Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(9):2702-2714. doi:10.7150/jca.55113 This issue Cite
Research Paper
Identification and validation of the clinical roles of the VHL-related LncRNAs in clear cell renal cell carcinoma
1. Department of Urology, Peking University First Hospital, Beijing 100034, P.R. China.
2. Hereditary Kidney Cancer Research Center, Peking University First Hospital, Beijing 100034, P.R. China.
3. Institute of Urology, Peking University, Beijing 100034, P.R. China.
4. National Urological Cancer Center, Beijing 100034, P.R. China.
Received 2020-10-27; Accepted 2021-2-14; Published 2021-3-5
Abstract
Accumulating evidence suggests that lncRNAs (long non-coding RNAs) function as oncogenes or tumor suppressor genes in ccRCC (clear cell renal cell carcinoma). Although VHL (Von Hippel-Lindau) gene inactivation is by far the most common carcinogenic driving event in ccRCC, the roles of VHL-related lncRNAs in ccRCC remain unknown. In this study, using RNA-seq and clinical data in TCGA-KIRC (the Cancer Genome Atlas-Kidney Renal Clear Cell Carcinoma), we identified VHL-related lncRNAs through WGCNA (Weighted Gene Co-expression Network Analysis), correlation analysis and catRAPID algorithm, and explored their clinical characteristics in ccRCC. Results showed that 10 lncRNAs (AC112220.2, AL391121.1, USP46-AS1, AL450326.1, MID1IP1-AS1, SUCLG2-AS1, RAP2C-AS1, FGD5-AS1, AC018647.2 and AC015922.2) were identified as VHL-related lncRNAs, and they were down-regulated in ccRCC tissues. Survival analysis results indicated that high expression groups of AC112220.2, AL391121.1, USP46-AS1, AL450326.1, SUCLG2-AS1, RAP2C-AS1, FGD5-AS1, AC018647.2 and AC015922.2 had significantly longer OS (Overall Survival) than their respective low expression groups. Meanwhile high AC112220.2, USP46-AS1, AL450326.1, SUCLG2-AS1, FGD5-AS1, AC018647.2 and AC015922.2 expression groups had remarkably longer DFS (Disease Free Survival) than their respective low expression groups. Besides, FGD5-AS1 and AL391121.1 expression were decreased in VHL mutant tissues compared with VHL non-mutant tissues. Moreover, high expression group of FGD5-AS1 had significantly longer OS and DFS than their respective low expression groups in VHL mutant ccRCC. In addition, we found that DNA hypermethylation may also play an important role in decreased FGD5-AS1 expression. Furthermore, we validated the expression of FGD5-AS1 in VHL mutant and non-mutant ccRCC tissues and cell lines. In conclusion, our results demonstrated that lncRNA FGD5-AS1 was significantly associated with VHL and can serve as a novel biomarker of ccRCC.
Keywords: VHL, lncRNAs, ccRCC, FGD5-AS1, survival
Introduction
RCC (Renal cell carcinoma) is the second leading cause of death in human urological malignancies, accounting for 2-3% of all cancer diagnoses and cancer deaths worldwide [1]. The global burden of RCC is approximately 403,262 new cases and 175,098 deaths per year, with an annual rate of increase of approximately 0.7% [2]. The most common histologic subtype ccRCC (clear cell RCC) represents about 75-80% of RCC, and up to 92% of these cancers have inactivated the VHL (Von Hippel-Lindau) gene [3]. Partial or radical nephrectomy represents the clinical protocol for localized RCC, and the 5-year cancers specific survival rates for patients with localized RCC (pathological stage pT1-2) after nephrectomy ranges from 71% to 97% [4, 5]. For patients with locally advanced tumors, 5-year cancer-specific survival rates after nephrectomy decrease to 20% to 53% [4]. Despite nephrectomy with curative intent, about 30% of patients with ccRCC with localized disease eventually develop metastases, and the 5-year survival rate of metastatic ccRCC dramatically drops down to 10% [3, 6]. Therefore, finding promising early detection markers and more effective therapeutic targets is imperative to improve the prognosis of ccRCC patients.
As a new class of ncRNAs (non-coding RNAs), lncRNAs (long ncRNAs) are characterized as non-coding transcripts greater than 200 base pairs in length transcribed by RNA Pol II from independent promoters [7]. In many cases, lncRNAs have been proven to be the main regulator of gene expression and thus, they can play a key role in a variety of biological functions and disease processes including cancers [8]. For example, lncRNA LBCS inhibits castration resistance of prostate cancer [9]; DANCR promotes cell metastasis and proliferation in bladder cancer [10]. Besides, recent accumulating evidence has indicated that lncRNAs, such as OTUD6B-AS1, URRCC, HOTAIRM1 and MRCCAT1, play important regulatory roles in diverse biological processes in ccRCC [11-14]. As for the molecular characteristics of RCC, VHL gene inactivation is by far the most common carcinogenic driving event in ccRCC [15]. However, there are currently few studies on the roles of VHL-related lncRNAs in ccRCC. Therefore, it is urgent to identify VHL-related lncRNAs and understand their roles in ccRCC.
In the present study, using RNA-seq and clinical data in TCGA-KIRC cohort, we identified VHL-related lncRNAs by WGCNA, correlation analysis and catRAPID algorithm, and explored their prognostic value and relationship with clinicopathological characteristics in ccRCC.
Materials and Methods
Ethics statement
This study was approved by the Biomedical Research Ethics Committee of Peking University First Hospital (Beijing, China, IRB00001052-18004). Written informed consents were also obtained from all patients.
TCGA and GEO data
Level-3 RNA-sequencing data, the clinicopathological and survival data of patients with ccRCC were downloaded from TCGA (https://portal.gdc.cancer.gov/). Briefly, 539 ccRCC and 72 adjacent normal renal tissues were included in this study. Their clinical and survival data, including tumor stage, lymph node, metastasis, pathological stage, histologic grade, OS (Overall Survival) and DFS (Disease Free Survival), were downloaded. The DNA methylation data of 325 ccRCC and 160 adjacent normal renal tissues were also downloaded. In addition the GSE105260 dataset which contained the methylation data of 485512 CpG sites in 9 normal kidney, 9 primary ccRCC and 26 metastasis ccRCC tumors was obtained from GEO datasets (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE105260).
