3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(13):3827-3833. doi:10.7150/jca.37437 This issue Cite
Research Paper
Prevalence and characteristics of PIK3CA mutation in mismatch repair-deficient colorectal cancer
Department of Pathology, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100021, China.
Received 2019-6-9; Accepted 2020-3-27; Published 2020-4-6
Abstract
Background: Chromosomal instability (CIN) and microsatellite instability (MSI) account for the major causes of colorectal cancer (CRC). As an important component of the CIN pathway, PIK3CA mutation is a negative prognostic factor in CRC. However, the relationship between PIK3CA mutation and mismatch repair (MMR) status has not been well clarified.
Methods: MMR status was determined by immunohistochemical assay. KRAS, NRAS, BRAF, PIK3CA and TP53 mutations were comparatively analyzed in 424 MMR-proficient (pMMR) and 104 MMR-deficient (dMMR) CRC tumors using next-generation sequencing (NGS).
Results: PIK3CA mutation was more commonly mutated in dMMR tumors. PIK3CA mutation less commonly coexisted with KRAS/NRAS/BRAF and TP53 mutations, but more likely coexisted with HER2 and PTCH1 mutations in dMMR tumors compared with pMMR tumors. In tumors with concurrent RAS/BRAF and PIK3CA mutations, PIK3CA and RAS/BRAF mutant allele frequencies (MAFs) were highly concordant in dMMR tumors, whereas PIK3CA MAFs were significantly lower than the corresponding RAS/BRAF MAFs in pMMR tumors, implying that PIK3CA mutation may occur in the early stage of dMMR CRC.
Conclusions: The molecular pathogenesis is different between dMMR and pMMR tumors with PIK3CA mutation in CRC. PIK3CA mutation may act as a clonally dominant truncal mutation in dMMR CRC. Thus, combination of PIK3CA mutation and MMR status might determine a specific group of CRC to select treatment or elevate prognosis.
Keywords: PIK3CA mutation, MMR, colorectal cancer, RAS/BRAF mutations, next-generation sequencing
Introduction
Colorectal cancer (CRC) is one of the most common malignancies in the world [1], and ranks as the fifth cause of cancer-related death in China [2]. It is a heterogenous disease evolving from diverse genetic pathways, which attribute to tumor development and progression [3]. Therefore, uncovering the molecular alterations of CRC may be helpful to develop potential new approaches for the diagnosis and treatment.
Chromosomal instability (CIN) and microsatellite instability (MSI) are two crucial pathways in CRC pathogenesis [4]. MSI is a hypermutable phenotype at the genomic level that is caused by deficient DNA mismatch repair (dMMR) mainly because of germline mutations (Lynch syndrome) or hypermethylation of MMR genes [5]. MMR status can be determined by immunohistochemical assay (IHC). The dMMR tumors show loss of expression in MLH1, MSH2, MSH6 or PMS2 protein, whereas the pMMR tumors have intact expression of all four MMR proteins. Studies have found that dMMR CRC cases exert some distinct differences in clinicopathologic features compared with pMMR CRC cases, such as preference of proximal colon, mucinous or signet ring differentiation, and a favorable prognosis [6]. Moreover, advanced dMMR/MSI-H CRC patients may benefit from immunotherapy, such as anti-PD-1 therapies [7].
PIK3CA, which encodes the catalytic p110-alpha subunit of PI3K and thus regulates PIK3CA/AKT pathway downstream of EGFR, has been described to be mutated in 10%-20% of CRC patients. More than 80% of PIK3CA mutations occur in the helical and kinase domains [8]. Basic studies have reported that mutation in PIK3CA can accelerate tumor progression, usually alongside with KRAS/BRAF mutations [9, 10]. Clinical studies have shown that PIK3CA mutation may be a biomarker for resistant to anti-EGFR therapy of CRC [11]. Moreover, PIK3CA-mutant CRC patients may benefit from adjuvant aspirin therapy or PI3K inhibitor treatment [12, 13]. Therefore, PIK3CA mutation plays an important role in CRC treatment.
