Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(3):644-651. doi:10.7150/jca.49391 This issue Cite
Research Paper
Genomic and clinical characteristics of MET exon14 alterations in a large cohort of Chinese cancer patients revealed distinct features and a novel resistance mechanism for crizotinib
1. Thoracic Medicine Department 1, Hunan Cancer Hospital, Affiliated Tumor Hospital of Xiangya Medical School of Central South University, Changsha, Hunan, China.
2. Department of Thoracic Surgery, The Second Affiliated Hospital, Air Force Military Medical University, Xi'an, China.
3. Department of Research and Development, Nanjing Geneseeq Technology Inc., Nanjing, Jiangsu, China.
4. Translational Medicine Research Institute, Geneseeq Technology Inc., Toronto, Ontario, Canada.
5. Department of Thoracic Surgery, Shanghai General Hospital, Shanghai, China.
6. Department of Medical Oncology, Cancer Hospital of the University of Chinese Academy of Sciences, Zhejiang Cancer Hospital, Hangzhou, China.
7. Institute of Cancer and Basic Medicine (IBMC), Chinese Academy of Sciences, Hangzhou, China.
*These authors contributed equally to this work.
Received 2020-6-12; Accepted 2020-11-1; Published 2021-1-1
Abstract
Background: Alterations in MET exon 14 (METex14) and its flanking intronic regions have been identified in a variety of cancers. Patients with METex14 alterations often benefit from MET inhibitors such as crizotinib. Given the unique mutation profiles of Chinese lung cancer patients, it is necessary to investigate the prevalence of METex14 alterations in a large cohort of cancer patients.
Patients and methods: Cases carrying METex14 alterations were screened from 26,391 Chinese cancer patients by next-generation sequencing (NGS), and the clinicopathologic and molecular characteristics were reviewed.
Results: Compared to Western population (~3%), the frequency of METex14 alterations is much lower in Chinese cancer patients (0.7%, n=184) and lung cancer patients (1.1%, n=175). Seventy-eight distinct METex14 alterations, including several novel alteration types were detected. Concurrent MET copy gain and non-exon14 MET mutations were also found. EGFR copy gain (11%) and mutations (8%), KRAS (5%) and PIK3CA (5%), appeared in a mutually exclusive pattern. Female patients contain much less TP53 mutations than male patients (65% vs. 24%, FDR = 0.01). Co-amplification of CDK4 and MDM2, CDK6 and EGFR were identified, which indicated cell cycle dysregulation and EGFR alteration are important co-occurring features in patients with METex 14 alteration. Of 9 tissue specimens having PD-L1 immunohistochemistry (IHC) results, 5 of them (55.5%) were found PD-L1 positive, which is comparable to other types of tumor. In 14 crizotinib-treated patients, the median progression free survival (mPFS) was 7 months. Upon resistance to crizotinib, two patients acquired secondary mutations in MET and one patient acquired BRAF p.K601E that can be a novel resistance mechanism.
Conclusion: Chinese cancer patients have a relatively lower frequency of METex14 alterations compared to Western patients. Patients with METex14 alterations showed distinct molecular characteristics and the representative case study showed responses to MET tyrosine kinase inhibitor (TKI).
Keywords: MET exon 14 alterations, lung cancer, next-generation sequencing, crizotinib-resistant mutation
Introduction
The hepatocyte growth factor (HGF) receptor, encoded by the MET oncogene, is a receptor tyrosine kinase that activates a wide range of cellular signaling pathways to mediate cell proliferation, survival and motility, and subsequently induces cancer development and progression [1, 2]. Pathologic activation of MET were frequently caused by point mutations in tyrosine kinase domain, gene copy gains and protein overexpression [3, 4], and less frequently, by alterations affected the splicing of exon14, which resulted in MET exon14 (METex14) skipping after translation. Besides, loss of p.Y1003 in exon 14, a critical binding site for Cbl (an E3 ubiquitin ligase) will also disrupt MET ubiquitination and degradation, leading to the overactivation of MET signaling [5, 6]. METex14 skipping has been identified in a wide variety of human cancers with an incidence of around 3% in all cancer types [7]. The alterations are highly diversified, including point mutations at splicing sites, in-frame deletion of intronic region around splicing sites at various lengths and the large fragment deletion to remove the entire exon 14 [8, 9].
Preclinical and clinical evidences suggest that tumors with METex14 are sensitive to small molecule tyrosine kinase inhibitors (TKIs), including non-selective inhibitor crizotinib [10], and several selective inhibitors, such as tepotinib, savilitinib and capmatinib [11-13]. In 2020, FDA approved the first targeted therapy capmatinib for METex14-positive patients with locally advanced or metastatic non-small cell lung cancer (NSCLC).