Weighted Gene Co-expression Network Analysis (WGCNA)
The R software package “WGCNA” was used for weighted gene co-expression network analysis. It is an algorithm for constructing co-expression networks, defined by the similarity of gene co-expression. Briefly, we first used paired Pearson correlations to evaluate the weighted co-expression relationships among the subjects in all data sets in the adjacency matrix. Then, a topological overlap matrix (TOM) similarity function was used to convert the matrix to a TOM. The resulting TOM was based on genetic similarity of biological significance and was used to measure the co-expression relationships between genes, and the defined weight threshold was 0.02.
catRAPID
catRAPID (http://s.tartaglialab.com/page/catrapid_group) was an algorithm to estimate the binding propensity of protein-RNA pairs. By combining secondary structure, hydrogen bonding and van der Waals contributions, catRAPID predicts protein-RNA associations with great accuracy.
Clinical samples and ccRCC cell lines
The paired tissues samples (adjacent normal renal tissue and ccRCC tissue) from 54 ccRCC patients including 12 ccRCC caused by VHL mutation were collected from Department of Urology, Peking University First Hospital for total RNA extraction, reverse transcription and q-RT-PCR analysis. The total RNA of HEK293, OSRC2, ACHN, A498, 769-P and 786-O was also extracted for reverse transcription and q-RT-PCR analysis. The detailed primer sequences was provided in Supplemental Table 1.
In addition, pLV-hef1a-mNeongreen-P2A-Puro-WPRE-CMV-VHL-3Xflag and its corresponding control plasmid vector were constructed by the SyngenTech Company (SyngenTech Co. Ltd., Beijing, China). Cells were transfected with the corresponding vector using Lipofectamine 3000 Transfection Reagent (Invitrogen, USA) according to the manufacturer's instructions. Lenti-virus was produced using three vectors system: transfer vector, viral packaging (psPAX2) and viral envelope (pMD2G) at 6:3:1 ratio transfected into 293T cells. VHL-overexpressed stable cell lines were selected with puromycin (5 μg/mL). The total protein and RNA of VHL-overexpressed 786-0 cells and the control 786-0 cells were extracted for western blot and q-RT-PCR analysis. For western blot assay, protein (30 μg per lane) was separated by SDS-PAGE and then were transblotted to PVDF membranes, and membranes were blocked in 5% nonfat milk powder and incubated overnight at 4 °C with anti-Flag (1:1000; CST, 14793S), anti-VHL (1:1000; CST, 68547S) and anti-GAPDH (1:10000, Proteintech, China). After incubated with horseradishperoxidase-conjugated goat anti-rabbit IgG, membranes were resolved by chemiluminescence.
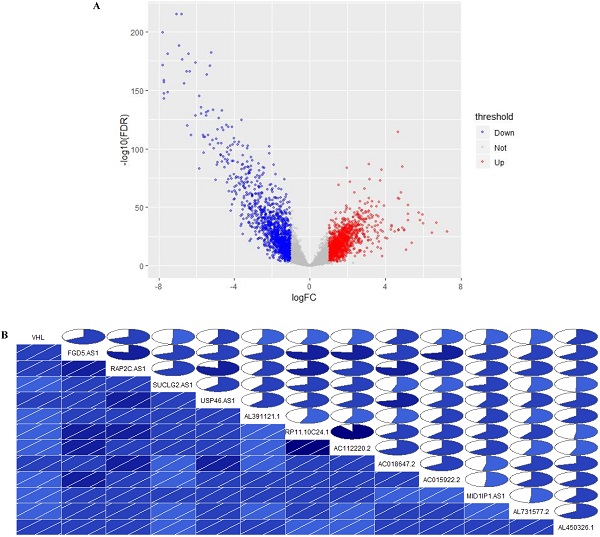
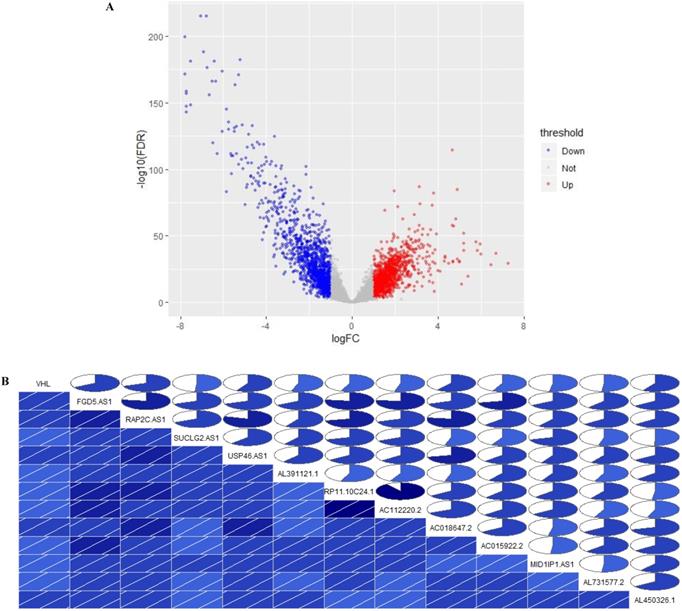
Identification of VHL co-expressed lncRNAs. (A) Volcano Plot, (B) The correlation between the expression of 12 lncRNAs and VHL. In the lower triangular cell, the blue color and the slash from lower left to upper right indicate that the two variables in the cell are positively correlated. The darker the color, the higher the saturation, and the greater the correlation between variables. Similarly, the upper triangle cell displays the same information in a pie chart. The color function is the same as above, and the degree of correlation is displayed by the size of the filled pie chart.
Statistical analysis
Non-parametric Mann-Whitney test was used to detect differences in continuous variables. The Pearson's correlation test was conducted to assess the correlations between lncRNAs and VHL. The prognostic roles of VHL co-expressed lncRNAs were examined with the Kaplan-Meier method, and the log-rank test was conducted to determine the significance of the difference between the survival curves. A P-value < 0.05 represented statistical significance. The statistical analyses were all carried out by R Studio and GraphPad Prism 7.00.
