3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(5):1445-1454. doi:10.7150/jca.50716 This issue Cite
Research Paper
Detection of Pancreatic Ductal Adenocarcinoma by A qPCR-based Normalizer-free Circulating Extracellular Vesicles RNA Signature
1. Department of Gastroenterology, Beijing Friendship Hospital, Capital Medical University, National Clinical Research Center for Digestive Disease, Beijing Digestive Disease Center, Beijing Key Laboratory for Precancerous Lesion of Digestive Disease, Beijing, 100050, P. R. China.
Abstract
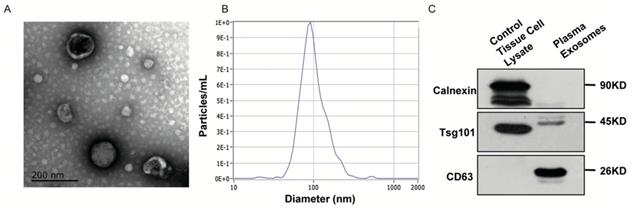
Background: Pancreatic ductal adenocarcinoma (PDAC) is difficult to diagnose and many efforts have been made to evaluate EVs-derived RNAs as biomarkers to predict PDAC. However, lack of robust internal references largely limited their clinical application. Here we proposed an RNA ratio-based, normalizer-free algorithm to quantitate EVs-derived RNAs in PDAC.
Methods: Differentially expressed RNAs in the training group were identified using “limma” package. The ratio of any two candidate RNAs in the same sample was calculated and used as a new biomarker. LASSO regression was performed to build prediction models based on those RNA ratios. RNA-seq data of 116 plasma samples and RT-qPCR data of 111 plasma samples were used for internal and external validation, separately. Three algorithms (lasso regression, logistic regression, and SVM) were compared to improve the performance of this RNA signature.
Results: We developed an RNA-ratio based prediction model which comprised eight EVs-derived RNAs, including FBXO7, MORF4L1, DDX17, TALDO1, AHNAK, TUBA1B, CD44, and SETD3. This model could well differentiate PDAC patients with a minimal AUC of 0.86 in internal verification using testing group. External validation using RT-qPCR data also exhibited a good classifier ability with an AUC of 0.89 when distinguishing PDAC from healthy controls.
Conclusion: We've developed a qPCR-based, normalizer-free circulating EVs RNA classifier, which could well distinguish PDAC patients from noncancerous controls.
Keywords: PDAC, EVs-derived RNAs, circulation biomarker, standardization, RT-qPCR

 Global reach, higher impact
Global reach, higher impact