With the advanced parallel sequencing technologies, it becomes much easier to screen for novel alterations in METex14 and comprehensively analyze the concurrent genomic alterations, thereby expand the number of druggable patients and uncover the intrinsic or acquired resistant mechanisms to TKI treatment. In this study, we retrospectively screened genomic data of 26,391 Chinese cancer patients and identified 184 cases with METex14 alterations in 10 different cancer types, among which 175 were lung cancer patients. Seventy-eight unique METex14 alterations were identified, including novel alterations c.2888-90_2899del, c.2888-55_2928del. comprehensively, genomic profiling also revealed concurrent and exclusive gene alterations in these patients. Clinical responses were observed in 14 patients who received crizotinib treatment and potential drug-resistance mechanisms were analyzed.
Materials and methods
Patients and samples
Genomic profiling results of 26,391 malignant tumors were screened and only samples with alterations in MET exon 14 or intron 13 and 14 that could potentially cause MET exon 14 skipping or the loss of MET p.Y1003 residue were analyzed. Genomic profiling of these samples was performed on formalin-fixed paraffin-embedded (FFPE) tumor/plasma biopsy specimens that were obtained from patients signed written informed consent.
Next-generation sequencing (NGS)
DNA extraction and sequencing library were prepared according to the protocols described previously [14, 15]. To be specific, 104 and 80 cancer samples were tested with 139-gene panel and 425-gene panel respectively, both of which cover the whole MET exon 14 and the adjacent intron regions, and all essential lung-cancer related genes. All samples were sequenced in a Clinical Laboratory Improvement Amendments (CLIA)- and College of American Pathologists (CAP)-certified genomic testing facility (Nanjing Geneseeq Technology Inc., Nanjing, China). Different types of genetic alterations were called using an internally-validated bioinformatics analysis pipeline [16]. Clinical information, including age at diagnosis, sex, disease stage, and treatment history was extracted from the medical records provided by physicians during the service order. Informed written consent was acquired from each patient at the time of sample submission. The study methodologies conformed to the standards set by the Declaration of Helsinki and was approved by the ethics committee.
Immunohistochemistry (IHC) staining of PD-L1
Eleven tissue biopsies of this study were performed IHC staining of PD-L1 expression with 22C3 anti-PD-L1 antibody (Dako) according to the protocol reported previously [17]. PD-L1 IHC was evaluated by a pathologist based on the tumor proportion score (TPS) with membranous and/or cytoplasmic staining, and divided into three groups: no-expression (<1% of tumor cells), low expression (1%-49%) and high expression (≥50%).
Data analysis and statistics
All statistical tests were conducted in R version 3.6.1. The concurrent mutations and exclusive/concurrent analysis were conducted with the SomaticInteraction function in Maftools package of R [18] and a p-value below 0.05 was considered as significant.
Results
Various METex14 alterations were identified in different cancer types
METex14 alterations were identified in 184 of 26,391 (0.7%) patients from 9 cancer types (Supplementary Fig. S1), including lung cancer (1.1%, 175/18112, Table 1), neuroendocrine tumor (NET, 1/85), bladder cancer (BLCA, 1/68), breast cancer (BRCA, 1/3639), colorectal cancer (CRC, 2/2551), pancreatic cancer (PACA, 1/661), esophageal cancer (ESCA, 1/549), cholangiocarcinoma (CHOL, 1/547) and melanoma (1/179). The frequency of METex14 alterations in Chinese patients is lower than 3% in Western countries [19]. In lung cancer patients with METex14 alterations, 79.4% (139/175) were non-small-cell lung cancer (NSCLC), including adenocarcinoma (ADC, 114/139, 82.0%), adenosquamous carcinoma (ASC, 9/139, 6.5%), squamous cell carcinoma (SCC, 8/139, 5.8%), large cell carcinoma (LCC, 1/139, 0.7%) and subtype-undetermined NSCLC (7/326, 2.1%) (Table 1). Thirty-six patients (36/175) were histologically undetermined.