Results
Identification of VHL-related lncRNAs in ccRCC
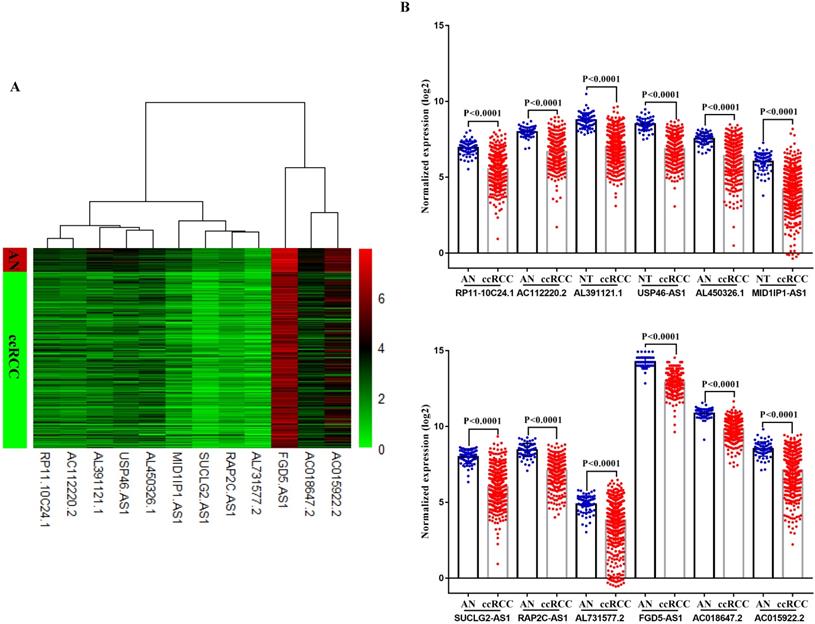
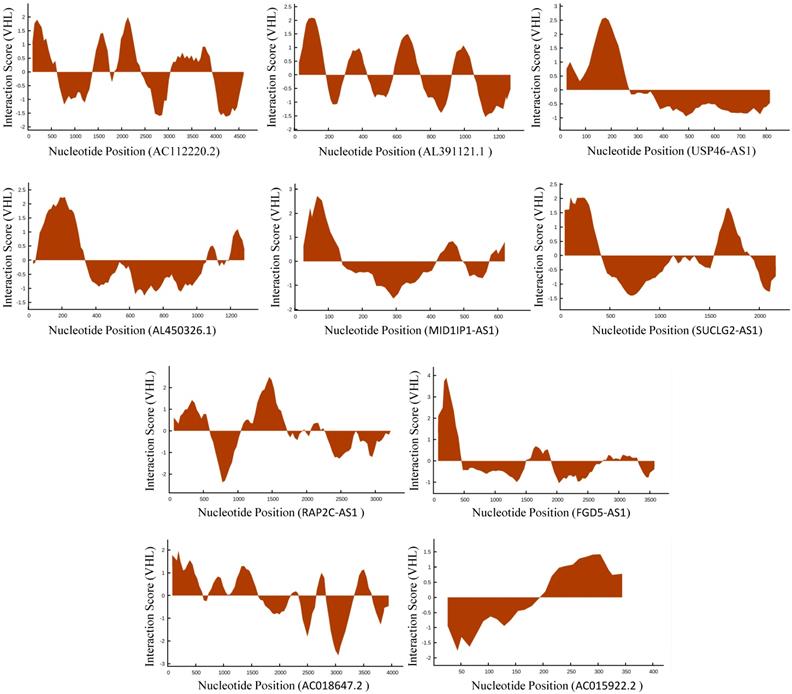
In TCGA-KIRC cohort, tumor tissues from 539 cases of ccRCC were subjected to RNA-seq study, among which 72 cases had matched adjacent normal tissues. The differential analysis was performed by “edgeR: a Bioconductor package for differential expression analysis of digital gene expression data” algorithm, results showed that 1333 lncRNAs were down-regulated and 1364 lncRNAs were up-regulated in ccRCC tissues compared to adjacent normal tissues (Figure 1A). Then the expression data of these 2697 lncRNAs and VHL was performed to WGCNA, results showed that 12 lncRNAs (FGD5-AS1, RAP2C-AS1, SUCLG2-AS1, USP46-AS1, AL391121.1, RP11-10C24.1, AC112220.2, AC018647.2, AC015922.2, MID1IP1-AS1, AL731577.2 and AL450326.1) were co-expressed with VHL (Table 1). Correlation analysis results showed that all of these 12 lncRNAs were highly positively correlated with VHL (Figure 1B). Moreover, all of these 12 lncRNAs were down-regulated in ccRCC tissues compared to adjacent normal tissues (Figure 2A and 2B). Furthermore, we used catRAPID to estimate the binding propensity of VHL protein with these 12 lncRNAs, and our results showed that AC112220.2, AL391121.1, USP46-AS1, AL450326.1, MID1IP1-AS1, SUCLG2-AS1, RAP2C-AS1, FGD5-AS1, AC018647.2 and AC015922.2 have a certain binding potential with VHL protein (Figure 3).
Comparison of VHL co-expressed lncRNAs expression in ccRCC tissues and adjacent normal tissues. (A) Heat map (B) Statistical comparison.
Prognostic value of VHL-related lncRNAs in ccRCC patients
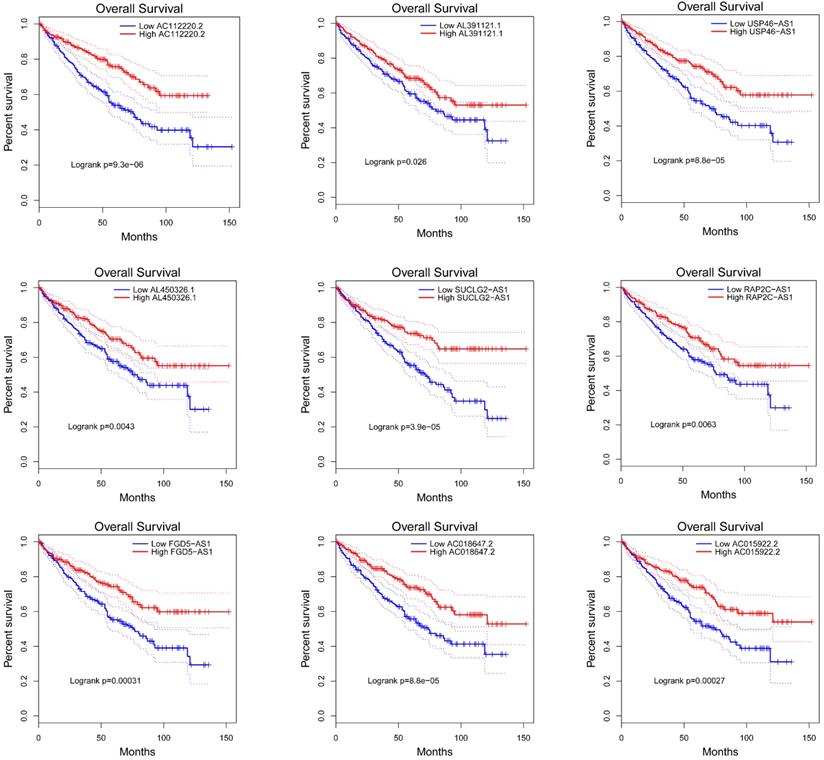
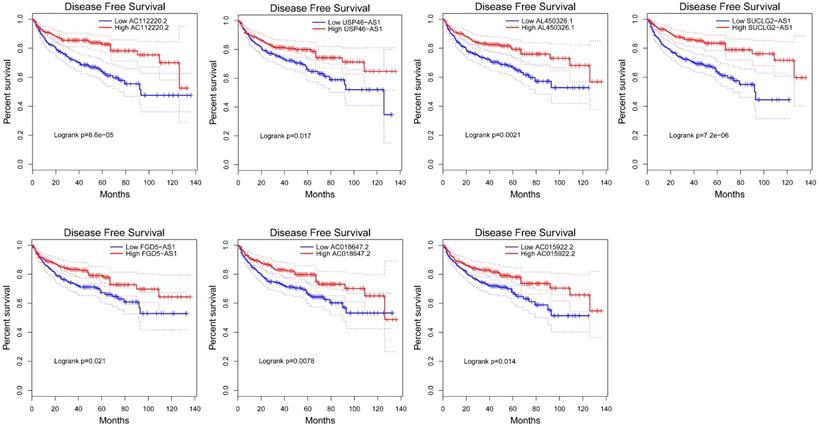
To explore the prognostic characteristics of VHL-related lncRNAs, we analyzed the effects of these lncRNAs on the OS and DFS among ccRCC patients by generating Kaplan-Meier survival curves. Results of the log-rank test showed that the high expression groups of AC112220.2, AL391121.1, USP46-AS1, AL450326.1, SUCLG2-AS1, RAP2C-AS1, FGD5-AS1, AC018647.2 and AC015922.2 had significantly longer OS than their respective low expression groups (Figure 4). Meanwhile, results of the log-rank test also showed that the high AC112220.2, USP46-AS1, AL450326.1, SUCLG2-AS1, FGD5-AS1, AC018647.2 and AC015922.2 expression groups had remarkably longer DFS than their respective low expression groups (Figure 5).
