Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(21):6376-6389. doi:10.7150/jca.46309 This issue Cite
Research Paper
PRELP has prognostic value and regulates cell proliferation and migration in hepatocellular carcinoma
Department of General Surgery, The Fifth People's Hospital of Shanghai, Fudan University, Shanghai, 200240, P.R. China.
*These authors contributed equally to this work.
Received 2020-3-24; Accepted 2020-8-11; Published 2020-9-9
Abstract
Purpose: Hepatocellular carcinoma (HCC) is an aggressive and prevalent tumor threatening human health. A previous study suggested low PRELP (proline/arginine-rich end leucine-rich repeat protein) expression was associated with poor patient survival in pancreatic ductal adenocarcinoma (PDAC). However, the role of PRELP in HCC has not yet been illuminated.
Methods: PRELP expression analyses were carried out using transcriptomic datasets from the Integrative Molecular Database of Hepatocellular Carcinoma (HCCDB). The correlations between PRELP expression and clinicopathological features, and prognostic analyses were performed with a tissue microarray (TMA) and immunohistochemistry (IHC). The endogenous expression and in vitro roles of PRELP were investigated in cultured HCC cell lines. The potential mechanisms were characterized by a Gene Set Enrichment Analysis (GSEA) and gene-gene correlation analyses.
Results: We found that PRELP mRNA expression was dramatically decreased in HCCs in comparison with that in adjacent normal tissues (NTs) or hepatic cirrhosis. IHC staining showed that PRELP was down-regulated in HCCs, which mainly located in cytoplasm, and was also found in nuclei. The correlation analyses revealed that PRELP expression was relevant to later p-stages (p= 0.028) and tumor size (p= 0.001). The overall survival (OS) and relapse free survival (RFS) time was shorter in HCC patients with lower PRELP expression levels than that with higher PRELP expression levels. Overexpression of PRELP inhibited, while knockdown of PRELP promoted proliferation and migration of HCC cells. For potential mechanisms, PRELP may inhibit progression of HCCs by interacting with integrin family members and the extracellular microenvironment.
Conclusion: Our findings demonstrated that overexpression of PRELP correlates with better patient survival and inhibits both cell proliferation and migration in HCC. Therefore, PRELP can serve as a potential prognostic biomarker and therapeutic target which deserves further investigation.
Keywords: PRELP, hepatocellular carcinoma, proliferation, prognosis, migration, integrin
Introduction
Hepatocellular carcinoma (HCC) comprises 75%-85% of liver cancer cases and is one of the most fatal solid malignancies worldwide, with an extremely low 5‑year survival rate [1]. The main causes are chronic infection with hepatic B virus (HBV) and exposure to aflatoxin in China. Although prevention measures and interventions like HBV vaccination, health education and surveillance have been put into practice, a considerable part of patients are diagnosed at later stages where surgical resection is not recommended [2]. The survival rate of these people has been slightly improved by applying targeted drugs such as sorafenib or regorafenib, a multikinase inhibitor mainly against tumor angiogenesis [3]. It is urgent to better know the molecular mechanisms of this malignancy, which would provide creative and effective therapeutic strategies to ease the burden of HCC.
PRELP (proline/arginine-rich end leucine-rich repeat protein), which is highly abundant in collagen-rich tissues, binds collagen to basement membranes and cartilage [4, 5]. PRELP belongs to the class II subfamily of the small leucine-rich proteoglycan (SLRP) family and is also known as MST161, MSTP161, prolargin, prolargin proteoglycan or SLRR2A[6]. The neopeptide, which is released from PRELP, is involved in progression and incidence of osteoarthritis (OA) [7, 8]. By modulating the β-catenin/connexin43 pathway, PRELP participates in the formation of new bone by osteoblasts [7]. In human congenital immunity processes, PRELP could bind with respiratory tract pathogens and impede their adherence to lung epithelial cells [9]. A previous study showed that fibroblast adhesion was enhanced on the basis of the binding between N-terminal of PRELP and integrin [10]. Furthermore, peptides corresponding to the N‐terminal heparin binding domain of PRELP act as antimicrobial molecules by binding bacterial surface [11]. The physiological roles of PRELP in liver still need to be elucidated, however, data from The Human Protein Atlas (HPA) shows that PRELP is secreted to the extracellular matrix and may anchor basement membranes to the underlying connective tissue [12], suggesting its potential function in maintaining normal cellular structure.
Studies of PRELP in human cancers have been reported previously. In a proteomic study, the authors demonstrated that PRELP overexpression was associated with poor patient survival in resectable pancreatic ductal adenocarcinomas (PDAC) [13]. In another study conducted in human brain tumors, the authors documented only limited potential of PRELP as a prognostic predictor [14]. Besides, data mining and bioinformatics analyses seemed unable to elicit the role of PRELP in bladder urothelial carcinoma [15]. While most of these are bioinformatics studies, the role of PRELP in HCC remains unknown. The Integrative Molecular Database of Hepatocellular Carcinoma (HCCDB), which collected fifteen public HCC expression datasets, is established to analyze the differential expression of genes conveniently and efficiently [16]. Hence, we utilized this database to investigate the expression of PRELP in HCC at the beginning.
In this study, we compared PRELP mRNA expression levels between HCCs and adjacent normal tissues (NTs) or hepatic cirrhosis by analyzing several public datasets. We utilized a tissue microarray (TMA) and immunohistochemistry (IHC) to investigate the prognostic value of PRELP in HCCs. We studied the in vitro activities of PRELP in cultured HCC cells using proliferation and migration assays. Besides, we explored the potential mechanisms of PRELP based on the Gene Set Enrichment Analysis (GSEA) and gene-gene correlation analyses.
Materials and Methods
Data collection
Fifteen transcriptome datasets were accessed from HCCDB [16], which is used to analyze the differential gene expression. These datasets include GSE14520 [17, 18], GSE63898 [19], GSE54236 [20], GSE64041 [21], GSE22058 [22], GSE112790 [23], GSE76427 [24], GSE10143 [25], GSE36376 [26], GSE46444 [27], GSE14323 [28], GSE25097 [29], GSE6764 [30], the Liver Hepatocellular Carcinoma Project of the Cancer Genome Atlas (TCGA-LIHC), and the Liver Cancer-RIKEN, JP Project from the International Cancer Genome Consortium (ICGC-LIRI-JP) [31] and were analyzed for PRELP mRNA expression between HCCs and hepatic NTs or cirrhosis.
Cell lines
A hepatic cell line (L02) and four HCC cell lines (HCCLM3, MHCC97-H, SMMC-7721, Alexander cells) were kindly provided by Dr. Lijie Ma at Zhongshan Hospital, Fudan university (Shanghai, China). HCC cell lines SK-HEP-1 and Hep3B2.1-7 were gifted from Prof. Yongzhong Liu at the Cancer Research Institute, Shanghai Jiao Tong University (Shanghai, China); Two HCC cell lines (HepG2 and PLC/PRF/5) were kindly provided by Prof. Xiuping Liu at Shanghai Cancer Hospital, Fudan university (Shanghai, China). All cell lines used in this study were regularly authenticated by morphological observation and tested for the absence of mycoplasma contamination (MycoAlert; Lonza Rockland, Rockland, ME, USA). All Cell lines were cultured at 37℃ with 5% CO2 in DMEM (Sangon Biotech, Shanghai, China) with 10% fetal bovine serum, 100 µg/mL penicillin, and 100 mg/mL streptomycin in a humidified incubator.