Recent study has reported that PIK3CA mutation is more commonly mutated in the MSI molecular subgroup of gastric cancer [14]. However, the relationship between PIK3CA mutation and MSI status in CRC patients remain elusive. In this retrospective study, we interrogated 424 pMMR and 104 dMMR CRC tumors by NGS to identify KRAS, NRAS, BRAF, PIK3CA and TP53 mutations. We further investigated the clinical and molecular differences of the PIK3CA-mutant tumors stratified by MMR status in Chinese CRC patients.
Patients and Methods
Patients and specimens
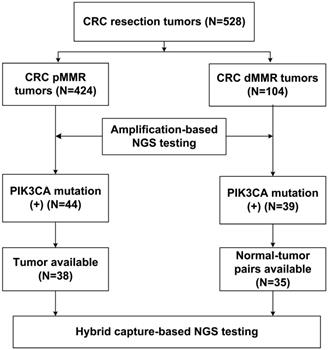
A total of 528 CRC patients who had undergone surgery at Cancer Hospital, Chinese Academy of Medical Sciences (CAMS) between 2013 and 2018 were enrolled. Among these 528 patients, 104 were dMMR CRC, whereas 424 were pMMR CRC. Clinicopathological characteristics were obtained from the medical records. None of the patients had received neoadjuvant therapy or radiotherapy before surgery. The study had been approved by the Institute Review Board of the Cancer Hospital, CAMS. Informed consents were obtained from all patients, and methods were carried out in accordance with the approved guidelines.
Immunohistochemical assay (IHC)
MLH1, PMS2, MSH2 and MSH6 expression was determined by IHC. Briefly, the tissue sections were deparaffinized in xylene, and then rehydrated in graded alcohol. After washing in distilled water, all tumor samples were stained in an autostainer (Autostainer Link 48, Dako, Denmark) with the antibodies of MLH1 (ES05, Dako) , MSH2 (FE11, Dako), MSH6 (EP49, Dako) and PMS2 (EP51, Dako), respectively.
Isolation of genomic DNA
All HE slides were observed by an expert pathologist (Dr J Ying) under the microscope, and formalin-fixed and paraffin-embedded (FFPE) tissue blocks with >20% tumor cellularity were used for further NGS testing. For PIK3CA-mutant tumors with dMMR, adjacent normal tissues were also selected to identify germline mutations. Genomic DNA was extracted from the selected blocks using QIAamp DNA FFPE Tissue Kits (Qiagen, Duesseldorf, Germany), following the manufacturer's protocols. Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Carlsbad, CA, USA) was used to determine DNA quantity.
Amplification-based NGS testing
The amplification-based NGS testing was performed to identify mutations in KRAS, NRAS, BRAF, PIK3CA and TP53. Briefly, multiplex PCR was performed with 10 ng of genomic DNA, and then adapters were ligated to each PCR product. The amplicon libraries were constructed after purifying with 75% ethanol. The quantity of amplicon libraries was determined using Ion Library Quantification Kit (Thermo Fisher, MA, USA). Each library was diluted to a concentration of 40 pM, and pooled in equal volumes. Template preparation was performed with Ion Chef, and sequencing was carried out on Ion S5 with 520 Chip. Finally, data were generated using the software of Torrent Suite. Variants with coverage depth> 500 and MAF >5% were identified as mutations using the Torrent Variant Caller.
Hybrid capture-based NGS testing
DNA from PIK3CA-mutant tumors was further interrogated using a capture-based targeted sequencing panel (Burning Rock Biotech, Guangzhou, China) to parallelly profile somatic mutations of 33 cancer-related genes and MSI status. DNA from adjacent normal tissues was also tested to determine germline mutations. Briefly, fragment genomic DNA was obtained by sonication (M220 Focused- Ultrasonicator, Covaris, Woburn, Massachusetts, USA). PCR amplification was performed after the adaptors were added on both ends. After purification with 75% ethanol, the PCR products were hybridized with the capture probes, and enrichmented with beads. Moreover, PCR amplification was performed to get the libraries. All the indexed libraries were mixed with proper concentration, and were then sequenced on Nextseq N500 (Illumina, San Diego, CA). Sequence data were generated and analyzed by GATK 3.2. Variants with coverage depth> 500 and MAF >5% were identified as mutations.