Association between VHL-related lncRNAs and the clinicopathological characteristics of ccRCC
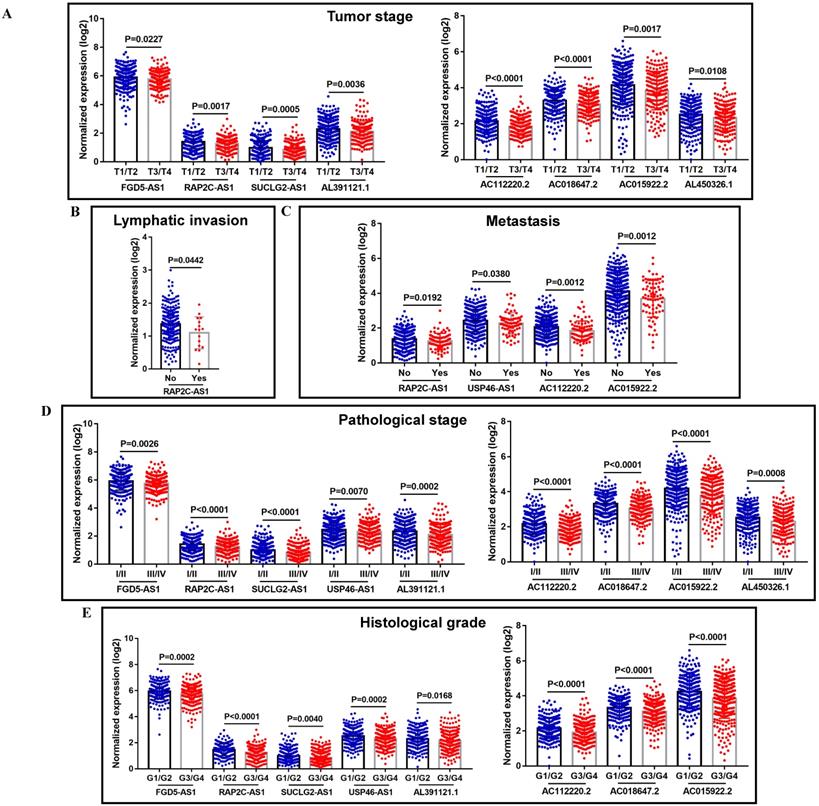
When concerning the correlation between these VHL-related lncRNAs and the progression of ccRCC, most of them were closely related to some clinical parameters of ccRCC, including tumor stage, lymphatic invasion, metastasis, pathological stage and histological grade. The detailed clinical information of TCGA-KIRC patients was summarized in Supplemental Table 2. Lower expression of FGD5-AS1, RAP2C-AS1, SUCLG2-AS1, AL391121.1, AC112220.2, AC018647.2, AC015922.2 and AL450326.1 were associated with advanced tumor stage (Figure 6A); lower expression of RAP2C-AS1 was related to lymphatic invasion (Figure 6B); lower expression of RAP2C-AS1, USP46-AS1, AC112220.2 and AC015922.2 tended to promote tumor metastasis (Figure 6C); lower FGD5-AS1, RAP2C-AS1, SUCLG2-AS1, USP46-AS1, AL391121.1, AC112220.2, AC018647.2, AC015922.2 and AL450326.1 expression levels were correlated with higher pathological stage (Figure 6D); lower FGD5-AS1, RAP2C-AS1, SUCLG2-AS1, USP46-AS1, AL391121.1, AC112220.2, AC018647.2 and AC015922.2 expression levels were also correlated with higher histological grade (Figure 6E).
Prognostic and clinicopathological characteristics of VHL-related lncRNAs in ccRCC with VHL mutation
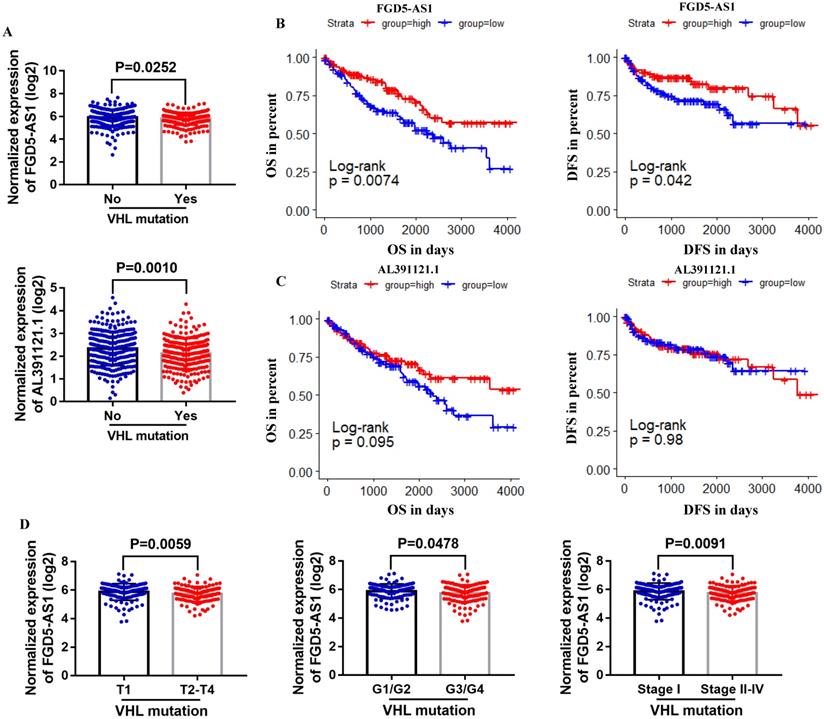
To further explore the correlation between these lncRNAs and VHL, we compared their expression in VHL mutant and non-mutant ccRCC tissues. Results suggested that only FGD5-AS1 and AL391121.1 expression were decreased in VHL mutant tissues compared with VHL non-mutant tissues (Figure 7A). Survival analysis results indicated that high expression group of FGD5-AS1 had significantly longer OS and DFS than their respective low expression groups in VHL mutant ccRCC (Figure 7B), while there was no difference in survival time between the high AL391121.1 expression group and its respective low expression group (Figure 7C). Besides, lower FGD5-AS1 expression was related to advanced tumor stage and higher histological grade and pathological stage (Figure 7D).