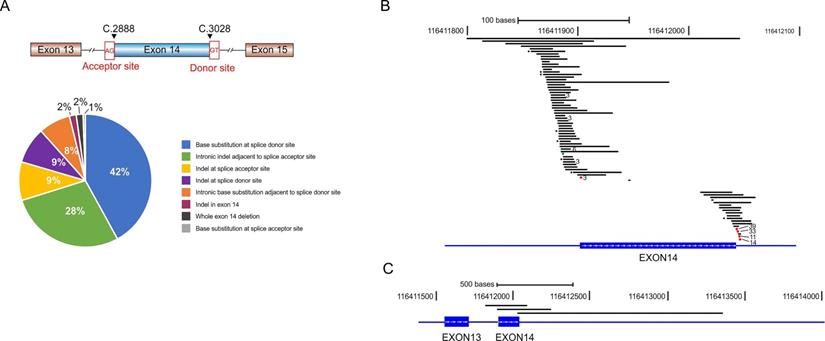
METex14 alterations comprise base substitution, insertion, and large fragment deletion spanning the entire or partial region of exon 14. We identified 78 distinct METex14 alterations and classified them into 8 subgroups according to the alteration types and locations (Fig. 1A and Supplementary Table S1). Base substitutions at the splicing donor sites were the most common alterations (42% of all METex14 alterations), represented by c.3082G > T/A/C (39/184, 21.2%), c.3082 + 1G > C/A/T (33/184, 17.9%), c.3082 + 2T > A/C/G (11/184, 6.0%) and c.3082 + 3A > T/C/G (14/184, 7.6%) (Fig. 1B, Supplementary Table S1). We also identified a great amount of indel alterations spanning the ~50 bp intronic upstream region of the splice acceptor site (28%), and base substitutions immediately adjacent to the splice donor site (8%, Fig. 1A and 1B). Three cases have large fragment deletion that removed the entire METex14 (Fig. 1C). There are also three insertion/deletions (indels) within METex14 that directly disrupt the ubiquitin ligase site (p.Y1003), including MET p.E999_P1008del, p.T1006_P1008del and p.D1002_Y1003del (Supplementary Table S1). Additionally, three patients harbored two different METex14 alteration types respectively (Supplementary Table S2).
Clinical characteristics and genomic profiling of lung cancer with METex14 alterations
The prevalence of METex14 alterations was much higher in ASC (6.25%) than other subtypes, including LCC (2.7%), ADC (1.0%) and SCC (0.7%) (Table 1). The median age of lung cancer patients with METex14 was 68 (range from 39 to 94), and females and males take up 48.6% (n=86) and 47.4% (n=84), respectively.
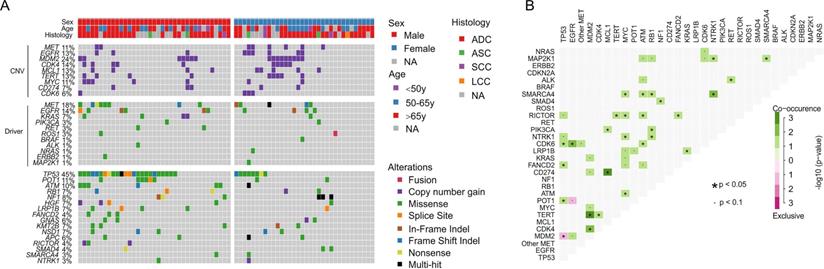
Genomic profiling of 175 lung cancer tumors revealed the high prevalence of TP53 alterations (43%) and relatively low co-occurrence of other cancer driver alterations, such as EGFR alterations (19%, Supplementary Fig. S2), which is much less than 40~60% in Asian lung cancer patients [20, 21]. The secondary MET alterations were detected in 15% of patients and 11% of them have MET copy gain (Supplementary Fig. S2). For 71 cases that were examined with the 425-gene panel, mutation frequencies of different genes were comparable between females and males, except that male patients were significantly more enriched with TP53 alterations than females (65% vs. 24%, FDR = 0.01, Fig. 2A). We observed a subpopulation that were prevalent with gene copy gains in MDM2 (24%), CDK4 (14%), MCL1 (13%), TERT (13%), MYC (11%) and CDK6 (6%), and showed a tendency of co-occurrence, e.g. MDM2 with TERT and CDK4 (p < 0.05, Fig. 2). Moreover, CD274 (PD-L1), the biomarker for immunotherapy, was also found to be amplified in 7% of cases and mostly co-existed with MCL1 copy gain (p < 0.05, Fig. 2B). EGFR copy gain and mutations were also identified in 13% and 15% patients, which is higher than previously reported frequency in Western lung cancer patients [9]. POT1, a component that binds and protects telomere [22], was mutated in 11% of cases and likely to co-occur with TP53 alterations (p < 0.05, Fig. 2B). The only significantly exclusive gene pair was TP53 and MDM2, a negative regulator of TP53 (p < 0.05). These data suggested the dysregulation of cell cycle and EGFR signaling pathway in tumorigenesis of patients with METex 14 alteration.