Short hairpin RNAs (shRNAs) construction and lentiviruses infection
Plasmids with PRELP-targeting shRNAs or a nontargeting scrambled RNA (scramble), and the recombined plasmids of PRELP or vector were synthesized by Shanghai GeneChem Co., Ltd. These plasmids were well packaged into lentiviruses as previously reported [32]. Four target shRNA sequences are as following: TTCGGCTTAACTACAACAA (shPRELP1), GTAACAAGATTGAGACCAT (shPRELP2), CTCTTAATTGCTCTAACAA (shPRELP3), and TGGTTTCTTTCAAGTTTAA (shPRELP4). All cells were infected and screened as described previously [33].
Samples and IHC
A TMA containing 90 cases of HCC patients was prepared by Shanghai Tufei Biotechnology Co., Ltd. The inclusion criteria for eligible patient selection were: Child-Pugh A liver function; histologically confirmed HCC; age >18 years; surgery with radical intent. The exclusion criteria were: tumor size <0.5 cm; simultaneous malignancies from other organs; previous systemic treatment; incomplete clinical data; lost follow-up. The clinicopathological characteristics of these patients are presented in Table 1. All slides were dewaxed in xylene and rehydrated in graded ethanol, then pretreatment in fresh 3% H2O2 to quench endogenous peroxidase activity was performed and washed in double-distilled water 3 times. Subsequently, antigen retrieval with citrate buffer pH 6.0 (1:50 dilution; Beyotime Biotechnology, Shanghai, China) was performed in an autoclave and incubated with goat serum (1:20 dilution; cat. no. C0265; Beyotime Biotechnology, Shanghai, China) at room temperature for 1 h, then slides were incubated with rabbit anti-PRELP polyclonal antibody (1:50 dilution; Absin-bio, Shanghai, China.) at 4°C overnight. After washing with PBS three times, the horseradish peroxidase HRP-conjugated goat anti-rabbit secondary antibody was covered on slides for 1h at room temperature washed with PBS 3 times. Then, these sections were reacted with 3,3′-diaminobenzidine (1:25 dilution; cat. no. GK500705; Gene Tech Co., Ltd.) at room temperature for 5 min and counterstained with 100% hematoxylin (Beyotime Biotechnology, Shanghai, China) at room temperature for 2 min. Finally, slides were washed in running water for 45 min before dehydration and clearing. Slides was scanned by the ultra-compact Aperio CS2 image capture device and processed by Aperio ImageScope (Leica Biosystems, Germany). The intensity of staining (0, negative; 1, weak; 2, moderate; and 3, strong) was multiplied by the percentage of positive tumor cells (0-100%) to generate the modified H-score in a range from 0 to 300. PRELP staining was categorized as high or low expression using the median H-score [33].
RNA extraction and quantitative real-time polymerase chain reaction (qPCR)
Total RNA was isolated from cell cultures using RNAiso Plus (Takara Bio, Kusatsu, Japan) as to the manufacturer's instructions. A 2-ΔΔCT method was utilized to count the relative expression of target sequences as we described in a previous report [34]. The sequences for RT-PCR primers were as following: PRELP forward primer 5'CAGCCAACAAGACGACCAAGA3' and PRELP reverse primer, 5'CAGTCAGGGAAGATAGATGGAGG3'. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as an internal control. Experiments were independently repeated three times, in duplicate.
Clinical and pathologic features of HCC patients* (n=90)
| Variable | No. of patients (%) |
|---|---|
| Sex | |
| Male | 70 (77.8) |
| Female | 20 (22.2) |
| Age | |
| ≤60 | 64 (71.1) |
| >60 | 26 (28.9) |
| Differentiation | |
| G1/2 | 54 (60) |
| G3 | 36 (40) |
| Tumor size (cm) | |
| ≤5 | 47 (52.2) |
| >5 | 43 (47.8) |
| HBV infection | |
| Negative | 19 (21.1) |
| Positive | 71 (78.9) |
| AFP (μg/L) | |
| ≤200 | 45 (50) |
| >200 | 45 (50) |
| Tumor stage | |
| T1 | 58 (64.5) |
| T2 | 17 (18.9) |
| T3 | 3 (3.3) |
| T4 | 12 (13.3) |
| Nodal stage | |
| N0 | 85 (94.4) |
| N1 | 5 (5.6) |
| M stage | |
| M0 | 88 (97.8) |
| M1 | 2 (2.2) |
| TNM stage | |
| I | 56 (62.2) |
| II | 17 (18.9) |
| III | 16 (17.8) |
| NA | 1 (1.1) |
| Vessel invasion | |
| No | 50 (55.6) |
| Yes | 25 (27.8) |
| NA | 15 (16.6) |
*Data shown here may be duplicated with those from other published resources that are based on the same cohorts. Abbreviations: NA: information not available HCC: Hepatocellular carcinoma; HBV: hepatic B virus.
Protein extraction and western blotting (WB)
After 48 hours of infection, total protein was extracted from cells using RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China), phenylmethylsulfonyl fluoride (Beyotime Biotechnology, Shanghai, China) and proteinase inhibitor cocktail solution (Roche, Basel, Switzerland). The whole extraction process was performed on ice. Then protein samples were centrifuged for 20 min at the speed of 12000 r/min at 4°C. The protein concentration was quantitated using the bicinchoninic acid protein assay (Beyotime Biotechnology, Shanghai, China) as recommended by the manufacturers. SDS-PAGE loading buffer (Beyotime Biotechnology, Shanghai, China) was add to the supernate at the proportion of 25%, then protein was heated at 95°C for 5 min. Western-blot assays were performed as our previous report [35] using rabbit anti-PRELP polyclonal antibody (1:1,000 dilution; Absin-bio, Shanghai, China). GAPDH (1:2,000 dilutions, rabbit anti-human; Beyotime Biotechnology, Shanghai, China) served as a loading control.
Proliferation assay
Cell suspensions of SMMC-7721 and HCCLM3 cells infected with corresponding lentiviruses were seeded in 96-well plates for 2 × 103 cells per well and cultured for 24, 48, 72 or 96 h. Then 10 uL Cell Counting Kit-8 (CCK-8) reagent [10% (v/v) in serum-free DMEM; Sangon biotech, Shanghai, China] was added to each well and cultured at 37°C for 1 h. A microplate reader (BioTek Synergy 2; BioTek, Winooski, VT, USA) was used to detect the optical density (OD) at 450 nm.
Colony formation assays
Infected cells were plated in 12-well plates (1 × 102 cells per well), and cultured for 14 days. The medium was changed every three days then cells were fixed in 4% phosphate-buffered paraformaldehyde and stained by 1% crystal violet when macroscopic colonies appeared in the plate. The plates were rinsed with running water then dried in the air before scanning. The colony number was counted on scanned pictures.
Transwell migration assay
Infected cells were added to 200 μL serum-free DMEM in the upper chamber of a transwell insert. The lower chamber was filled with the medium containing 20% FBS. After 24 h incubation at 37°C, the cells on the upper side of the membrane were removed with a cotton swab, and those on the lower surface were fixed in 4% phosphate-buffered paraformaldehyde for 20 min and stained with crystal violet (0.1% in PBS) for 15 min. Cells were photographed in six randomly selected fields per well with an inverted microscope and the numbers of migrated cells were counted.
Scratch wound healing assay
Infected cells (4 × 105 cells/mL) were cultured in 12-well plates until confluence. After 24 h of serum-free cultivation, a cross wound was scratched using a 200 μL pipette tip. After washing twice with PBS, the wound areas were photographed under an inverted microscope at 0 and 24 h. Then the wound areas were compared and analyzed as previously described [35].