Flow chart of the pMMR and dMMR CRC samples subjected to NGS testing.
Patient characteristics in 528 CRC patients, including 424 pMMR and 104 dMMR patients.
| Clinicopathologic characteristics | N | pMMR | dMMR | P | |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 320 | 260 (61.3%) | 60 (57.7%) | 0.497 | |
| Female | 208 | 164 (38.7%) | 44 (42.3%) | ||
| Age | |||||
| Median (range) | 58 (25-83) | 58 (26-83) | 56 (25-83) | ||
| <60 | 304 | 238 (56.1%) | 66 (63.5%) | 0.175 | |
| ≥60 | 224 | 186 (43.9%) | 38 (36.5%) | ||
| Tumor site | |||||
| Left colon | 157 | 132 (31.1%) | 25 (24.0%) | <0.001 | |
| Right colon | 156 | 88 (20.8%) | 68 (65.4%) | ||
| Rectum | 215 | 204 (48.1%) | 11 (10.6%) | ||
| Histological differentiation | |||||
| Well/Moderate | 341 | 274 (64.6%) | 67 (64.4%) | 0.970 | |
| Poor | 187 | 150 (35.4%) | 37 (35.6%) | ||
| pT stage | |||||
| pT1-2 | 54 | 38 (9.0%) | 16 (15.4%) | 0.053 | |
| pT3-4 | 474 | 386 (91.0%) | 88 (84.6%) | ||
| pN stage | |||||
| pN0 | 192 | 112 (26.4%) | 80 (76.9%) | <0.001 | |
| pN1-2 | 336 | 312 (73.6%) | 24 (23.1%) | ||
| Lymphovascular invasion | |||||
| Yes | 231 | 204 (48.1%) | 27 (26.0%) | <0.001 | |
| No | 297 | 220 (51.9%) | 77 (74.0%) | ||
| Cancerous node | |||||
| Yes | 119 | 113 (26.7%) | 6 (5.8%) | <0.001 | |
| No | 409 | 311 (73.3%) | 98 (94.2%) | ||
Statistical analysis
Molecular and clinicopathological differences between pMMR and dMMR cases were investigated by Chi-square test or Fisher's exact test. The relationship between concurrent PIK3CA and RAS/BRAF MAFs was determined by paired Student's t-test. All analyses were performed with SPSS 18.0 Software. Statistically significance was identified when a two-sided P-value was less than 0.05.
Results
Patient characteristics
With respect to the MMR status, a total of 528 CRC resection cases were classified into 424 pMMR cases and 104 dMMR cases. The clinicopathological characteristics were listed in Table 1, based on the MMR status. The results showed that dMMR cases were significantly associated with right colon location (65.4% vs. 20.8%) and reduced lymph node metastasis (23.1% vs. 73.6%) compared with pMMR cases. In addition, lymphovascular invasion (26.0% vs. 48.1%) and cancerous nodes (5.8% vs. 26.7%) were less frequently observed in dMMR cases than pMMR cases. The study population was subjected to NGS- based molecular testing, as summarized in Figure 1.
Mutation alterations in pMMR and dMMR tumors
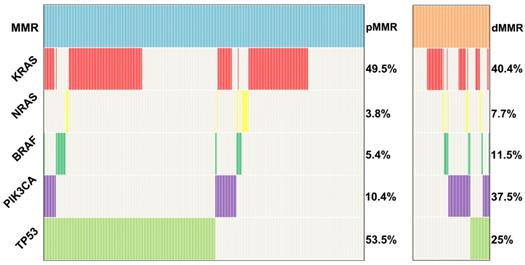
KRAS, NRAS, BRAF, PIK3CA and TP53 mutations were tested in 424 pMMR tumors and 104 dMMR tumors using the amplification-based NGS testing. The results showed that KRAS, NRAS, BRAF, PIK3CA and TP53 mutations were observed in 49.5% (210/424), 3.8% (16/424), 5.4% (23/424), 10.4% (44/424) and 53.5% (227/424) of pMMR tumors, respectively. However, KRAS, NRAS, BRAF, PIK3CA and TP53 mutations were observed in 40.4% (42/104), 7.7% (8/104), 11.5% (12/104), 37.5% (39/104) and 25% (26/104) of dMMR tumors, respectively. TP53 mutation was more frequently to be observed in pMMR tumors than dMMR tumors (53.5% vs. 25%, P<0.001), whereas PIK3CA mutation was more likely to be observed in dMMR tumors compared with pMMR tumors (37.5% vs. 10.4%, P<0.001) (Figure 2). To further validated our conclusion, we also investigated the association of MSI status and PIK3CA mutation in TCGA databases. A total of 1611 CRC samples detected by MSKCC were included in the analysis as an independent cohort. Interestingly, we found that PIK3CA mutation was also more likely to be observed in dMMR tumors compared with pMMR tumors (26.7% vs. 9.4%, P<0.001).