Comparatively, gene alterations in 9 non-lung cancer cases revealed a different mutational spectrum, including a much higher occurrence of TP53 alterations (89%), and the absence of MDM2/CDK4 amplification. Moreover, KRAS mutations and non-METex14 alterations were found in 44% of cases (Supplementary Fig. S3), suggesting the companion mutations might be different across cancer types.
Potential treatment strategies for patients with METex14 alterations
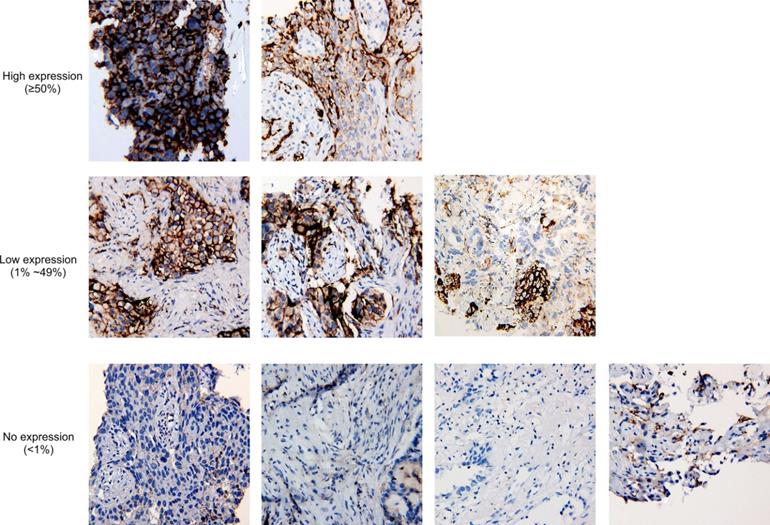
It has been reported that MET amplification was associated with increased PD-L1 expression [23]. For 9 tissue biopsies having available PD-L1 IHC staining results, 5 samples were PD-L1 positive, including two high-expression samples and three low-expression samples (Fig. 3), and the frequency is comparable to the overall positive ratio in lung cancer [24], which shows no correlation between METex14 alteration and PD-L1 overexpression.
Various METex14 alterations and their schematic locations around exon 14. A) Pie chart shows the frequency of different METex14 alterations that were grouped by the locations and mutation types. B-C) Schematic view of each METex14 alteration on human genome build GRCh37/hg19. The frequency of each METex14 are indicated with * for two and the number of cases for greater than two, as labeled behind.
Comprehensive genomic profiling of 71 lung cancer patients sequenced by 425-gene panel. A) Co-mutation plot of clinically relevant and frequently altered genes. B) Concurrence and exclusive analysis of gene alterations. A p-value < 0.05 is considered statistically significant.
Clinical and molecular characteristics of lung cancer patients harboring METex14 skipping
| Total lung cancer | NSCLC | Lung cancer with unknown histological subtype | |||||
|---|---|---|---|---|---|---|---|
| ADC | SCC | ASC | LCC | Subtype-undetermined NSCLC | |||
| Cases with MET Exon14 skipping, n/N (%) a | 175/18112 (1.1) | 114/11208 (1.0) | 8/1133 (0.7) | 9/144 (6.25) | 1/37 (2.7) | 7/326 (2.1) | 36/4851 (0.7) |
| Median age (range), y | 67 (39-94) | 68 (43-94) | 74 (55-81) | 70 (39-83) | 61 (61-61) | 64 (56-73) | 74 (49-87) |
| Sex, n (%) | |||||||
| Male | 71 (51.1) | 58 (50.9) | 4 (50.0) | 5 (55.6) | 0 | 4 (57.1) | 15 (41.7) |
| Female | 68 (48.9) | 56 (49.1) | 4 (50.0) | 4 (44.4) | 1 (100.0) | 3 (42.9) | 21 (58.3) |
| Stage, n (%) | |||||||
| I-II | 9(6.5) | 8(7.0) | 0 | 1 (11.1) | 0 | 0 | 1 (2.7) |
| III | 7 (5.0) | 5 (4.4) | 0 | 1 (11.1) | 0 | 1 (14.3) | 0 |
| IV | 29 (20.9) | 24 (21.1) | 1 (12.5) | 2 (22.2) | 0 | 3 (42.9) | 4 (11.1) |
| Unknown | 94 (67.6) | 77 (67.5) | 7 (87.5) | 5 (55.5) | 1 (100.0) | 3 (42.9) | 31 (86.1) |
| Smoking history, n (%) | |||||||
| Yes | 5 (3.6) | 3 (2.6) | 0 | 1 (11.1) | 0 | 1 (14.3) | 2 (5.6) |
| No | 17 (12.2) | 15 (13.2) | 0 | 1 (11.1) | 0 | 1 (14.3) | 3 (8.3) |
| Unknown | 117 (84.2) | 96 (84.2) | 8 (100.0) | 7 (77.7) | 1 (100.0) | 5 (71.4) | 31 (86.1) |
a, n/N represents the occurrence of MET exon 14 skipping and the total number of patients in each subcategory.