Gene set enrichment analysis
The Gene set enrichment analysis (GSEA) is developed by the Broad Institute website, which is normally used to analysis the distribution of the predefined genes in ranked genes associated with complex phenotypes. After the classification of high or low PRELP expression, we managed to investigate whether there were potential mechanisms of PRELP in HCC progression. The TCGA-LIHC dataset was analyzed using the module of the Kyoto Encyclopedia of Genes and Genomes (KEGG) in GSEA [35].
Statistical analysis
Statistical analyses were conducted by using the SPSS statistical software for Windows, version 22 (IBM Corp., Armonk, NY, USA), Microsoft Excel 2010 (Microsoft, Redmond, WA, USA) and GraphPad Prism7 (GraphPad, San Diego, CA, USA). An independent-samples t test was used to analysis the difference of PRELP mRNA expression between HCCs and NTs or hepatic cirrhosis. Comparisons among multiple groups were performed by one-way analysis of variance. The Bonferroni's post hoc test was performed after pairwise comparisons. The Pearson's χ2 test and Fisher's exact test were utilized to analyze the correlation between PRELP protein expression and clinicopathological features. Univariate and multivariate Cox proportional hazards models were used to explore key factors that are significantly associated with patient prognoses. The survival curves were graphed using Kaplan-Meier method and the difference between two curves was analyzed by log-rank test. The p value less than 0.05 was defined as statistically significant. All statistical tests were two-sided.
Results
PRELP is down-regulated in HCC tissues
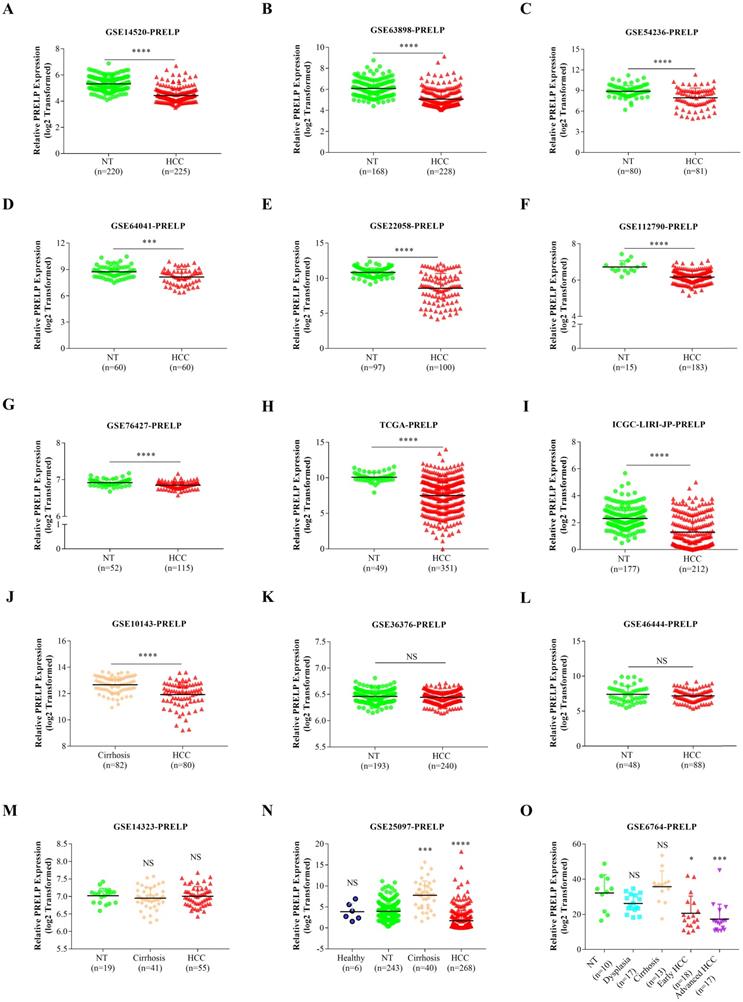
In order to explore the expression pattern of PRELP mRNA, we conducted a comparison between HCCs and adjacent tissues based on the transcriptomic data acquired from the public database HCCDB. PRELP mRNA expression was significantly down-regulated in HCCs compared with that in adjacent NTs or hepatic cirrhosis in 10 out of the 15 datasets (Figure 1A-1J). However, the result showed no significant difference between HCCs and adjacent NTs with regard to PRELP expression in three datasets (Figure 1K-1M). Analysis of a dataset which provided healthy liver tissues demonstrated that PRELP expression was dramatically reduced in HCCs compared with that in healthy liver tissues or NTs (Figure 1N). In another dataset that listed different disease stages during HCC development, the expression of PRELP decreased gradually as disease became more invasive (Figure 1O). Generally, PRELP transcription was diminished in HCCs and may be associated with cancer progression.
PRELP was downregulated in HCCs. The comparisons of PRELP mRNA expression between HCCs and adjacent NTs or hepatic cirrhosis in 12 transcriptomic datasets (A-L). The comparison of PRELP mRNA expression among HCCs, adjacent NTs and hepatic cirrhosis (M), or together with healthy hepatic tissues (N). PRELP mRNA expression levels in different stages during HCC progression (O). Abbreviations: HCC: hepatocellular carcinoma; NTs: normal tissues; NS: not significant; *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001.
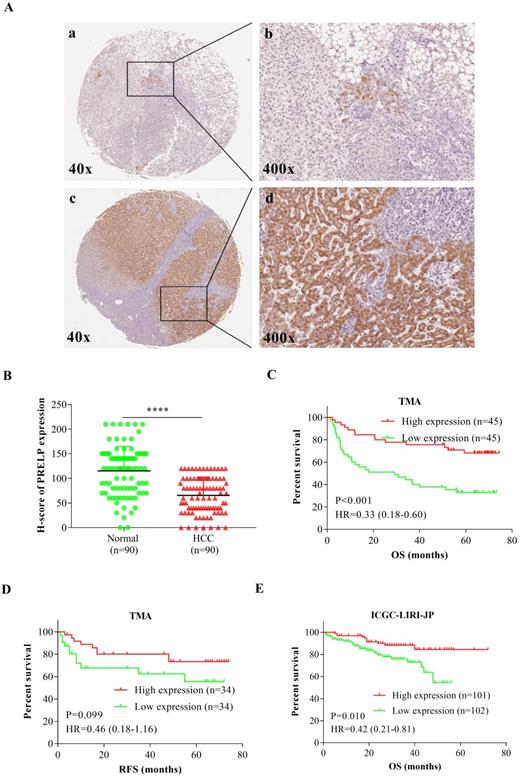
Down-regulation of PRELP was associated with poor prognoses. Representative IHC staining images of PRELP in adjacent NTs and HCCs (200 x and 400x magnification) (A). The intensity of PRELP expression in IHC staining were evaluated by H-score (B) and categorized as low or high protein expression. Patients with low PRELP expression tend to have shorter OS time (C), as well as RFS time (D), which was confirmed by analysis of ICGC-LIRI-JP (E). Abbreviations: IHC: immunohistochemistry; OS: overall survival; RFS: relapse free survival; HR: hazard ratio; ****: p < 0.0001; ICGC-LIRI-JP: the Liver Cancer-RIKEN, JP Project from the International Cancer Genome Consortium.