Estimation of the binding propensity of VHL protein and lncRNAs.
Kaplan-Meier curves of OS in patients with ccRCC. Patients were grouped according to the median cutoff of AC112220.2, AL391121.1, USP46-AS1, AL450326.1, SUCLG2-AS1, RAP2C-AS1, FGD5-AS1, AC018647.2 and AC015922.2 expression for OS detection. Patients were separated into two groups (low expression group (n=268) and high expression group (269)) according to the median cutoff of lncRNAs expression.
The co-expressed lncRNAs of VHL identified by WGCNA
| Gene 1 | Gene 2 | Weight | |
|---|---|---|---|
| ensembl | gene symbol | ||
| VHL | ENSG00000225733 | FGD5-AS1 | 0.051624798 |
| VHL | ENSG00000232160 | RAP2C-AS1 | 0.043959581 |
| VHL | ENSG00000241316 | SUCLG2-AS1 | 0.032784519 |
| VHL | ENSG00000248866 | USP46-AS1 | 0.027462711 |
| VHL | ENSG00000272933 | AL391121.1 | 0.026646185 |
| VHL | ENSG00000271020 | RP11-10C24.1 | 0.025374632 |
| VHL | ENSG00000271643 | AC112220.2 | 0.024199759 |
| VHL | ENSG00000271122 | AC018647.2 | 0.023547268 |
| VHL | ENSG00000265519 | AC015922.2 | 0.023450544 |
| VHL | ENSG00000238123 | MID1IP1-AS1 | 0.021209168 |
| VHL | ENSG00000249456 | AL731577.2 | 0.020339211 |
| VHL | ENSG00000230555 | AL450326.1 | 0.020245942 |
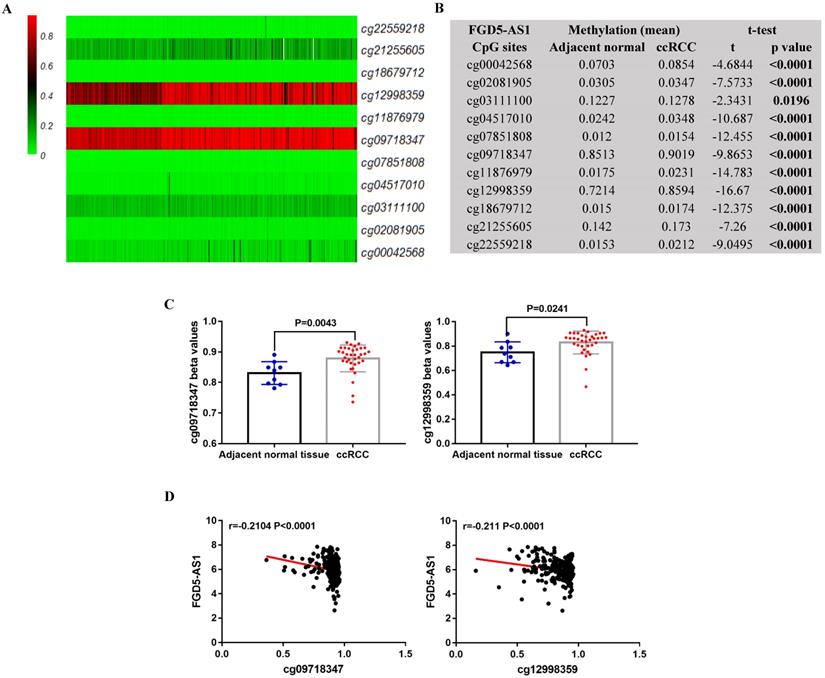
FGD5-AS1 expression were negatively correlated with their DNA methylation status in ccRCC
In TCGA-KIRC, 325 ccRCC tissues samples and 160 adjacent normal tissues samples were subjected to DNA methylation analysis simultaneously. Using the methylation data, we compared the methylation status of 11 CpG sites (cg00042568, cg02081905, cg03111100, cg04517010, cg07851808, cg09718347, cg11876979, cg12998359, cg18679712, cg21255605 and cg22559218) in FGD5-AS1 DNA, and the detailed information of these CpG sites was provided in Table 2. Results showed that these CpG sites were significantly hypermethylated in the ccRCC tissues compared to adjacent normal tissues (Figure 8A and 8B). Moreover, the data from GEO validated the higher methylation levels of cg09718347 and cg12998359 in the ccRCC tissues compared to adjacent normal tissues (Figure 8C). To further examine the potential regulatory effect of DNA methylation on FGD5-AS1, we analyzed the correlation between FGD5-AS1 expression and the methylation status of their CpG sites. Correlation analysis results showed that the expression of FGD5-AS1 were negatively associated with the methylation levels of cg09718347 and cg12998359 (Figure 8D).
Kaplan-Meier curves of DFS in patients with ccRCC. Patients were grouped according to the median cutoff of AC112220.2, USP46-AS1, AL450326.1, SUCLG2-AS1, FGD5-AS1, AC018647.2 and AC015922.2 expression for DFS detection. Patients were separated into two groups (low expression group (n=245) and high expression group (n=246)) according to the median cutoff of lncRNAs expression.