Tumor proportion score of PD-L1 in nine cases with PD-L1 immunohistochemistry staining results available. At 20x magnification, no expression of PD-L1 is defined as <1% of tumor cells in the sample, while low expression and high expression are defined by 1%-49% of tumor cells and ≥50% of tumor cells, respectively.
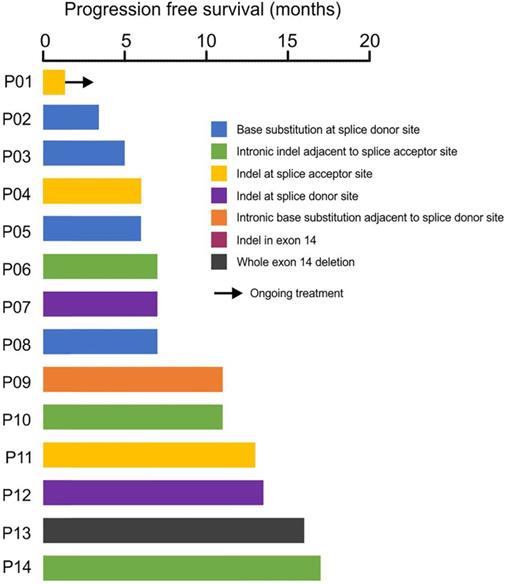
Clinical treatment records showed that 14 patients have received crizotinib treatment, and the median progress-free survival (mPFS) was 7 months with the longest PFS of 17 months (Fig. 4). Four patients (P06, P09, P10, P14) with METex14 alterations in the intronic region adjacent to splice acceptor site showed durable response to crizotinib treatment, suggesting the loss of exon 14 by these alterations (Fig. 4). Acquired secondary mutations, including MET p.Y1230N, p.D1228N and BRAF p.K601E were identified in the available post-treatment tumors from three patients (P05, P12, P14, Fig. 4, Supplementary Table S3).
Patient P05 was a 60-year-old female diagnosed with advanced lung adenocarcinoma, and was carrying a METex14 skipping mutation (c.3028+1G>C). After 4 months of crizotinib treatment, the disease progressed and a BRAF p.K601E mutation was identified. BRAF p.K601E is an activating mutation occurred in 0.15% of all NSCLC patients and showed low sensitivity to BRAF-targeted therapy in clinical studies [25-29], which could mediate crizotinib resistance in treatment.
Patient P12 was a 65-year-old female diagnosed with stage IV lung adenocarcinoma and underwent surgery after diagnosis. Disease was relapsed with bone metastasis after 12 months and a METex14 skipping alteration (c.2888-88_2901del) was detected. The patient was subjected to crizotinib treatment for 13 months, and a secondary mutation MET p.Y1230N was identified in plasma sample after resistance.
Patient P14 was a 61-year-old female and diagnosed with stage IV non-small cell lung cancer. She harbored MET c.2888-22_2888-8del and was subjected to crizotinib for 17 months. Post-treatment sample was detected with MET p.D1228N mutation.
Discussion
This is the largest cohort study of METex14 alterations in a variety of cancer types. The incidence of METex14 alterations in Chinese lung cancer patients was 1.1%, which is lower than 3% in Western countries [8, 9], but its frequency in lung adenosquamous carcinoma is higher (6.5% vs 2.8%) [8], suggesting different mutation frequency of METex14 between Asian and Western lung cancer patients. We also identified an LCC patient carried METex14 alteration, which has not been reported. There is no significant difference of sex in our data, differing from the Western people (male vs. female=39.6% vs 60.4%) [9].
Response of crizotinib-treatment in 14 patients with METex14 skipping. The x-axis showed the patient ID and the y-axis showed the progression-free survival (PFS) in months.
Seventy-eight unique METex14 alterations were identified and some of them have not been reported before. Aside from base substitutions of splice acceptor and donor sites [9, 19], insertion/deletions within/across intron13, exon 14 and intron 15 at different lengths were observed. In our data, the frequency of patients with deletion of entire METex14 is higher than the Western countries (2% vs. 0.7%) [8, 9] . A number of patients were also carrying other driver mutations, such as copy gain of MET and EGFR, non-exon14 MET mutations and EGFR mutations, suggesting that other driving forces in addition to METex14 alteration might be required for tumor initiation and evolvement.