Low PRELP expression is associated with poor patient survival
Then we performed IHC staining to analyze PRELP expression on a paraffin-embedded, archived TMA. Figure 2A showed that PRELP was located in both cytoplasm and nuclei and its protein expression was dramatically down-regulated in HCC tissues versus controls. The IHC scoring was based on tumor cell staining and each observed tissue component was counted. H-Score plot was graphed to show the quantitative results (p<0.0001) (Figure 2B). The clinicopathological data was acquired from the TMA including 90 HCC patient cases. All patients were divided into two groups according to high or low PRELP protein expression using the median H-score. The Pearson's χ2 test and Fisher's exact test analyses showed that PRELP expression had no significant correlation with age, sex, histological grade, HBV infection, AFP (ug/L), pT stage, pN stage, pM stage, and vessel invasion (Table 2). However, cases with low PRELP expression tended to have advanced p-stages (p=0.028) and larger tumor size (p=0.001). By adjustments of histological grade, tumor size, AFP, p-stage, and vessel invasion, multivariate Cox proportional hazards analyses revealed that overall survival (OS) in HCC patients was independently correlated with PRELP expression [hazard ratio (HR), 0.43; 95% confidence interval (CI), 0.19-0.99; p=0.048], along with pstages [HR, 2.95; 95% CI, 1.24-7.02; p=0.015] (Table 3). Kaplan-Meier survival curves and Log-Rank tests indicated that OS of high PRELP expression cases was longer than that of low PRELP expression with an HR of 0.33 [estimated mean OS, 58.13 months; 95% CI, 50.48-65.78 months; vs. 34.82 months; 95% CI, 26.10-34.54 months; log-rank test, p=0.001, Figure 2C]. There were 68 patients who provided relapse information during follow-up, the analysis of which demonstrated that there was a trend toward significance that low PRELP expression was correlated with shorter relapse free survival (RFS) time with an HR of 2.17 [estimated mean RFS, 47.34 months; 95%CI, 35.64-59.64 months; vs. 59.66 months; 95%CI, 51.23-68.08 months; log-rank test, p=0.099, Figure 2D]. To further verify the prognostic value of PRELP, the transcriptomic data from an outside cohort was accessed and analyzed, supporting that patients with low PRELP expression had significantly shorter survival time with an HR of 2.38 [estimated mean OS, 43.62 months; 95% CI, 39.47-47.77 months; vs. 64.39 months; 95% CI, 60.04-68.74 months; log-rank test, p=0.010, Figure 2E]. To sum up, these findings revealed that low PRELP protein expression in HCCs is associated with poor patient survival.
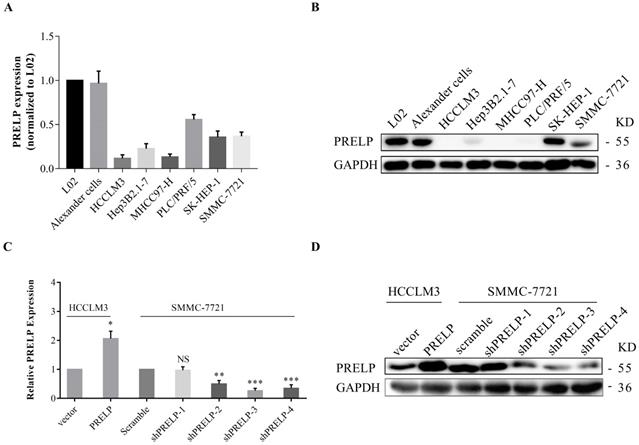
Endogenous PRELP expression in a normal hepatic cell line and seven HCC cell lines. PRELP mRNA and protein were respectively examined by qPCR and WB in L02 and 7 HCC cell lines (A and B). PRELP expression was normalized by L02 cell line. The mRNA and protein expression of PRELP was respectively determined by qPCR and WB in SMMC-7721and HCCLM3 cells (C and D), which were both infected by corresponding lentiviruses. Abbreviations: NS: not significant; *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001 vs. vector control or scramble control; qPCR: quantitative real-time polymerase chain reaction; WB: western-blotting; shRNA: short hairpin RNA.
Downregulation of PRELP promotes proliferation and migration of cultured HCC cells
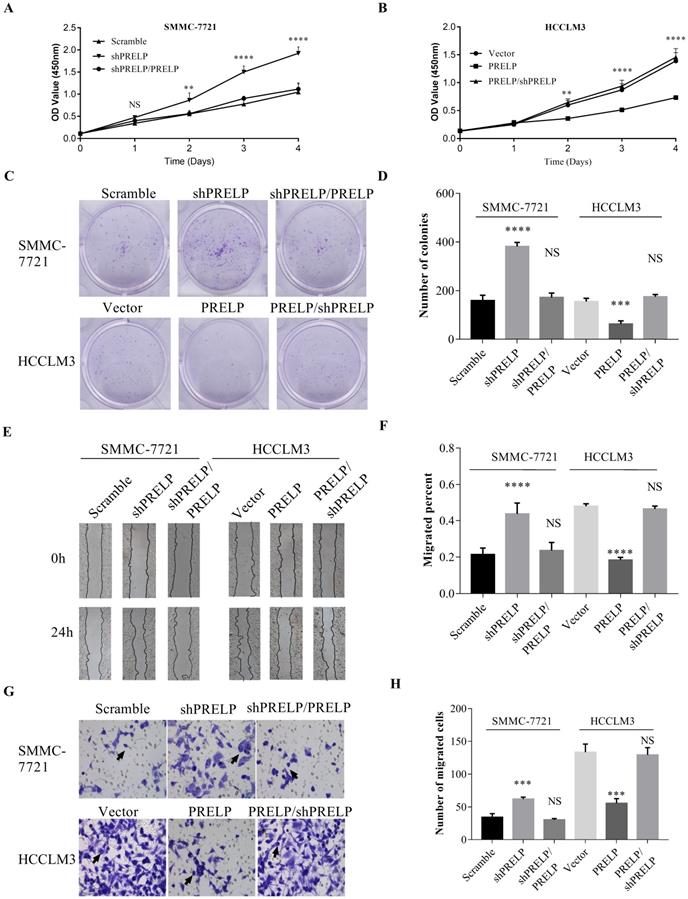
In order to understand the in vitro roles of PRELP in HCC cells, we utilized qPCR and WB to evaluate the endogenous expression of PRELP in a hepatic cell line L02 and seven HCC cell lines from the start. As shown in Figure 3A and 3B, PRELP was downregulated in all HCC cell lines except Alexander cells compared to that in L02 cells. Referring to foregoing results, we then ectopically overexpressed PRELP in HCCLM3 cells and knocked down PRELP in SMMC-7721 cells through lentiviruses infection (Figure 3C and 3D). We performed CCK-8 assay in these two cells. Knocking down of PRELP dramatically accelerated the growth of SMMC-7721 cells (Figure 4A), by contrast, overexpression of PRELP decelerated that of HCCLM3 cells (Figure 4B), compared to respective controls. Similarly, after cultivation for 14 consecutive days, knock down of PRELP markedly increased the mean colony number in the colony formation assay and vice versa (Figure 4C and 4D). Next, we performed wound healing assay on these two cell types, and the migratory areas were measured after culturing for 24 hours. The results showed that PRELP knockdown significantly enhanced the migratory capacity of SMMC-7721 cells and PRELP overexpression impeded that in the HCCLM3 cells, in comparison with that of respective control cells (Figure 4E and 4F). To further validate the roles of PRELP in migration, we then planted aforesaid cell lines into tiny insert chambers, and calculated cell numbers migrating through the chamber after 24h of cultivation. The transwell migration assay further confirmed that PRELP-knockdown cells had enhanced migratory potential than scramble control, while PRELP overexpression cells had diminished migratory potential than vector control (Figure 4G and 4H). In summary, PRELP may inhibit proliferation and migration of cultured HCC cells.