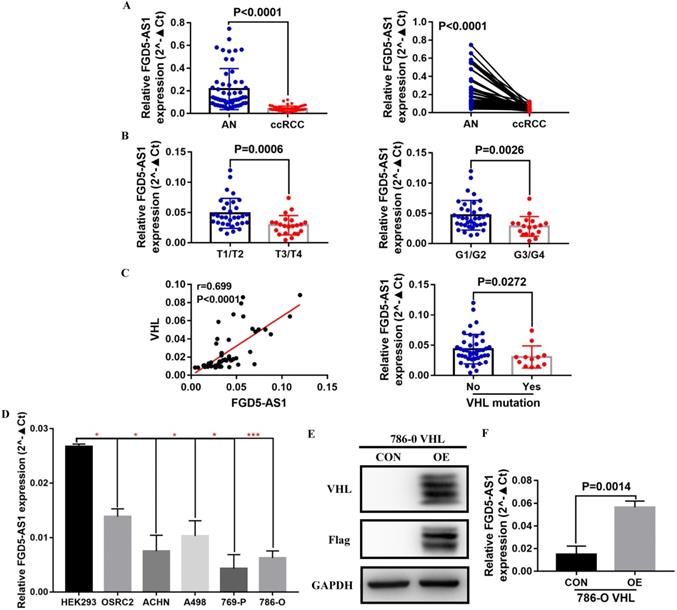
Validation of the expression of FGD5-AS1 in VHL mutant and non-mutant ccRCC tissues and cell lines
To validate the correlation between FGD5-AS1 and VHL expression, we examined the expression of FGD5-AS1 and VHL in 12 VHL mutant and 42 non-mutant ccRCC tissues, and detailed clinical and pathological data of these 54 ccRCC patients were provided in Table 3. Our results confirmed that FGD5-AS1 expression was decreased in these 54 ccRCC tissues compared with their adjacent normal tissues (Figure 9A). Besides, FGD5-AS1 expression was lower in advanced tumor stage and higher histological grade (Figure 9B). Moreover, FGD5-AS1 expression was highly positively correlated with VHL expression, and FGD5-AS1 expression was significantly reduced in VHL mutant ccRCC tissues compared to VHL non-mutant ccRCC tissues (Figure 9C). In addition, we examined the expression of FGD5-AS1 in HEK293 cell line, VHL wild-type RCC cell lines (OSRC2, ACHN and A498) and VHL mutant RCC cell lines (769-P and 786-O). Results indicated that FGD5-AS1 expression was decreased in these RCC cell lines compared with HEK293 cell line, especially in 769-P and 786-O cell lines (Figure 9D). Furthermore, we found that FGD5-AS1 expression was increased in VHL-overexpressed 786-O cells compared with the control 786-O cells (Figure 9E, F).
Discussion
It is all known that VHL play a critical tumor suppressor role in ccRCC, and VHL gene inactivation is by far the most common carcinogenic driving event in ccRCC. Although lncRNAs play important roles in the development and progression of ccRCC [16, 17], there are currently few literatures on the role of VHL-related lncRNAs in ccRCC.
In this study, we found that several lncRNAs, including FGD5-AS1, RAP2C-AS1, SUCLG2-AS1, USP46-AS1, AL391121.1, ENSG00000271020, AC112220.2, AC018647.2, AC015922.2, MID1IP1-AS1, AL731577.2 and AL450326.1 were co-expressed with VHL, and their expression were positively correlated with VHL expression. Intriguingly, all of them were down-regulated in ccRCC tissues compared to adjacent normal tissues. Besides, the analysis results of catRAPID showed that AC112220.2, AL391121.1, USP46-AS1, AL450326.1, MID1IP1-AS1, SUCLG2-AS1, RAP2C-AS1, FGD5-AS1, AC018647.2 and AC015922.2 have a certain binding potential with VHL protein. Moreover, results of the log-rank test showed that high expression groups of AC112220.2, AL391121.1, USP46-AS1, AL450326.1, SUCLG2-AS1, RAP2C-AS1, FGD5-AS1, AC018647.2 and AC015922.2 had significantly longer OS than their respective low expression groups, and high AC112220.2, USP46-AS1, AL450326.1, SUCLG2-AS1, FGD5-AS1, AC018647.2 and AC015922.2 expression groups had remarkably longer DFS than their respective low expression groups. In addition, they were also closely associated with some clinical parameters of ccRCC, including tumor stage, metastasis, pathological stage and histological grade.
The correlation between VHL-related lncRNAs expression and the clinicopathological characteristics of ccRCC patients. The correlation between VHL-related lncRNAs expression and tumor stage (A), lymphatic invasion (B), metastasis (C), pathological stage (D) and histological grade (E).
To further explore the correlation between these lncRNAs and VHL, we compared their expression in VHL mutant and non-mutant ccRCC tissues. Results showed that FGD5-AS1 and AL391121.1 expression were remarkably decreased in VHL mutant tissues compared with VHL non-mutant tissues. However, only high expression group of FGD5-AS1 had significantly longer OS and DFS than its respective low expression groups in VHL mutant ccRCC. Besides, we validated the lower expression of FGD5-AS1 in VHL mutant and non-mutant ccRCC tissues and cell lines. Moreover, we found that FGD5-AS1 expression was increased in VHL-overexpressed 786-O cells compared with the control 786-O cells.
Comparison of lncRNAs expression in VHL mutant and non-mutant ccRCC tissues. (A) FGD5-AS1 and AL391121.1 expression in VHL mutant and non-mutant ccRCC tissues. (B and C) Patients were grouped according to the median cutoff of FGD5-AS1 and AL391121.1 expression for OS and DFS detection. (D) Comparison of FGD5-AS1 expression in different tumor stage, historical grade and pathological stage.
The detailed information of CpG sites of FGD5-AS1 DNA
| Composite Element REF | Chromosome | Start | End | CGI_Coordinate | Feature_Type |
|---|---|---|---|---|---|
| cg00042568 | chr3 | 14946432 | 14946433 | CGI:chr3:14947105-14948477 | N_Shore |
| cg02081905 | chr3 | 14947162 | 14947163 | CGI:chr3:14947105-14948477 | Island |
| cg03111100 | chr3 | 14948476 | 14948477 | CGI:chr3:14947105-14948477 | Island |
| cg04517010 | chr3 | 14948461 | 14948462 | CGI:chr3:14947105-14948477 | Island |
| cg07851808 | chr3 | 14947328 | 14947329 | CGI:chr3:14947105-14948477 | Island |
| cg09718347 | chr3 | 14933436 | 14933437 | CGI:chr3:14947105-14948477 | . |
| cg11876979 | chr3 | 14947322 | 14947323 | CGI:chr3:14947105-14948477 | Island |
| cg12998359 | chr3 | 14944016 | 14944017 | CGI:chr3:14947105-14948477 | N_Shelf |
| cg18679712 | chr3 | 14948165 | 14948166 | CGI:chr3:14947105-14948477 | Island |
| cg21255605 | chr3 | 14947624 | 14947625 | CGI:chr3:14947105-14948477 | Island |
| cg22559218 | chr3 | 14947874 | 14947875 | CGI:chr3:14947105-14948477 | Island |
FGD5-AS1 expression was associated with its DNA methylation status. Heatmap (A) and statistical comparison (B) of the difference in methylation levels in 11 CpG sites of FGD5-AS1. (C) Validation of the methylation levels of cg09718347 and cg12998359 based on GSE105260 dataset. (D) The correlation between FGD5-AS1 expression and the methylation status of its CpG sites.