Similar as previous reports, we observed copy gain of MDM2, CDK4, TERT, MYC and MCL1 at different frequencies, but with a tendency of co-occurring. MDM2 is a negative regulator of TP53 by mediating TP53's degradation [30], and its amplification has been identified in a variety of cancers at a frequency of ~3%, which might have a potential role in treatment resistance of prostate cancer, neuroblastoma and lung cancer [31-33]. Here, we found that MDM2 amplification is likely to be exclusive with TP53 alterations, possibly because the functional overlap between these two genes. We also observed co-occurrent of copy number gain of MDM2 with cell cycle-dependent kinase CDK4. MDM2 and CDK4 are frequently co-amplified in NSCLC [34] and sarcomas, and play crucial roles in tumorigenesis via increasing cell growth and migration [35]. There are statistically significant associations between copy number gain of EGFR and CDK6, which is consistent with a previous study in glioma [36]. These data suggest cell cycle dysregulation and EGFR alteration are important co-occurring features in patients with METex 14 alteration. Since inhibitors to cell cycle are approved or clinically tested for therapy, patients with METex 14 alteration may have better outcome from treatment targeting both MET and cell cycle. Studies have shown that the down-regulation of CD274 (PD-L1) and the apoptosis gene MCL1 is synchronized [37]. Our data shows here a co-occurrence of copy number amplification between PDL1 and MCL1, suggesting that there may be some positive regulatory relationship between the two genes.
Lastly, in 14 patients who were treated with crizotinib, the PFS varied from 4 months to as long as 17 months. Two patients acquired secondary MET mutations, including MET p.Y1230N and p.D1228N, which have been reported as the potential resistance mechanisms after treating METex14 skipping with crizotinib [38-40]. Additionally, acquired BRAF p.K601E was found in another crizotinib-resistance patient with only 4-month PFS. Similar to BRAF p.V600E, BRAF p.K601E is also an activating mutation of BRAF resulting in continuous activation of MEK/ERK signaling pathway [41, 42], therefore can be considered as a bypass resistance mechanism.
Conclusion
The incidence of METex14 skipping was lower in Chinese cancer patients than Western cancer patients, but its prevalence in lung adenocarcinoma is higher than Western patients. The alteration is highly diversified and deep into the intron region. Therefore, it requires precaution when choosing the right test for it. We observed treatment efficacy of crizotinib in some patients and reported potential resistance mechanisms in a few cases. Several MET inhibitors, including crizotinib, are currently under evaluation for the treatment of NSCLC patients with METex14 skipping and capmatinib was recently approved by FDA to treat this alteration in NSCLC. Along with the increased use of these small molecule TKIs in treatment, more acquired resistance mechanisms will be investigated.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
Funding
This work was supported by Zhejiang Provincial Natural Science Foundation of China [grant number: LY17H160040].
Data Availability Statement
All data generated or used during the study are available from the corresponding author by reasonable request.
Competing Interests
Danni Song and Sisi Liu are the employees of Nanjing Geneseeq Technology Inc., China; Xiaoling Tong and Xue Wu are the employees of Geneseeq Technology Inc., Canada. Remaining authors declare no conflict of interest.
References
1. Organ SL, Tsao MS. An overview of the c-MET signaling pathway. Therapeutic advances in medical oncology. 2011;3:S7-S19
2. Van Der Steen N, Giovannetti E, Pauwels P, Peters GJ, Hong DS, Cappuzzo F. et al. cMET exon 14 skipping: from the structure to the clinic. Journal of Thoracic Oncology. 2016;11:1423-32
3. Maroun CR, Rowlands T. The Met receptor tyrosine kinase: a key player in oncogenesis and drug resistance. Pharmacology & therapeutics. 2014;142:316-38
4. Ma PC, Tretiakova MS, MacKinnon AC, Ramnath N, Johnson C, Dietrich S. et al. Expression and mutational analysis of MET in human solid cancers. Genes, chromosomes & cancer. 2008;47:1025-37