Overexpression of PRELP inhibited HCC cell proliferation and migration. CCK8 assay was used to examine proliferation ability of HCC cells (SMMC-7721 and HCCLM3) infected with lentiviruses carrying scramble control or shPRELPs vector control or PRELP (A and B). Overexpression of PRELP decreased the mean number of colonies in the colony formation assay while down-regulation of endogenous PRELP increased colony formation ability of HCC cells (C and D). Wound healing assay was used to evaluate the migratory ability of these two cells. The representative images (left) and quantitative results(right) of wound healing assay both indicated that overexpressed PRELP inhibited cell migration (E and F). The same results of HCC cells were showed by transwell migration assay. Representative micrographs (left) and quantitative results (right) of transwell migration assay both indicated that overexpressed PRELP could limit the migratory ability of HCC cells. Abbreviations: CCK8: Cell counting Kit‑8; NS: not significant; **: p < 0.01; ***: p < 0.001; ****: p < 0.0001 vs. vector control or scramble control; shRNA: short hairpin RNA.
Association between PRELP expression and clinicopathological variables in HCC patients (n=90)
| Clinicopathological features | N | PRELP expression | ||
|---|---|---|---|---|
| Low (45) | High (45) | P-value | ||
| Sex | ||||
| Male | 70 | 35 (50.0) | 35 (50.0) | |
| Female | 20 | 10 (50.0) | 10 (50.0) | 1.000 |
| Age | ||||
| ≤60 | 64 | 34 (53.1) | 30 (46.9) | |
| >60 | 26 | 11 (42.3) | 15 (57.7) | 0.486 |
| Histological grade | ||||
| G1/G2 | 54 | 23 (42.6) | 31 (57.4) | |
| G3 | 36 | 22 (61.1) | 14 (38.9) | 0.132 |
| Tumor size (cm) | ||||
| ≤5 | 47 | 15 (31.9) | 32 (68.1) | |
| >5 | 43 | 30 (69.8) | 13 (30.2) | 0.001 |
| HBV infection | ||||
| Negative | 19 | 10 (52.6) | 9 (47.4) | |
| Positive | 71 | 35 (49.3) | 36 (50.7) | 1.000 |
| AFP (μg/L) | ||||
| ≤200 | 45 | 18 (40.0) | 27 (60.0) | |
| >200 | 45 | 27 (60.0) | 18 (40.0) | 0.091 |
| pT stage | ||||
| T1 | 58 | 25 (43.1) | 33 (56.9) | |
| T2/T3/T4 | 32 | 20 (62.5) | 12 (37.5) | 0.123 |
| pN stage | ||||
| N0 | 85 | 42 (49.4) | 43 (50.6) | |
| N1-N3 | 5 | 4 (80.0) | 1 (20.0) | 0.616 |
| pM stage | ||||
| M0 | 88 | 43 (49.4) | 45 (50.6) | |
| M1 | 2 | 2 (100.0) | 0 (0.0) | 0.494 |
| pStage* | ||||
| I | 56 | 23 (41.1) | 33 (58.9) | |
| II/III/IV | 33 | 22 (66.7) | 11 (33.3) | 0.028 |
| Vessel invasion# | ||||
| No | 50 | 22 (44.0) | 28 (56.0) | |
| Yes | 25 | 17 (68.0) | 8 (32.0) | 0.085 |
*stages are grouped into I and II/III/IV, #vessel invasion is grouped into No and Yes. Bold type indicates significance. Abbreviations: HCC: Hepatocellular carcinoma; HBV: hepatic B virus.
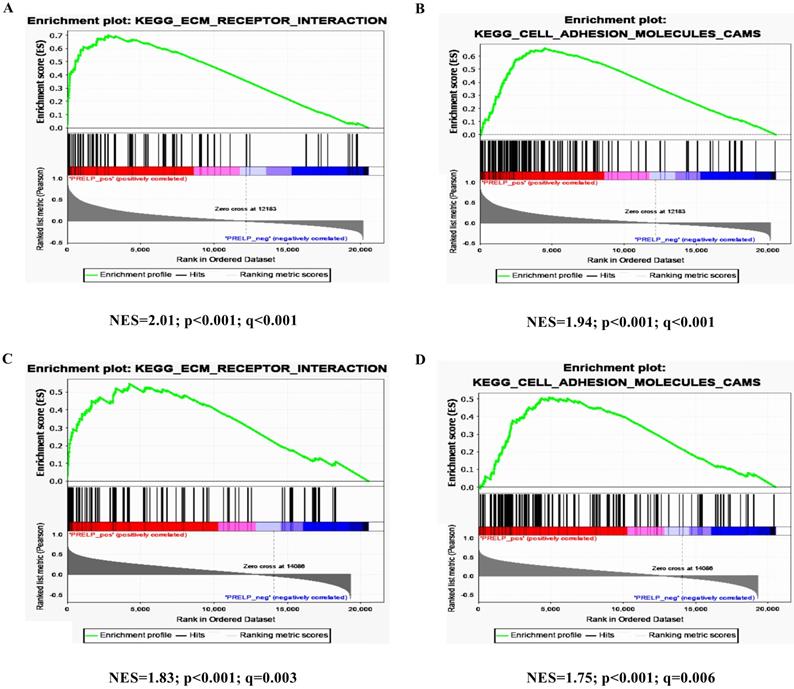
PRELP is associated with extracellular matrix (ECM) receptors and cell adhesion pathways
To further investigate the potential mechanisms of PRELP in the tumorigenesis and progression of HCC, we performed KEGG pathway analyses on the GSEA platform using the TCGA-LIHC dataset. The analyses of transcriptomic data from both HCCs (Figure 5A and 5B) and NTs (Figure 5C and 5D) revealed that high PRELP expression was positively correlated with gene sets of ECM receptors and cell adhesion. Since previous studies suggested that integrin family members mediated cell proliferation, migration and invasion, as well as cross-talks between cells and the ECM [36], while integrins are included in both gene sets mentioned above, we further conducted gene-gene correlation analyses between PRELP and integrin family members using the TCGA-LIHC dataset. The results demonstrated that PRELP expression was significantly and positively correlated with the expression of several integrin family members, including Integrin A3 (ITGA3), ITGA4, ITGA8, ITGA9, ITGA10, ITGA11, ITGAM, ITGB1, ITGB2, ITGB4, ITGB6 and ITGB8 (Figure 6A-L). Collectively, these analyses manifested that PRELP may interact with integrins and play critical roles in ECM and cell adhesion of HCC.
Univariate and multivariate Cox proportional hazard models for overall survival in HCC patients (n=90)
| Clinicopathological features | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR [95% CIs] | P-value | HR [95% CIs] | P-value | |
| Sex | ||||
| Male | 1 [Reference] | |||
| Female | 1.47[0.76-2.85] | 0.258 | ||
| Age | ||||
| ≤60 | 1 [Reference] | |||
| >60 | 0.78[0.40-1.51] | 0.462 | ||
| Histological grade | ||||
| G1/2 | 1 [Reference] | 1 [Reference] | ||
| G3 | 2.01[1.11-3.63] | 0.022 | 0.94 [0.47-1.89] | 0.861 |
| Tumor size (cm) | ||||
| ≤5 | 1 [Reference] | 1 [Reference] | ||
| >5 | 2.32[1.26-4.27] | 0.007 | 1.59[0.72-3.49] | 0.250 |
| HBV infection | ||||
| Negative | 1 [Reference] | |||
| Positive | 1.06[0.51-2.21] | 0.872 | ||
| AFP (μg/L) | ||||
| ≤200 | 1 [Reference] | 1 [Reference] | ||
| >200 | 3.17[1.67-6.00] | <0.001 | 2.35 [1.00-5.55] | 0.051 |
| pStage | ||||
| I | 1 [Reference] | 1 [Reference] | ||
| II/III/IV | 5.11[2.73-9.53] | <0.001 | 2.95 [1.24-7.02] | 0.015 |
| Vessel invasion | ||||
| No | 1 [Reference] | 1 [Reference] | ||
| Yes | 4.15[2.12-8.10] | <0.001 | 1.59 [0.73-3.50] | 0.246 |
| PRELP expression | ||||
| Low | 1 [Reference] | 1 [Reference] | ||
| High | 0.33[0.17-0.62] | 0.001 | 0.43[0.19-0.99] | 0.048 |
Bold type indicates significance. Abbreviations: CI: confidence interval; HR: hazard ratio.