Clinicopathologic characteristics of pMMR and dMMR cases with PIK3CA mutation
The association of clinicopathologic characteristics and PIK3CA mutation was investigated in pMMR and dMMR cases, respectively. In pMMR CRC cases, PIK3CA mutation was more frequent in older age (59.1% vs. 42.1%, P=0.032) and right colon cancer (45.5% vs. 17.9%, P<0.001). In dMMR CRC cases, no association of gender, age, tumor site, histological differentiation, pT stage, pN stage, lymphovascular invasion and cancerous node with PIK3CA mutation was observed (Table 2).
Distribution of KRAS, NRAS, BRAF, PIK3CA and TP53 mutations in 424 pMMR and 104 dMMR CRCs.
Clinicopathological features of CRC by PIK3CA mutation and MMR status.
| Clinicopathologic characteristics | pMMR | PIK3CA | P | dMMR | PIK3CA | P | ||
|---|---|---|---|---|---|---|---|---|
| Mutation | Wild-type | Mutation | Wild-type | |||||
| Gender | ||||||||
| Male | 260 | 28 (63.6%) | 232 (61.1%) | 0.740 | 60 | 23 (59.0%) | 37 (56.9%) | 0.838 |
| Female | 164 | 16 (36.4%) | 148 (38.9%) | 44 | 16 (41.0%) | 28 (43.1%) | ||
| Age | ||||||||
| <60 | 238 | 18 (40.9%) | 220 (57.9%) | 0.032 | 66 | 28 (71.8%) | 38 (58.5%) | 0.172 |
| ≥60 | 186 | 26 (59.1%) | 160 (42.1%) | 38 | 11 (28.2%) | 27 (41.5%) | ||
| Tumor site | ||||||||
| Left colon | 132 | 8 (18.2%) | 124 (32.6%) | <0.001 | 25 | 10 (25.6%) | 15 (23.1%) | 0.412 |
| Right colon | 88 | 20 (45.5%) | 68 (17.9%) | 68 | 23 (59.0%) | 45 (69.2%) | ||
| Rectum | 204 | 16 (36.4%) | 188 (49.5%) | 11 | 6 (15.4%) | 5 (7.7%) | ||
| Histological differentiation | ||||||||
| Well/Moderate | 274 | 27 (61.4%) | 247 (65.0%) | 0.633 | 67 | 28 (71.8%) | 39 (60.0%) | 0.224 |
| Poor | 150 | 17 (38.6%) | 133 (35.0%) | 37 | 11 (28.2%) | 26 (40.0%) | ||
| pT stage | ||||||||
| pT1-2 | 38 | 3 (6.8%) | 35 (9.2%) | 0.805 | 16 | 7 (17.9%) | 9 (13.8%) | 0.575 |
| pT3-4 | 386 | 41 (93.2%) | 345 (90.8%) | 88 | 32 (82.1%) | 56 (86.2%) | ||
| pN stage | ||||||||
| pN0 | 112 | 15 (34.1%) | 97 (25.5%) | 0.223 | 80 | 33 (84.6%) | 47 (72.3%) | 0.149 |
| pN1-2 | 312 | 29 (65.9%) | 283 (74.5%) | 24 | 6 (15.4%) | 18 (27.7%) | ||
