238:42-51
33. Hansen TB, Jensen TI, Clausen BH. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
34. Riccardo T, Cristian L, Pier Paolo P. From pseudo-ceRNAs to circ-ceRNAs: a tale of cross-talk and competition. Nat Struct Mol Biol. 2013;20:541-3
35. Hitoshi S, Toshifumi T. A View of Pre-mRNA Splicing from RNase R Resistant RNAs. Int J Mol Sci. 2014;15:9331-42
36. Hansen TB, Wiklund ED, Bramsen JB. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. Embo J. 2014;30:4414-22
37. Capel B, Swain A, Nicolis S. et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019-30
38. Salzman J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016;32:309-16
39. Zhang HD, Jiang LH, Sun DW. et al. CircRNA: a novel type of biomarker for cancer. Breast cancer. 2018;25:1-7
40. Zhang Y, Zhang X-O, Chen T. et al. Circular intronic long noncoding RNAs. Mol cell. 2013;51:792-806
41. Li Z, Huang C, Bao C. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-64
42. Zhang Y, Yang L, Chen LL. Life without A tail: New formats of long noncoding RNAs. Int J Biochem Cell Biol. 2014;54:338-49
43. Li Z, Huang C, Bao C. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-64
44. Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415-7
45. Yang Q, Du WW, Wu N. et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609-20
46. Dang RY, Liu FL, Li Y. Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1α axis. Biochem Biophys Res Commun. 2017;490:104-10
47. Hua K, Jin J, Zhang H. et al. MicroRNA-7 inhibits proliferation, migration and invasion of thyroid papillary cancer cells via targeting CKS2. Int J Oncol. 2016;49:1531-40
48. Yue K, Wang X, Wu Y. et al. microRNA-7 regulates cell growth, migration and invasion via direct targeting of PAK1 in thyroid cancer. Mol Med Rep. 2016;14:2127-34
49. Cui YX, Bradbury R, Flamini V. et al. MicroRNA-7 suppresses the homing and migration potential of human endothelial cells to highly metastatic human breast cancer cells. Br J Cancer. 2017;117:89-101
50. Xia W, Qiu M, Rui C. et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576
51. Shang X, Li G, Liu H. et al. Comprehensive Circular RNA Profiling Reveals That hsa_circ_0005075, a New Circular RNA Biomarker, Is Involved in Hepatocellular Crcinoma Development. Medicine. 2016;95:e3811
52. Xu L, Zhang M, Zheng X. et al. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17-27
53. Li GF, Li L, Yao ZQ. et al. Hsa_circ_0007534/miR-761/ZIC5 regulatory loop modulates the proliferation and migration of glioma cells. Biochem Biophys Res Commun. 2018;499:765-71
54. Zhang R, Xu J, Zhao J. et al. Silencing of hsa_circ_0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells. Eur Rev Med Pharmacol Sci. 2018;22:118-26
55. Chi Y, Luo Q, Song Y. et al. Circular RNA circPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through miR-600/HIF-1α regulation. J Cell Biochem. 2019;120:19019-30
56. Zhan W, Liao X, Chen Z. et al. Circular RNA hsa_circRNA_103809 promoted hepatocellular carcinoma development by regulating miR-377-3p/FGFR1/ERK axis. J Cell Physiol. 2020;235:1733-45
57. Liu W, Ma W, Yuan Y. et al. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500:846-51
58. Bian L, Zhi X, Ma L. et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p / FOXO4 axis. Biochem Biophys Res Commun. 2018;505:346-52
59. Vannini I, Fanini F, Fabbri M. Emerging roles of microRNAs in cancer. Curr Opin Genet Dev. 2018;48:128-33
60. Qu S, Liu Z, Yang X. et al. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2017;414:301-9
61. Jiang PC, Bu SR. Clinical value of circular RNAs and autophagy-related miRNAs in the diagnosis and treatment of pancreatic cancer. Hepatobiliary Pancreat Dis Int. 2019;18:511-6
62. Fathizadeh H, Hallajzadeh J, Asemi Z. Circular RNAs as diagnostic biomarker in pancreatic cancer. Pathol Res Pract. 2020;216:153075
63. Rong Z, Xu J, Shi S. et al. Circular RNA in pancreatic cancer: a novel avenue for the roles of diagnosis and treatment. Theranostics. 2021;11:2755-69
64. Limb C, Liu DSK, Veno MT. et al. The Role of Circular RNAs in Pancreatic Ductal Adenocarcinoma and Biliary-Tract Cancers. Cancers. 2020;12:3250
65. Wang YZ, An Y, Li BQ. et al. Research progress on circularRNAs in pancreatic cancer: emerging but promising. Cancer Biol Ther. 2019;20:1163-71