5. Drilon A. MET exon 14 alterations in lung cancer: exon skipping extends half-life. Clinical Cancer Research. 2016;22:2832-4
6. Kong-Beltran M, Seshagiri S, Zha J, Zhu W, Bhawe K, Mendoza N. et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer research. 2006;66:283-9
7. Reungwetwattana T, Liang Y, Zhu V, Ou SI. The race to target MET exon 14 skipping alterations in non-small cell lung cancer: The why, the how, the who, the unknown, and the Inevitable. Lung cancer. 2017;103:27-37
8. Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, Bauer TM. et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer discovery. 2015;5:850-9
9. Schrock AB, Frampton GM, Suh J, Chalmers ZR, Rosenzweig M, Erlich RL. et al. Characterization of 298 patients with lung cancer harboring MET exon 14 skipping alterations. Journal of Thoracic Oncology. 2016;11:1493-502
10. Drilon A, Clark JW, Weiss J, Ou SI, Camidge DR, Solomon BJ. et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nature medicine. 2020;26:47-51
11. Paik PK, Felip E, Veillon R, Sakai H, Cortot AB, Garassino MC. et al. Tepotinib in non-small-cell lung cancer with MET exon 14 skipping mutations. The New England journal of medicine. 2020;383:931-43
12. Lu S, Fang J, Cao L, Li X, Guo Q, Zhou J. et al. Abstract CT031: Preliminary efficacy and safety results of savolitinib treating patients with pulmonary sarcomatoid carcinoma (PSC) and other types of non-small cell lung cancer (NSCLC) harboring MET exon 14 skipping mutations. Cancer research. 2019;79:CT031
13. Wolf J, Seto T, Han J-Y, Reguart N, Garon EB, Groen HJM. et al. Capmatinib (INC280) in METΔex14-mutated advanced non-small cell lung cancer (NSCLC): Efficacy data from the phase II GEOMETRY mono-1 study. Journal of Clinical Oncology. 2019;37:9004
14. Yang Z, Yang N, Ou Q, Xiang Y, Jiang T, Wu X. et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clinical Cancer Research. 2018;24:3097-107
15. Fang W, Ma Y, Yin JC, Hong S, Zhou H, Wang A. et al. Comprehensive genomic profiling identifies novel genetic predictors of response to anti-PD-(L)1 therapies in non-small cell lung cancer. Clinical Cancer Research. 2019;25:5015-26
16. Tong L DN, Tong X. et al. Tumor-derived DNA from pleural effusion supernatant as a promising alternative to tumor tissue in genomic profiling of advanced lung cancer. Theranostics. 2019;9:5532-41
17. Roach C, Zhang N, Corigliano E, Jansson M, Toland G, Ponto G. et al. Development of a companion diagnostic PD-L1 immunohistochemistry assay for pembrolizumab therapy in non-small-cell lung cancer. Applied Immunohistochemistry & Molecular Morphology. 2016;24:392-7
18. Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome research. 2018;28:1747-56
19. Awad MM, Oxnard GR, Jackman DM, Savukoski DO, Hall D, Shivdasani P. et al. MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. Journal of Clinical Oncology. 2016;34:721-30
20. Shigematsu H, Lin L, Takahashi T. et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. Journal of the National Cancer Institute. 2005;97:339-46
21. Thomas C. et al. A genomics-based classification of human lung tumors: The Clinical Lung Cancer Genome Project (CLCGP) and Network Genomic Medicine (NGM). Science translational medicine. 2013;5:209ra153
22. Palm W, de Lange T. How shelterin protects mammalian telomeres. Annual review of genetics. 2008;42:301-34
23. Martin V, Chiriaco C, Modica C, Acquadro A, Cortese M, Galimi F. et al. Met inhibition revokes IFNgamma-induction of PD-1 ligands in MET-amplified tumours. British journal of cancer. 2019;120:527-36
24. Sabari JK, Leonardi GC, Shu CA, Umeton R, Montecalvo J, Ni A. et al. PD-L1 expression, tumor mutational burden, and response to immunotherapy in patients with MET exon 14 altered lung cancers. Annals of Oncology. 2018;29:2085-91
25. Tabernero J, Shapiro GI, LoRusso PM, Cervantes A, Schwartz GK, Weiss GJ. et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer discovery. 2013;3:406-17
26. Sheikine Y, Pavlick D, Klempner SJ, Trabucco SE, Chung JH, Rosenzweig M. et al. BRAF in lung cancers: analysis of patient cases reveals recurrent BRAF mutations, fusions, kinase duplications, and concurrent alterations. JCO Precision Oncology. 2018;2:1-15