| Lymphovascular invasion | ||||||||
| Yes | 204 | 19 (43.2%) | 185 (48.7%) | 0.489 | 27 | 8 (20.5%) | 19 (29.2%) | 0.326 |
| No | 220 | 25 (56.8%) | 195 (51.3%) | 77 | 31 (79.5%) | 46 (70.8%) | ||
| Cancerous node | ||||||||
| Yes | 113 | 11 (25.0%) | 102 (26.8%) | 0.794 | 6 | 0 | 6 (9.2%) | 0.128 |
| No | 311 | 33 (75.0%) | 278 (73.2%) | 98 | 39 (100%) | 59 (90.8%) | ||
Concomitant mutations in PIK3CA-mutant tumors
Paired tumor-normal tissues from 35 PIK3CA-mutant dMMR patients were subjected to the hybrid capture-based NGS testing to parallelly profile somatic mutations of 33 cancer-related genes, MSI status and germline mutations. Moreover, 38 PIK3CA-mutant tumors with pMMR were also tested by the hybrid capture-based NGS testing to determine the somatic mutations of 33 cancer-related genes and MSI status. All 35 dMMR tumors were identified as MSI-H, whereas 38 pMMR tumors were MSS/MSI-L. The concordance rate was 100% between NGS-MSI and IHC testing.
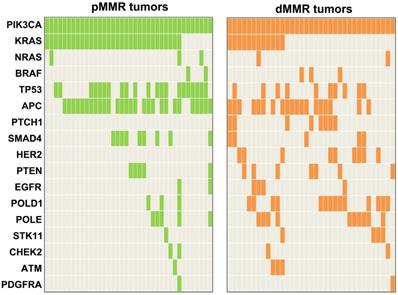
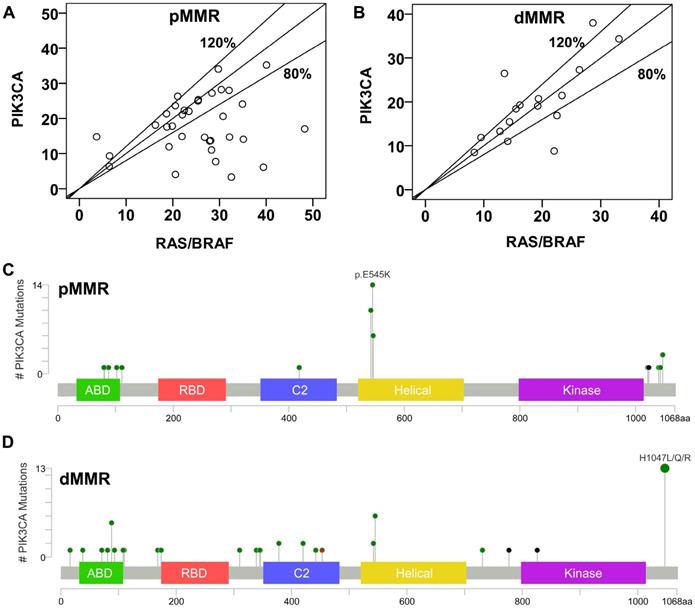
Concomitant mutations were compared between pMMR and dMMR tumors with PIK3CA mutation. The results showed that PIK3CA mutation was more commonly to coexist with KRAS/NRAS/ BRAF (pMMR vs. dMMR, 89.5% vs. 45.7%, P<0.001) and TP53 mutations (pMMR vs. dMMR, 55.3% vs. 28.6%, P=0.021) in pMMR tumors, but was more likely to coexist with HER2 (pMMR vs. dMMR, 0% vs. 25.7%, P<0.001) and PTCH1 mutations (pMMR vs. dMMR, 0% vs. 22.9%, P=0.006) in dMMR tumors (Figure 3). Moreover, we found that the ratio of PIK3CA/RAS or BRAF MAFs was below 80% in 18.8% (3/16) of dMMR tumors (Figure 4B), but in 52.9% (18/34) of pMMR tumors (P=0.022) (Figure 4A). There was no statistically significant difference between PIK3CA (mean ± SD, 19.4 ± 8.6; 95% CI, 14.9-24.0) and concurrent RAS/BRAF MAFs (mean ± SD, 18.7 ± 6.9; 95% CI, 15.0-22.4, P=0.612) in dMMR tumors. However, PIK3CA MAFs (mean ± SD, 18.1 ± 8.2; 95% CI, 15.2-20.9) were significantly lower than RAS/BRAF MAFs (mean ± SD, 26.0 ± 10.4; 95% CI, 22.4-30.0, P = 0.001) in pMMR tumors.