66. Qu S, Hao X, Song W. et al. Circular RNA circRHOT1 is upregulated and promotes cell proliferation and invasion in pancreatic cancer. Epigenomics. 2019;11:53-63
67. Yang Dong-Yan, Jin-Tao et al. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23:8345-54
68. Arne S, Sandra H, Detjen KM. et al. Activated signal transducer and activator of transcription 3 (STAT3) supports the malignant phenotype of human pancreatic cancer. Gastroenterology. 2003;125:891-905
69. Bromberg JF, Wrzeszczynska MH, Devgan G. et al. Stat3 as an Oncogene. Cell. 1999;98:295-303
70. Xiong X, Feng J, Yang X. et al. Circular RNA CDR1as promotes tumor progression by regulating miR-432-5p/E2F3 axis in pancreatic cancer. Cancer cell Int. 2021;21:112
71. Liu L, Liu FB, Huang M. et al. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat Dis Int. 2019;18:580-6
72. Huang WJ, Wang YC, Liu SS. et al. Silencing circular RNA hsa_circ_0000977 suppresses pancreatic ductal adenocarcinoma progression by stimulating miR-874-3p and inhibiting PLK1 expression. Cancer Lett. 2018;422:70-80
73. Hao L, Rong W, Bai L. et al. Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR-892b. J Cell Biochem. 2018;120:3780-9
74. An Y, Cai H, Zhang Y. et al. CircZMYM2 competed endogenously with miR-335-5p to regulate JMJD2C in pancreatic cancer. Cell Physiol Biochem. 2018;51:2224-36
75. Ye Z, Zhu Z, Xie J. et al. Hsa_circ_0000069 Knockdown Inhibits Tumorigenesis and Exosomes with Downregulated hsa_circ_0000069 Suppress Malignant Transformation via Inhibition of STIL in Pancreatic Cancer. Int J Nanomedicine. 2020;15:9859-73
76. Zhang X, Xue C, Cui X. et al. Circ_0075829 facilitates the progression of pancreatic carcinoma by sponging miR-1287-5p and activating LAMTOR3 signalling. J Cell Mol Med. 2020;24:14596-607
77. Guo W, Zhao L, Wei G. et al. Blocking circ_0013912 Suppressed Cell Growth, Migration and Invasion of Pancreatic Ductal Adenocarcinoma Cells in vitro and in vivo Partially Through Sponging miR-7-5p. Cancer Manag Res. 2020;12:7291-303
78. Han X, Fang Y, Chen P. et al. Upregulated circRNA hsa_circ_0071036 promotes tumourigenesis of pancreatic cancer by sponging miR-489 and predicts unfavorable characteristics and prognosis. Cell Cycle. 2021;20:369-82
79. Xu S, Lei SL, Liu KJ. et al. circSFMBT1 promotes pancreatic cancer growth and metastasis via targeting miR-330-5p/PAK1 axis. Cancer gene Ther. 2020
80. Shi H, Li H, Zhen T. et al. hsa_circ_001653 Implicates in the Development of Pancreatic Ductal Adenocarcinoma by Regulating MicroRNA-377-Mediated HOXC6 Axis. Mol Ther Nucleic Acids. 2020;20:252-64
81. Xing C, Ye H, Wang W. et al. Circular RNA ADAM9 facilitates the malignant behaviours of pancreatic cancer by sponging miR-217 and upregulating PRSS3 expression. Artif Cells Nanomed Biotechnol. 2019;47:3920-8
82. Chen Y, Li Z, Zhang M. et al. Circ-ASH2L promotes tumor progression by sponging miR-34a to regulate Notch1 in pancreatic ductal adenocarcinoma. J Exp Clin cancer Res. 2019;38:466
83. Wong CH, Lou UK, Li Y. et al. CircFOXK2 Promotes Growth and Metastasis of Pancreatic Ductal Adenocarcinoma by Complexing with RNA-Binding Proteins and Sponging MiR-942. Cancer Res. 2020;80:2138-49
84. Guo X, Zhou Q, Su D. et al. Circular RNA circBFAR promotes the progression of pancreatic ductal adenocarcinoma via the miR-34b-5p/MET/Akt axis. Mol cancer. 2020;19:83
85. Zhang T, Li M, Lu H. et al. Up-Regulation of circEIF6 Contributes to Pancreatic Cancer Development Through Targeting miR-557/SLC7A11/PI3K/AKT Signaling. Cancer Manag Res. 2021;13:247-58
86. Shao F, Cai M, Fan FF. et al. Overexpression of circRNA chr7:154954255-154998784+ in cancer-associated pancreatic stellate cells promotes the growth and metastasis of pancreatic cancer by targeting the miR-4459/KIAA0513 axis. Am J Transl Res. 2020;12:5048-63
87. Huang L, Han J, Yu H. et al. CircRNA_000864 Upregulates B-cell Translocation Gene 2 Expression and Represses Migration and Invasion in Pancreatic Cancer Cells by Binding to miR-361-3p. Front Oncol. 2020;10:547942
88. Zhang X, Tan P, Zhuang Y. et al. hsa_circRNA_001587 upregulates SLC4A4 expression to inhibit migration, invasion, and angiogenesis of pancreatic cancer cells via binding to microRNA-223. Am J Physiol Gastrointest Liver Physiol. 2020;319:G703-G17