27. Gautschi O, Milia J, Cabarrou B, Bluthgen M-V, Besse B, Smit EF. et al. Targeted therapy for patients with BRAF-mutant lung cancer results from the European EURAF cohort. Journal of Thoracic Oncology. 2015;10:1451-7
28. Voskoboynik M, Mar V, Mailer S, Colebatch A, Fennessy A, Logan A. et al. Clinicopathological characteristics associated with BRAF(K601E) and BRAF(L597) mutations in melanoma. Pigment cell & melanoma research. 2016;29:222-8
29. Falchook GS, Long GV, Kurzrock R, Kim KB, Arkenau TH, Brown MP. et al. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: a phase 1 dose-escalation trial. Lancet. 2012;379:1893-901
30. Bose I, Ghosh B. The p53-MDM2 network: from oscillations to apoptosis. Journal of biosciences. 2007;32:991-7
31. Bianco R, Caputo R, Caputo R, Damiano V, De Placido S, Ficorella C. et al. Combined targeting of epidermal growth factor receptor and MDM2 by gefitinib and antisense MDM2 cooperatively inhibit hormone-independent prostate cancer. Clinical cancer research. 2004;10:4858-64
32. Wang HQ, Halilovic E, Li X, Liang J, Cao Y, Rakiec DP. et al. Combined ALK and MDM2 inhibition increases antitumor activity and overcomes resistance in human ALK mutant neuroblastoma cell lines and xenograft models. eLife. 2017;6:e17137
33. Kim Y, Lee B, Shim JH, Lee SH, Park WY, Choi YL. et al. Concurrent genetic alterations predict the progression to target therapy in EGFR-mutated advanced NSCLC. Journal of thoracic oncology. 2019;14:193-202
34. Yamaura T, Muto S, Mine H, Takagi H, Watanabe M, Ozaki Y. et al. Genetic alterations in epidermal growth factor receptor-tyrosine kinase inhibitor-naïve non-small cell lung carcinoma. Oncology Letters. 2020;19:4169-76
35. Kim YJ, Kim M, Park HK, Yu DB, Jung K, Song K. et al. Co-expression of MDM2 and CDK4 in transformed human mesenchymal stem cells causes high-grade sarcoma with a dedifferentiated liposarcoma-like morphology. Laboratory Investigation. 2019;99:1309-20
36. Zhou J, Wu Z, Wong G, Pectasides E, Nagaraja A, Stachler M. et al. CDK4/6 or MAPK blockade enhances efficacy of EGFR inhibition in oesophageal squamous cell carcinoma. Nature communications. 2017;8:1-12
37. Tremblay-LeMay R, Rastgoo N, Chang H. Modulating PD-L1 expression in multiple myeloma: an alternative strategy to target the PD-1/PD-L1 pathway. Journal of hematology & oncology. 2018;11:46
38. Heist RS, Sequist LV, Borger D, Gainor JF, Arellano RS, Le LP. et al. Acquired resistance to crizotinib in NSCLC with MET exon 14 skipping. Journal of Thoracic Oncology. 2016;11:1242-5
39. Ou S-HI, Young L, Schrock AB, Johnson A, Klempner SJ, Zhu VW. et al. Emergence of preexisting MET Y1230C mutation as a resistance mechanism to crizotinib in NSCLC with MET exon 14 skipping. Journal of Thoracic Oncology. 2017;12:137-40
40. Dong H-J, Li P, Wu C-L, Zhou X-Y, Lu H-J, Zhou T. Response and acquired resistance to crizotinib in Chinese patients with lung adenocarcinomas harboring MET Exon 14 splicing alternations. Lung cancer. 2016;102:118-21
41. Tissot C, Couraud Sb, Tanguy R, Bringuier P-P, Girard N, Souquet P-J. Clinical characteristics and outcome of patients with lung cancer harboring BRAF mutations. Lung cancer. 2016;91:23-8
42. Dahlman KB, Xia J, Hutchinson K, Ng C, Hucks D, Jia P. et al. BRAF L597 mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer discovery. 2012;2:791-7
Author contact
![]() Corresponding authors: Dr Wei Hong, Department of Medical Oncology, Zhejiang Cancer Hospital, Guangji Road 38, Hangzhou, China 310022. Tel: +86-0571-88122215; Fax: +86-0571-88122215; E-mail: hongweiorg.cn; Dr Zhifeng Lin, Department of Thoracic Surgery, Shanghai General Hospital, Xinsongjiang Road 650, Shanghai, China 201620. Tel: +86-18017690870; Fax: +86-021-63240090; E-mail: linzhifeng2611com.
Corresponding authors: Dr Wei Hong, Department of Medical Oncology, Zhejiang Cancer Hospital, Guangji Road 38, Hangzhou, China 310022. Tel: +86-0571-88122215; Fax: +86-0571-88122215; E-mail: hongweiorg.cn; Dr Zhifeng Lin, Department of Thoracic Surgery, Shanghai General Hospital, Xinsongjiang Road 650, Shanghai, China 201620. Tel: +86-18017690870; Fax: +86-021-63240090; E-mail: linzhifeng2611com.

 Global reach, higher impact
Global reach, higher impact