3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(10):2921-2932. doi:10.7150/jca.51851 This issue Cite
Research Paper
Bioinformatics analysis of lncRNA‑associated ceRNA network in melanoma
1. Department of Histology and Embryology, School of Basic Medical Sciences, Xinjiang Medical University, Urumqi, Xinjiang, China.
2. Department of Pharmacology, Pharmacy College, Xinjiang Medical University, Urumqi, China.
3. Xinjiang Urumqi City Center Blood Station, Urumqi, China.
Abstract
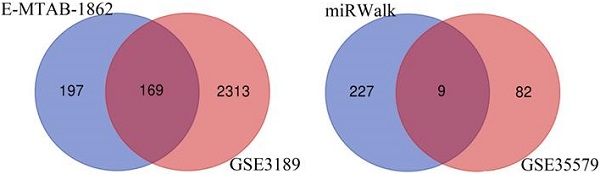
Melanoma is an extremely malignant tumor with early metastasis and high mortality. Little is known about the process of by which melanoma occurs, as its mechanism is very complex and only limited data are available on its long non-coding RNA (lncRNA)-associated competing endogenous RNAs (ceRNAs). The purpose of this study was to screen out potential prognostic molecules and identify a ceRNA network related to the occurrence of melanoma. We screened 169 differentially expressed mRNAs (DEmRNAs) from E-MTAB-1862 and GSE3189; gene ontology (GO) enrichment analysis showed that these genes were closely related to the development of skin. In the protein-protein interaction network, we screened out a total of 19 hub genes. Furthermore, we predicted the microRNAs (miRNAs) that regulate hub genes using the miRWalk database and then intersected these with GSE35579, resulting in nine DEmiRNAs. We also predicted the lncRNAs that regulate the miRNAs using the LncBasev.2 database. According to the ceRNA hypothesis, and based on the intersection of the DElncRNAs with merged GTEx and TCGA data, we obtained 20 DElncRNAs. A total of four DEmRNAs, nine DEmiRNAs, and 20 DElncRNAs were included in the ceRNA network. Based on Cox stepwise regression and survival analysis, we identified five biomarkers, ZSCAN16-AS1, LINC00520, XIST, DTL, and let-7a-5p, and obtained risk scores. The results showed that most of the differentially expressed genes were related to epithelial-mesenchymal transition (EMT) in melanoma. Finally, we obtained a LINC00520/let-7a-5p/DTL molecular regulatory network. These results suggest that ceRNA networks have an important role in evaluating the prognosis of patients with melanoma and provide a new experimental basis for exploring the EMT process in the development of melanoma.
Keywords: Bioinformatics analysis, ceRNA, differentially expressed gene, melanoma, survival analysis.

 Global reach, higher impact
Global reach, higher impact