Further, double somatic PIK3CA mutations were observed in 37.1% (13/35) of dMMR tumors, which were significantly higher than those in pMMR tumors (4/38, 10.5%, P=0.007) (Table S1). The distribution of PIK3CA mutations was shown in Figure 4C and 4D. The results showed that 35 of 38 PIK3CA mutations located in exon 9 and 20 in pMMR tumors, whereas 21 of 35 PIK3CA mutations located in exon 9 and 20 in dMMR tumors (92.1% vs. 60%, P=0.001). Moreover, PIK3CA exon 9 mutation was more common in pMMR tumors, whereas PIK3CA exon 20 mutation was more frequent in dMMR tumors (P<0.001). However, no predilection of concurrent RAS/BRAF and PIK3CA exon 9 mutations was observed in dMMR (3/8, 37.5% vs. 6/13, 46.2%; P=0.948) or pMMR tumors (26/30, 86.7% vs. 6/6, 100%, P=0.813), as compared to concurrent RAS/BRAF and PIK3CA exon 20 mutations.
In 35 dMMR cases with PIK3CA mutation, 6 cases (17.1%) were identified as Lynch syndrome. However, no significant difference was observed between these Lynch syndrome cases and non-Lynch syndrome cases in molecular characteristics (data not shown).
Mutation profiling of PIK3CA-mutant tumors by MMR status.
Molecular characteristics of PIK3CA-mutant tumors. (A) Correlation of MAFs in pMMR tumors with concurrent PIK3CA and RAS/BRAF mutations. (B) Correlation of MAFs in dMMR tumors with concurrent PIK3CA and RAS/BRAF mutations. (C) and (D) The subtype mutations of PIK3CA in (C) pMMR tumors and (D) dMMR tumors. Mutations were plotted using the cBioPortal visualization engine.
Discussion
PIK3CA mutation is the most common alteration in PI3K pathway, which plays a pivotal role in tumor development [15]. PIK3CA mutation has been revealed to be a poor prognostic factor, and a negative predictive marker of anti-EGFR therapies in CRC [16]. In this study, we interrogated 528 Chinese CRC resection samples using NGS, and retrospectively investigated the molecular and clinicopathologic characteristics of PIK3CA-mutant dMMR CRCs stratified by MMR status.
It is reported that PIK3CA mutation is related to older age, proximal tumors, mucinous histology, and KRAS mutation [17, 18]. However, these studies do not stratify CRC cases by MMR status, since pMMR and dMMR tumors show distinct differences in clinicopathologic and molecular characteristics. Our previous study has found that intratumor heterogeneity is more likely to occur in PIK3CA-mutant tumors compared with RAS-mutant tumors [19]. However, the effect of MMR status is not considered, and most of the samples we collect are pMMR tumors in the previous study. Here, we found that PIK3CA MAFs were lower than the corresponding RAS/BRAF MAFs in pMMR tumors with concurrent RAS/BRAF and PIK3CA mutations, suggesting that PIK3CA mutation may usually occur in the later stage of CRC in pMMR tumors. However, there was no statistically significant difference between PIK3CA and concurrent RAS/BRAF MAFs in dMMR tumors, indicating that PIK3CA mutation may acts as a clonally dominant truncal mutation, and intratumor heterogeneity of PIK3CA mutation is uncommon in dMMR tumors.