Discussion
Studies for PRELP in human diseases are limited at present, most of which focus on rheumatic diseases. There are a few oncological studies shed light on the roles of PRELP. By glycosylation, PRELP could transform into a 38 kDa core protein which was specifically expressed in chronic lymphocytic leukemia (CLL) cells [37]. A previous study based on proteomic identification suggested that high PRELP expression was relevant to better patient prognosis compared to low PRELP expression in resectable PDAC [13]. However, Iuga et al. had drew a completely opposite conclusion that high PRELP expression was relevant to poor prognosis of patients in resectable PDAC [38]. These seemingly contradictory findings may be due to the influence of different conditions of experiments. In our study, we demonstrated that PRELP was generally down-regulated in HCC tissues in comparison with matched NTs or hepatic cirrhosis. However, analyses of some of the datasets (GSE36376, GSE46444, GSE14323, GSE25097, and GSE6764) presented no significant results, which may be because of their relatively smaller sample sizes and the cohort effect. Besides, PRELP expression status was significantly associated with prognoses of patients. Patients with low expression of PRELP tended to possess shorter OS time and RFS time. Furthermore, patients who had low tissue expression levels of PRELP tended to have larger tumor size and advanced pathological stage. All these show that PRELP is down-regulated in HCCs and has the potential to be a prognostic predictor. Although timely biopsy and tissue fixation had been done after surgical resection, the variant intervals from biopsy to IHC staining may have impact on PRELP expression, thus inevitably bringing bias to the evaluation of prognostic value of PRELP.
Tumor proliferation and metastasis are significant hallmarks of HCC; hence, studies on these aspects would find therapeutic targets to control disease progression [39]. It is widely known that continuous activation of multiple pathways leads to uncontrolled cell proliferation and promotes tumor metastasis [40, 41]. We found that PRELP knockdown promoted, while PRELP overexpression inhibited cell proliferation and migration. In a word, PRELP may act as a therapeutic target against HCC progression. It has been reported that PRELP could inhibit osteoclastogenesis via impeding transcriptional activity of NF-kB signaling pathway [42], and regulate β-catenin/connexin43 pathway in osteoblast differentiation [7]. We assume that the inhibitory role of PRELP in HCC cells may be implemented by modulations of multiple signaling pathways.
PRELP was associated with ECM Receptors and cell adhesion pathways. High PRELP expression was positively associated with cell-adhesion-activated gene signatures (ECM_RECEPTOR_INTERACTION and CELL_ADHESION_ MOLECULER_CAMS) in GSEA of transcriptomic data from HCCs (A and B) and adjacent NTs (C and D) in TCGA-LIHC. Abbreviations: TCGA-LIHC: The Liver Hepatocellular Carcinoma Project of the Cancer Genome Atlas; ES: enrichment score; FDR: false discovery rate; GSEA: gene set enrichment analysis.
Expression of PRELP positively correlated with most integrin family members. Gene-gene correlation analyses between PRELP and selected integrin family members (A-L). Abbreviations: ITG: integrin.
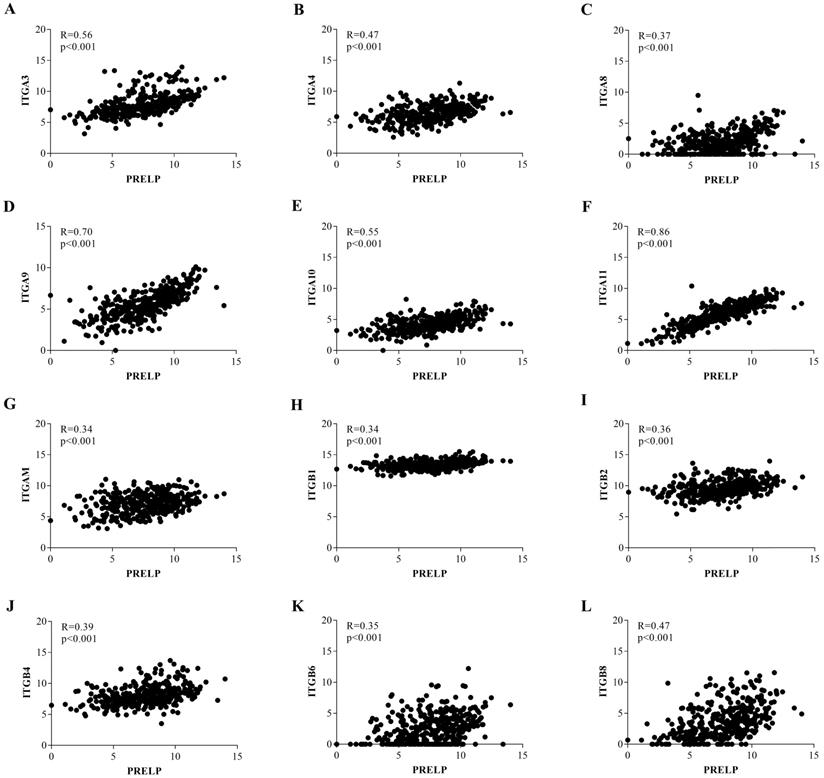
ECM, which is mainly comprised of collagens, elastin, fibronectin, laminins, glycoproteins, proteoglycans (PGs) and glycosaminoglycans (GAGs), is an essential and dynamic part of tumor microenvironment [43]. PRELP was originally isolated from bovine articular cartilage, which is an essential territorial matrix [44]. Electron microscopy showed that PRELP can closely bind to perlecan and procollagen [44], probably because of its special amino-terminal region and the glycosaminoglycan-binding domain [42, 45]. There is a study which certified that the amino-terminal region of PRELP increased cell adhesion and regulated their interaction with GAGs [10]. Basement membranes (BMs) serve as a net shaped structure of ECM to separate cells from matrix [46]. Bengtsson et al. had confirmed that localization of PRELP was adjacent to BMs by confocal immunohistochemistry. These findings verified the collaboration between PRELP and ECM [4]. The integrin family, which is wildly reported in HCC, is involved in multiple molecular mechanisms within and outside tumor cells [47]. Integrins, which normally bind to ECM, promote cell invasion and metastasis by regulating cell adhesion to basement membrane and altering cell malignant features [47, 48]. A previous study reported that activation of ITGB1 signaling dramatically enhanced cell proliferation and invasion in HCC [49]. The similar effect has been examined on ITGB3 [50], ITGB4 [51], ITGB5 [52], ITGA1 [53], ITGA2 [54], ITGA5 [55], ITGA6 [56], ITGA7 [57] in HCC. However, it was worth noting that a recent study found that overexpression of ITGA3 can attenuate the proliferation of HCC cells regulated by LAMB3, and the inhibitory roles have also been reported on ITGA9 in HCC [58]. From former studies we made a conjecture that PRELP inhibits HCC progression by interacting with integrin family members within the tumor microenvironment. To confirm it, we performed KEGG pathway analyses on the GSEA platform and the results suggested that high expression of PRELP was positively related to gene sets extracellular matrix receptors and cell adhesion which are main pathways in tumor microenvironment. By further exploration of these two enriched pathways we found that PRELP expression was positively relevant to most integrin family members. Taken together, PRELP may activate extracellular matrix receptors and accelerate cell adhesion by interacting with certain integrin members, then attenuate cell proliferation and invasion. All these observed results need further confirmation both in vitro and in vivo.