The clinical and pathological characteristics of the 54 ccRCC patients that used for validation
| Clinicopathologic characteristics | N (%) |
|---|---|
| Age | |
| <60 | 31 (57.4) |
| ≥60 | 23 (42.6) |
| Gender | |
| Male | 32 (59.3) |
| Female | 22 (40.7) |
| Tumor stage | |
| T1 | 20 (37.0) |
| T2 | 12 (22.2) |
| T3 | 21 (38.9) |
| T4 | 1 (1.9) |
| Lymph node stage | |
| N0 | 52 (96.3) |
| N1 | 2 (3.7) |
| Pathological stage | |
| Stage I | 22 (40.7) |
| Stage II | 17 (31.5) |
| Stage III | 12 (22.2) |
| Stage IV | 3 (5.6) |
| Histological grade | |
| G1 | 18 (33.3) |
| G2 | 18 (33.3) |
| G3 | 17 (31.5) |
| G4 | 1 (1.9) |
| VHL mutation | |
| No | 42 (77.8) |
| Yes | 12 (22.2) |
Epigenetic alterations have been identified as one of the hallmarks of tumorigenesis [18, 19], and epigenetic regulation is one of the main mechanisms utilized to control lncRNAs expression and tissue specificity [20-22]. For instance, survival-related lncRNAs SNHG12 and MINCR were epigenetically activated in multiple cancer types, including breast, bladder, endometrial, colorectal, and lung cancer [23]; EPIC1 was epigenetically activated and correlated with poor survival in breast cancer [24]; DNA-methylation-mediated activating of lncRNA SNHG12 promoted temozolomide resistance in glioblastoma [25]. Moreover, a recent study indicated that SNHG3 and SNHG15 were valuable prognostic markers for ccRCC, and DNA hypomethylation might play an important role in increased SNHG3 and SNHG15 expression in ccRCC [26]. In this study, we also examined the correlation between FGD5-AS1 expression and its DNA methylation status. Results showed that the methylation status of cg09718347 and cg12998359 might substantially influence FGD5-AS1 expression in ccRCC.
Validation of the expression of FGD5-AS1 in VHL mutant and non-mutant ccRCC tissues and cell lines. (A) FGD5-AS1 expression in 54 paired clinical ccRCC tissues. (B) Comparison of FGD5-AS1 expression in different tumor stage and histological grade. (C) Correlation between FGD5-AS1 expression and VHL expression. (D) FGD5-AS1 expression in VHL mutant and non-mutant RCC cell lines, *p<0.05, ***p<0.001 vs. HEK293. (E) VHL protein was successfully overexpressed in 786-O cells. (F) FGD5-AS1 expression was increased in VHL-overexpressed 786-O cells.
Previous studies have identified the important roles of FGD5-AS1 in several types of human cancer, including colorectal cancer [27], oral cancer [28], non-small cell lung cancer [29] and esophageal squamous cell carcinoma [30]. FGD5-AS1 was aberrantly up-regulated in these tumors compared to adjacent normal tissues. Moreover, increased FGD5-AS1 expression manifested a close association with tumor size, TNM stage, and lymph node metastasis in esophageal squamous cell carcinoma [30]; FGD5-AS1 may promote NSCLC cell proliferation [29]; FGD5-AS1 knockdown inhibited colorectal cancer cell proliferation, migration, and invasion, and promoted cell apoptosis [27]. However, in this study, we found that FGD5-AS1 expression was significantly lower in ccRCC than in adjacent normal tissues, and increased FGD5-AS1 was associated with of longer OS and DFS. Moreover, we found that FGD5-AS1 was significantly associated with VHL, and DNA methylation might play an important role in altered FGD5-AS1 expression.
The loss of pVHL (VHL protein) function affects several cellular processes, of which the activation of HIFs (hypoxia inducible factors) pathway is the best-known function. Constitutive activation of HIFs signaling in turn activates hundreds of genes involved in numerous oncogenic pathways that contribute to the development or progression of ccRCC [17]. Previous studies reported that hypoxia/HIF-1α-induced lincRNA-p21 is able to bind HIF-1α and pVHL and thus disrupts the VHL-HIF-1α interaction; SNHG11 binds to the pVHL recognition sites on HIF-1α, thus blocking the interaction of pVHL with HIF-1α and preventing its ubiquitination and degradation, resulting in the increased expression of HIF-1α target genes such as AK4, ENO1, HK2 and Twist1 in colorectal cancer [31]. Based on these findings, we can infer that VHL binding lncRNAs may affect the interaction between pVHL and HIFs, thereby affecting HIFs and its target genes expression. Therefore, identification of VHL-related lncRNAs and further exploration of their regulatory mechanisms can provide new theoretical basis for tumor therapy.
In summary, we for the first time identified VHL-related lncRNAs through WGCNA, correlation analysis and catRAPID algorithm, and explored their prognostic and clinicopathological characteristics in ccRCC. As a result, we found that FGD5-AS1 was significantly associated with VHL expression and DNA hypermethylation might play an important role in decreased FGD5-AS1 expression. Moreover, FGD5-AS1 can serve as a valuable diagnostic and prognostic markers in ccRCC. To further confirm our findings, more studies are needed to explore the specific relationship between FGD5-AS1 and VHL, and to verify its prognostic value.
Abbreviations
ccRCC: clear cell renal cell carcinoma; lncRNAs: long non-coding RNAs; TCGA-KIRC: the Cancer Genome Atlas-Kidney Renal Clear Cell Carcinoma; WGCNA: Weighted Gene Co-expression Network Analysis; OS: Overall Survival; DFS: Disease Free Survival; VHL: Von Hippel-Lindau; pVHL: VHL protein; HIFs: hypoxia inducible factors.
Supplementary Material
Supplementary tables.
Acknowledgements
We would like to express our sincere gratitude to TCGA and GEO programs, which provide the high quality genomic data.
Author Contributions
W.Y: design, analysis and interpretation of data, drafting of the manuscript; J.Z, K.Z and L.L: statistical analysis; Y.X, K.M, H.X and L.C: acquisition of data; W.Y, Y.G and K.G: administrative support, obtaining funding, supervision, wrote and edited the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant 81872081), and the Fundamental Research Funds for the Central Universities (grant BMU2018JI002).