89. Kong Y, Li Y, Luo Y. et al. circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR-486-5p/PIK3R1/VEGF-C axis in pancreatic cancer. Mol Cancer. 2020;19:82
90. Jiang Y, Tong W, Li Y. et al. A novel prognostic biomarker for pancreatic ductal adenocarcinoma: hsa_circ_0001649. Gene. 2018;675:88-93
91. Liu Y, Xia L, Dong L. et al. CircHIPK3 Promotes Gemcitabine (GEM) Resistance in Pancreatic Cancer Cells by Sponging miR-330-5p and Targets RASSF1. Cancer Manag Res. 2020;12:921-9
92. Xu C, Yu Y, Ding F. Microarray analysis of circular RNA expression profiles associated with gemcitabine resistance in pancreatic cancer cells. Oncol Rep. 2018;40:395-404
93. Feng S, Mei H, Futao M. et al. Circular RNA Signature Predicts Gemcitabine Resistance of Pancreatic Ductal Adenocarcinoma. Front Pharmacol. 2018;9:584
94. Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163-7
95. Leprivier G, Rotblat B, Khan D. et al. Stress-mediated translational control in cancer cells. Biochim Biophys Acta. 2015;1849:845-60
96. Legnini I, Timoteo GD, Rossi F. et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66:22-37.e9
97. Peng L, Chen G, Zhu Z. et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung's disease. Oncotarget. 2017;8:808-18
98. Wu J, Li J, Liu H. et al. Circulating plasma circular RNAs as novel diagnostic biomarkers for congenital heart disease in children. J Clin Lab Anal. 2019;33:e22998-e
99. Chen J, Li Y, Zheng Q. et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208-19
100. Westholm J, Miura P, Olson S. et al. Genome-wide Analysis of Drosophila Circular RNAs Reveals Their Structural and Sequence Properties and Age-Dependent Neural Accumulation. Cell Rep. 2014;9:1966-80
101. Jae Hoon B, Qing Z, Feng L. et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221-30
102. Li P, Chen S, Chen H. et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132-6
103. Yang J, Cong X, Ren M. et al. Circular RNA hsa_circRNA_0007334 is Predicted to Promote MMP7 and COL1A1 Expression by Functioning as a miRNA Sponge in Pancreatic Ductal Adenocarcinoma. J Oncol. 2019;2019:7630894
104. Li J, Li Z, Jiang P. et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37:177
105. Li Z, Wu Y, Jie L. et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237-50
106. Xu Y, Yao Y, Gao P. et al. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun. 2018;509:138-42
107. Shen X, Chen Y, Li J. et al. Identification of Circ_001569 as a Potential Biomarker in the Diagnosis and Prognosis of Pancreatic Cancer. Technol Cancer Res Treat. 2021;20:1533033820983302
108. Li H, Hao X, Wang H. et al. Circular RNA Expression Profile of Pancreatic Ductal Adenocarcinoma Revealed by Microarray. Cell Physiol Biochem. 2016;40:1334-44
109. Zhao R, Ni J, Lu S. et al. CircUBAP2-mediated competing endogenous RNA network modulates tumorigenesis in pancreatic adenocarcinoma. Aging. 2019;11:8484-501
110. Wang Y, Liu J, Ma J. et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol cancer. 2019;18:116
111. Ren GL, Zhu J, Li J. et al. Noncoding RNAs in acute kidney injury. J Cell physiol. 2019;234:2266-76
112. Shi X, Wang B, Feng X. et al. circRNAs and Exosomes: A Mysterious Frontier for Human Cancer. Mol Ther Nucleic Acids. 2020;19:384-92
113. Preußer C, Hung LH, Schneider T. et al. Selective release of circRNAs in platelet-derived extracellular vesicles. J Extracell Vesicles. 2018;7:1424473
114. Louis C, Desoteux M, Coulouarn C. Exosomal circRNAs: new players in the field of cholangiocarcinoma. Clin Sci (Lond). 2019;133:2239-44
115. Zhang L, Sanagapalli S, Stoita A. Challenges in diagnosis of pancreatic cancer. World J Gastroenterol. 2018;24:2047-60
Author contact
![]() Corresponding author: Jin Wang, Ph.D. Scientific Research Center, Shanghai Public Health Clinical Center, Fudan University, 2901 Caolang Road, Jinshan District, Shanghai 201508, China. Ph: 86-21-57036495; Fax: 86-21-57247094; E-mail: wjincityucom.
Corresponding author: Jin Wang, Ph.D. Scientific Research Center, Shanghai Public Health Clinical Center, Fudan University, 2901 Caolang Road, Jinshan District, Shanghai 201508, China. Ph: 86-21-57036495; Fax: 86-21-57247094; E-mail: wjincityucom.

 Global reach, higher impact
Global reach, higher impact