3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(7):1867-1883. doi:10.7150/jca.48979 This issue Cite
Research Paper
Screening and bioinformatical analysis of differentially expressed genes in nasopharyngeal carcinoma
1. Department of Radiation Oncology, Anhui Provincial Hospital Affiliated to Anhui Medical University, Hefei 230001, China.
2. Xinxiang Medical University, Xinxiang 453000, China.
3. Department of Radiation Oncology, the First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230031, China.
Abstract
Objective: To identify differentially expressed genes via bioinformatical analysis for nasopharyngeal carcinoma (NPC) and explore potential biomarkers for NPC.
Methods: We downloaded the NPC gene expression datasets (GSE40290, GSE53819) and obtained differentially expressed genes (DEGs) via GEO2R. Functional analysis of DEGs was performed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. In order to explore the interaction of DEGs and screen the core genes, we established protein-protein interaction (PPI) network. Then the expression level, prognostic and diagnostic analysis of the core genes in NPC were performed to reveal their potential effects on NPC. Furthermore, we obtained the transcription factors (TF) and microRNAs of core genes to construct the coregulatory network.
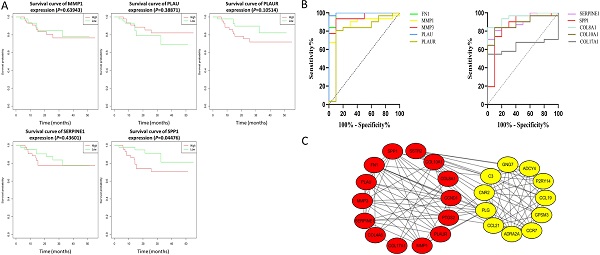
Results: We obtained 124 up-regulated genes and 190 down-regulated genes in total. These genes were found to be related to signal transduction, extracellular matrix organization and cell adhesion based on GO analysis. KEGG analysis revealed that the NF-kappa B (NF-κB) signaling pathway, pathways in cancer were mainly enriched signaling pathways. 25 core genes were obtained by constructing PPI network. Then the high expression of 10 core genes in NPC were verified via GEPIA, Oncomine databases and laboratory experiments. The TF-microRNA coregulatory network of the 10 core genes was built. Survival and diagnostic analysis indicated that SPP1 had negative influence on the prognosis of NPC patients based on two datasets and nine up-regulated core genes (FN1, MMP1, MMP3, PLAU, PLAUR, SERPINE1, SPP1, COL8A1, COL10A1) might be diagnostic markers for NPC.
Conclusions: Core genes of NPC were screened out by bioinformatical analysis in the present study and these genes may serve as prognostic and diagnostic biomarkers for NPC.
Keywords: nasopharyngeal carcinoma, core genes, bioinformatical analysis, prognosis, microarray

 Global reach, higher impact
Global reach, higher impact