Data Availability
The data used to support the findings of this study are available on request to the corresponding author.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87-108
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68:394-424
3. Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M. et al. Renal cell carcinoma. Nature reviews Disease primers. 2017;3:17009
4. Frank I, Blute ML, Leibovich BC, Cheville JC, Lohse CM, Zincke H. Independent validation of the 2002 American Joint Committee on cancer primary tumor classification for renal cell carcinoma using a large, single institution cohort. The Journal of urology. 2005;173:1889-92
5. Choueiri TK, Motzer RJ. Systemic Therapy for Metastatic Renal-Cell Carcinoma. The New England journal of medicine. 2017;376:354-66
6. Barata PC, Rini BI. Treatment of renal cell carcinoma: Current status and future directions. CA: a cancer journal for clinicians. 2017;67:507-24
7. Slack FJ, Chinnaiyan AM. The Role of Non-coding RNAs in Oncology. Cell. 2019;179:1033-55
8. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661-7
9. Gu P, Chen X, Xie R, Xie W, Huang L, Dong W. et al. A novel AR translational regulator lncRNA LBCS inhibits castration resistance of prostate cancer. Molecular cancer. 2019;18:109
10. Chen Z, Chen X, Xie R, Huang M, Dong W, Han J. et al. DANCR Promotes Metastasis and Proliferation in Bladder Cancer Cells by Enhancing IL-11-STAT3 Signaling and CCND1 Expression. Molecular therapy: the journal of the American Society of Gene Therapy. 2019;27:326-41
11. Wang G, Zhang ZJ, Jian WG, Liu PH, Xue W, Wang TD. et al. Novel long noncoding RNA OTUD6B-AS1 indicates poor prognosis and inhibits clear cell renal cell carcinoma proliferation via the Wnt/beta-catenin signaling pathway. Molecular cancer. 2019;18:15
12. Hamilton MJ, Young M, Jang K, Sauer S, Neang VE, King AT. et al. HOTAIRM1 lncRNA is downregulated in clear cell renal cell carcinoma and inhibits the hypoxia pathway. Cancer letters. 2020;472:50-8
13. Zhai W, Zhu R, Ma J, Gong D, Zhang H, Zhang J. et al. A positive feed-forward loop between LncRNA-URRCC and EGFL7/P-AKT/FOXO3 signaling promotes proliferation and metastasis of clear cell renal cell carcinoma. Molecular cancer. 2019;18:81
14. Li JK, Chen C, Liu JY, Shi JZ, Liu SP, Liu B. et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Molecular cancer. 2017;16:111
15. Carril-Ajuria L, Santos M, Roldán-Romero JM, Rodriguez-Antona C, de Velasco G. Prognostic and Predictive Value of PBRM1 in Clear Cell Renal Cell Carcinoma. Cancers. 2019 12
16. Barth DA, Slaby O, Klec C, Juracek J, Drula R, Calin GA. et al. Current Concepts of Non-Coding RNAs in the Pathogenesis of Non-Clear Cell Renal Cell Carcinoma. Cancers. 2019 11
17. Mehdi A, Riazalhosseini Y. Epigenome Aberrations: Emerging Driving Factors of the Clear Cell Renal Cell Carcinoma. International journal of molecular sciences. 2017 18
18. Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature reviews Genetics. 2002;3:415-28
19. Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38-55
20. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223-7
21. Wu SC, Kallin EM, Zhang Y. Role of H3K27 methylation in the regulation of lncRNA expression. Cell research. 2010;20:1109-16
22. Amin V, Harris RA, Onuchic V, Jackson AR, Charnecki T, Paithankar S. et al. Epigenomic footprints across 111 reference epigenomes reveal tissue-specific epigenetic regulation of lincRNAs. Nature communications. 2015;6:6370
23. Wang G, Zhang ZJ, Jian WG, Liu PH, Xue W, Wang TD. et al. Novel long noncoding RNA OTUD6B-AS1 indicates poor prognosis and inhibits clear cell renal cell carcinoma proliferation via the Wnt/β-catenin signaling pathway. Molecular cancer. 2019;18:15
24. Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y. et al. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer cell. 2018;33:706-20.e9
25. Lu C, Wei Y, Wang X, Zhang Z, Yin J, Li W. et al. DNA-methylation-mediated activating of lncRNA SNHG12 promotes temozolomide resistance in glioblastoma. Molecular cancer. 2020;19:28
26. Yang W, Zhang K, Li L, Ma K, Hong B, Gong Y. et al. Discovery and validation of the prognostic value of the lncRNAs encoding snoRNAs in patients with clear cell renal cell carcinoma. Aging. 2020;12:4424-44
27. Li D, Jiang X, Zhang X, Cao G, Wang D, Chen Z. Long noncoding RNA FGD5-AS1 promotes colorectal cancer cell proliferation, migration, and invasion through upregulating CDCA7 via sponging miR-302e. In vitro cellular & developmental biology Animal. 2019;55:577-85
28. Liu L, Zhan Y, Huang Y, Huang L. LncRNA FGD5-AS1 can be predicted as therapeutic target in oral cancer. Journal of oral pathology & medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 2020;49:243-52
29. Fan Y, Li H, Yu Z, Dong W, Cui X, Ma J. et al. Long non-coding RNA FGD5-AS1 promotes non-small cell lung cancer cell proliferation through sponging hsa-miR-107 to up-regulate FGFRL1. Bioscience reports. 2020 40
30. Gao J, Zhang Z, Su H, Zong L, Li Y. Long Noncoding RNA FGD5-AS1 Acts as a Competing Endogenous RNA on microRNA-383 to Enhance the Malignant Characteristics of Esophageal Squamous Cell Carcinoma by Increasing SP1 Expression. Cancer management and research. 2020;12:2265-78
31. Xu L, Huan L, Guo T, Wu Y, Liu Y, Wang Q. et al. LncRNA SNHG11 facilitates tumor metastasis by interacting with and stabilizing HIF-1α. Oncogene. 2020;39:7005-18
Author contact
![]() Corresponding authors: Dr. Yanqing Gong, No. 8, Xishiku Street, Xicheng District, Beijing 100034, China. E-mail: yqgongedu.cn; Dr. Kan Gong, No. 8, Xishiku Street, Xicheng District, Beijing 100034, China. E-mail: gongkan_pkucom; Tel: +86-010-83575101; Fax: +86-010-66551122.
Corresponding authors: Dr. Yanqing Gong, No. 8, Xishiku Street, Xicheng District, Beijing 100034, China. E-mail: yqgongedu.cn; Dr. Kan Gong, No. 8, Xishiku Street, Xicheng District, Beijing 100034, China. E-mail: gongkan_pkucom; Tel: +86-010-83575101; Fax: +86-010-66551122.

 Global reach, higher impact
Global reach, higher impact