Conclusion
To sum up, we found that PRELP may serve as a potential prognostic marker and a treatment target against HCC progression. PRELP may cross-talk with integrin-mediated ECM and cell adhesion pathways which require further validation.
Abbreviations
HCC: Hepatocellular carcinoma; PRELP: proline/arginine-rich end leucine-rich repeat protein; HCCDB: the Integrative Molecular Database of Hepatocellular Carcinoma; TMA: tissue microarray; GSEA: Gene Set Enrichment Analysis; NTs: normal tissues; HCCs: HCC tissues; OS: overall survival; RFS: relapse free survival; HBV: hepatic B virus; SLRP: small leucine-rich proteoglycan; OA: osteoarthritis; PDAC: pancreatic ductal adenocarcinomas; HPA: The Human Protein Atlas; NX: normalized expression; TCGA-LIHC: the Liver Hepatocellular Carcinoma Project of the Cancer Genome Atlas; ICGC-LIRI-JP: the Liver Cancer-RIKEN, JP Project from the International Cancer Genome Consortium; qPCR: quantitative real-time polymerase chain reaction; WB: western-blotting; shRNA: short hairpin RNA; KEGG: the Kyoto Encyclopedia of Genes and Genomes; ANOVA: analysis of variance; HR: hazard ratio; CI: confidence interval; NS: not significant; GAPDH: Glyceraldehyde 3-phosphate dehydrogenase; CCK-8: Cell Counting Kit-8; CLL: chronic lymphocytic leukemia; PGs: proteoglycans; GAGs: glycosaminoglycans; ECM: extracellular matrix; BMs: Basement membranes; Integrin: ITG.
Acknowledgements
We thank the constructive suggestions given by Professor He Meng at Shanghai Jiao Tong University. We greatly appreciate the technological help from the Department of Pathology of our hospital for the IHC staining and data analysis. We also appreciate the valuable work carried out by Dr. Lijie Ma at Zhongshan Hospital (Shanghai, China). We would like to thank Prof. Yongzhong Liu and Prof. Xiuping Liu for providing the cell lines.
Funding
This work was supported by the Medical System of Shanghai Minhang District (grant number 2017MWDXK01); the Shanghai Minhang District Health and Family Planning Commission (grant number 2016MW03); and the Shanghai Minhang District Science and Technology Commission (grant number 2017MHZ02 and 2019MHZ054). The funding sources were not involved in the study design; in the collection, analysis, or interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Availability of data and materials
The datasets generated and analyzed in this article are included within this article and its additional images. Raw data will be available from the corresponding author if reasonable request is presented.
Authors' contributions
Ke CW and Hong L designed this study. Hong RQ, Niu GM and Gu JW collected and analyzed the transcriptomic data. Hong RQ performed the IHC assay and analyzed the results. Song T, Hu ZQ, Zhang XT, Han SL and Hong RQ were responsible for the in vitro experiments. Hong RQ was responsible for manuscript drafting. Ke CW, Hong L revised the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Yang JD, Hainaut P, Gores GJ. et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589-604
3. Bruix J, Qin S, Merle P. et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56-66
4. Bengtsson E, Morgelin M, Sasaki T. et al. The leucine-rich repeat protein PRELP binds perlecan and collagens and may function as a basement membrane anchor. J Biol Chem. 2002;277:15061-15068
5. Bengtsson E, Neame PJ, Heinegard D. et al. The primary structure of a basic leucine-rich repeat protein, PRELP, found in connective tissues. J Biol Chem. 1995;270:25639-25644
6. Matsushima N, Ohyanagi T, Tanaka T. et al. Super-motifs and evolution of tandem leucine-rich repeats within the small proteoglycans-biglycan, decorin, lumican, fibromodulin, PRELP, keratocan, osteoadherin, epiphycan, and osteoglycin. Proteins. 2000;38:210-225
7. Li H, Cui Y, Luan J. et al. PRELP (proline/arginine-rich end leucine-rich repeat protein) promotes osteoblastic differentiation of preosteoblastic MC3T3-E1 cells by regulating the beta-catenin pathway. Biochem Biophys Res Commun. 2016;470:558-562
8. Fernandez-Puente P, Gonzalez-Rodriguez L, Calamia V. et al. Analysis of Endogenous Peptides Released from Osteoarthritic Cartilage Unravels Novel Pathogenic Markers. Mol Cell Proteomics. 2019;18:2018-2028
9. Liu G, Ermert D, Johansson ME. et al. PRELP Enhances Host Innate Immunity against the Respiratory Tract Pathogen Moraxella catarrhalis. J Immunol. 2017;198:2330-2340
10. Bengtsson E, Lindblom K, Tillgren V. et al. The leucine-rich repeat protein PRELP binds fibroblast cell-surface proteoglycans and enhances focal adhesion formation. Biochem J. 2016;473:1153-1164
11. Malmsten M, Davoudi M, Schmidtchen A. Bacterial killing by heparin-binding peptides from PRELP and thrombospondin. Matrix Biol. 2006;25:294-300
12. Uhlen M, Fagerberg L, Hallstrom BM. et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419
13. Chen R, Dawson DW, Pan S. et al. Proteins associated with pancreatic cancer survival in patients with resectable pancreatic ductal adenocarcinoma. Lab Invest. 2015;95:43-55
14. Castells X, Acebes JJ, Boluda S. et al. Development of a predictor for human brain tumors based on gene expression values obtained from two types of microarray technologies. OMICS. 2010;14:157-164
15. Ning X, Deng Y. Identification of key pathways and genes influencing prognosis in bladder urothelial carcinoma. Onco Targets Ther. 2017;10:1673-1686
16. Lian Q, Wang S, Zhang G. et al. HCCDB: A Database of Hepatocellular Carcinoma Expression Atlas. Genomics Proteomics Bioinformatics. 2018;16:269-275
17. Roessler S, Long EL, Budhu A. et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957-966 e912
18. Roessler S, Jia HL, Budhu A. et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202-10212
19. Losic B, Craig AJ, Villacorta-Martin C. et al. Intratumoral heterogeneity and clonal evolution in liver cancer. Nat Commun. 2020;11:291
20. Villa E, Critelli R, Lei B. et al. Neoangiogenesis-related genes are hallmarks of fast-growing hepatocellular carcinomas and worst survival. Results from a prospective study. Gut. 2016;65:861-869
21. Makowska Z, Boldanova T, Adametz D. et al. Gene expression analysis of biopsy samples reveals critical limitations of transcriptome-based molecular classifications of hepatocellular carcinoma. J Pathol Clin Res. 2016;2:80-92
22. Burchard J, Zhang C, Liu AM. et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol. 2010;6:402
23. Shimada S, Mogushi K, Akiyama Y. et al. Comprehensive molecular and immunological characterization of hepatocellular carcinoma. EBioMedicine. 2019;40:457-470
24. Grinchuk OV, Yenamandra SP, Iyer R. et al. Tumor-adjacent tissue co-expression profile analysis reveals pro-oncogenic ribosomal gene signature for prognosis of resectable hepatocellular carcinoma. Mol Oncol. 2018;12:89-113
25. Hoshida Y, Villanueva A, Kobayashi M. et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995-2004
26. Lim HY, Sohn I, Deng S. et al. Prediction of disease-free survival in hepatocellular carcinoma by gene expression profiling. Ann Surg Oncol. 2013;20:3747-3753
27. Kojima K, April C, Canasto-Chibuque C. et al. Transcriptome profiling of archived sectioned formalin-fixed paraffin-embedded (AS-FFPE) tissue for disease classification. PLoS One. 2014;9:e86961
28. Mas VR, Maluf DG, Archer KJ. et al. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med. 2009;15:85-94
29. Tung EK, Mak CK, Fatima S. et al. Clinicopathological and prognostic significance of serum and tissue Dickkopf-1 levels in human hepatocellular carcinoma. Liver Int. 2011;31:1494-1504
30. Wurmbach E, Chen YB, Khitrov G. et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938-947
31. Fujimoto A, Furuta M, Totoki Y. et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48:500-509
32. Shi JY, Ma LJ, Zhang JW. et al. FOXP3 Is a HCC suppressor gene and Acts through regulating the TGF-beta/Smad2/3 signaling pathway. BMC Cancer. 2017;17:648
33. Niu G, Yang Y, Ren J. et al. Overexpression of CPXM2 predicts an unfavorable prognosis and promotes the proliferation and migration of gastric cancer. Oncol Rep. 2019;42:1283-1294
34. Zhao S, Zhou L, Niu G. et al. Differential regulation of orphan nuclear receptor TR3 transcript variants by novel vascular growth factor signaling pathways. FASEB J. 2014;28:4524-4533
35. Hu Z, Niu G, Ren J. et al. TAFA5 promotes proliferation and migration in gastric cancer. Mol Med Rep. 2019;20:4477-4488
36. Cooper J, Giancotti FG. Integrin Signaling in Cancer: Mechanotransduction, Stemness, Epithelial Plasticity, and Therapeutic Resistance. Cancer Cell. 2019;35:347-367
37. Mikaelsson E, Osterborg A, Jeddi-Tehrani M. et al. A proline/arginine-rich end leucine-rich repeat protein (PRELP) variant is uniquely expressed in chronic lymphocytic leukemia cells. PLoS One. 2013;8:e67601
38. Iuga C, Seicean A, Iancu C. et al. Proteomic identification of potential prognostic biomarkers in resectable pancreatic ductal adenocarcinoma. Proteomics. 2014;14:945-955
39. Craig AJ, von Felden J, Garcia-Lezana T. et al. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17:139-152
40. Jiang Y, Han QJ, Zhang J. Hepatocellular carcinoma: Mechanisms of progression and immunotherapy. World J Gastroenterol. 2019;25:3151-3167
41. Chen Y, Wang D, Peng H. et al. Epigenetically upregulated oncoprotein PLCE1 drives esophageal carcinoma angiogenesis and proliferation via activating the PI-PLCepsilon-NF-kappaB signaling pathway and VEGF-C/ Bcl-2 expression. Mol Cancer. 2019;18:1
42. Rucci N, Rufo A, Alamanou M. et al. The glycosaminoglycan-binding domain of PRELP acts as a cell type-specific NF-kappaB inhibitor that impairs osteoclastogenesis. J Cell Biol. 2009;187:669-683
43. Theocharis AD, Skandalis SS, Gialeli C. et al. Extracellular matrix structure. Adv Drug Deliv Rev. 2016;97:4-27
44. Heinegard D, Larsson T, Sommarin Y. et al. Two novel matrix proteins isolated from articular cartilage show wide distributions among connective tissues. J Biol Chem. 1986;261:13866-13872
45. Bengtsson E, Aspberg A, Heinegard D. et al. The amino-terminal part of PRELP binds to heparin and heparan sulfate. J Biol Chem. 2000;275:40695-40702
46. Randles MJ, Humphries MJ, Lennon R. Proteomic definitions of basement membrane composition in health and disease. Matrix Biol. 2017;57-58:12-28
47. Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91-100
48. Marsico G, Russo L, Quondamatteo F. et al. Glycosylation and Integrin Regulation in Cancer. Trends Cancer. 2018;4:537-552
49. Li Y, Ren Z, Wang Y. et al. ADAM17 promotes cell migration and invasion through the integrin beta1 pathway in hepatocellular carcinoma. Exp Cell Res. 2018;370:373-382
50. Zhang N, Ma D, Wang L. et al. Insufficient Radiofrequency Ablation Treated Hepatocellular Carcinoma Cells Promote Metastasis by Up-Regulation ITGB3. J Cancer. 2017;8:3742-3754
51. Li XL, Liu L, Li DD. et al. Integrin beta4 promotes cell invasion and epithelial-mesenchymal transition through the modulation of Slug expression in hepatocellular carcinoma. Sci Rep. 2017;7:40464
52. Lin Z, He R, Luo H. et al. Integrin-beta5, a miR-185-targeted gene, promotes hepatocellular carcinoma tumorigenesis by regulating beta-catenin stability. J Exp Clin Cancer Res. 2018;37:17
53. Liu X, Tian H, Li H. et al. Derivate Isocorydine (d-ICD) Suppresses Migration and Invasion of Hepatocellular Carcinoma Cell by Downregulating ITGA1 Expression. Int J Mol Sci. 2017 18
54. Yu J, Zhang C, Yu Q. et al. ADAR1 p110 Enhances Adhesion of Tumor Cells to Extracellular Matrix in Hepatocellular Carcinoma via Up-Regulating ITGA2 Expression. Med Sci Monit. 2019;25:1469-1479
55. Chen J, Ji T, Wu D. et al. Human mesenchymal stem cells promote tumor growth via MAPK pathway and metastasis by epithelial mesenchymal transition and integrin alpha5 in hepatocellular carcinoma. Cell Death Dis. 2019;10:425
56. Lv G, Lv T, Qiao S. et al. RNA interference targeting human integrin alpha6 suppresses the metastasis potential of hepatocellular carcinoma cells. Eur J Med Res. 2013;18:52
57. Ge JC, Wang YX, Chen ZB. et al. Integrin alpha 7 correlates with poor clinical outcomes, and it regulates cell proliferation, apoptosis and stemness via PTK2-PI3K-Akt signaling pathway in hepatocellular carcinoma. Cell Signal. 2020;66:109465
58. Zhang YL, Xing X, Cai LB. et al. Integrin alpha9 Suppresses Hepatocellular Carcinoma Metastasis by Rho GTPase Signaling. J Immunol Res. 2018;2018:4602570
Author contact
![]() Corresponding author: Dr. Chongwei Ke and Dr. Liang Hong, Department of General Surgery, The Fifth People's Hospital of Shanghai, Fudan University, 801 Heqing Road, Minhang District, Shanghai, 200240, P.R. China. Tel: +86 021 24289021; Fax: +86 021 64300477; E-mail: kechongweicom; doctor_hlcom.
Corresponding author: Dr. Chongwei Ke and Dr. Liang Hong, Department of General Surgery, The Fifth People's Hospital of Shanghai, Fudan University, 801 Heqing Road, Minhang District, Shanghai, 200240, P.R. China. Tel: +86 021 24289021; Fax: +86 021 64300477; E-mail: kechongweicom; doctor_hlcom.

 Global reach, higher impact
Global reach, higher impact