Consistent with previous studies [6, 20], we found that lymph node metastasis, lymphovascular invasion, left colon/rectum location and TP53 mutation were more prevalent in pMMR tumors compared with dMMR tumors. Some studies have reported that less consistent or no correlations about the association of PIK3CA mutation and MMR/MSI status in CRC [17, 21]. However, these studies restrict to hotspot mutation analysis of PIK3CA exon 9 and 20 in western CRC population. We here screened PIK3CA mutation in all exon using NGS, and identified that PIK3CA mutation was more frequent in dMMR tumors than pMMR tumors in Chinese CRC patients.
PIK3CA mutation frequently coexists with RAS/RAF/MEK/MAPK pathway activation (often because of RAS or BRAF mutations) in CRC, leading to poor prognosis [21, 22]. In our study, we found that concomitant RAS/BRAF and PIK3CA mutations were less frequently observed in dMMR tumors than pMMR tumors, further support the notion that PIK3CA mutation may act its role as a truncal alteration in the progression of dMMR CRCs. Recent study has shown that agents targeting PIK3CA mutation demonstrate encouraging preliminary activity in PIK3CA-mutant patients. However, concomitant RAS/BRAF mutations may be a potential mechanism of resistance to PI3K inhibitors [14, 23]. Thus, our findings suggest that PIK3CA-mutant dMMR CRC patients may benefit from PI3K inhibitor because of RAS/BRAF independence. It is reported that PIK3CA exon 9, but not exon 20 mutation, is related to RAS mutation in CRC [24]. Therefore, the association of concurrent RAS/BRAF mutations and PIK3CA exon 9 and 20 mutations were further analyzed, since the high frequency of PIK3CA exon 20 mutation in dMMR tumors in our cohort. However, no significant difference of concurrent RAS/BRAF mutations was observed between PIK3CA exon 9 and 20 mutations, no matter in pMMR or dMMR tumors. These findings indicate that the RAS/BRAF independent PIK3CA mutation has no correlation with the distribution of PIK3CA exon mutation in dMMR tumors.
There are some limitations in our study. First, although no significant difference of molecular characteristics was observed between the Lynch syndrome cases and non-Lynch syndrome cases in PIK3CA-mutant dMMR tumors, the number of Lynch syndrome cases was relatively small. Our findings need to be validated with future larger studies. Second, higher frequency of double PIK3CA mutations was observed in dMMR tumors than pMMR tumors, which we had to attribute to as-yet-unknown covariables. Further basic studies are needed to uncover the function of double PIK3CA mutations in dMMR CRCs.
In summary, our study demonstrates that PIK3CA mutation is more frequent in dMMR tumors than pMMR tumors. Concomitant PIK3CA and RAS/BRAF mutations are more likely to be observed in pMMR tumors than dMMR tumors. In tumors with concurrent RAS/BRAF and PIK3CA mutations, PIK3CA MAFs are highly concordant with the corresponding RAS/BRAF MAFs in dMMR tumors, whereas PIK3CA MAFs are significantly lower than the corresponding RAS/BRAF MAFs in pMMR tumors. These data suggest that PIK3CA mutation may act as a critical player in the development of dMMR CRC. Therapies targeting PIK3CA mutation may gain favorable outcomes in dMMR CRC.
Supplementary Material
Supplementary table.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81702436), WU JIEPING Medical Foundation (320675018156) and CAMS Innovation Fund for Medical Sciences (2017-I2M-1-006).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A. et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177-93
2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32
3. Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479-507
4. Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-7
5. Duval A, Hamelin R. Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Res. 2002;62:2447-54
6. Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609-18
7. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409-13
8. Cathomas G. PIK3CA in Colorectal Cancer. Front Oncol. 2014;4:35
9. Halilovic E, She QB, Ye Q, Pagliarini R, Sellers WR, Solit DB. et al. PIK3CA mutation uncouples tumor growth and cyclin D1 regulation from MEK/ERK and mutant KRAS signaling. Cancer Res. 2010;70:6804-14
10. Trejo CL, Green S, Marsh V, Collisson EA, Iezza G, Phillips WA. et al. Mutationally activated PIK3CA(H1047R) cooperates with BRAF(V600E) to promote lung cancer progression. Cancer Res. 2013;73:6448-61
11. Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S. et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851-7
12. Paleari L, Puntoni M, Clavarezza M, DeCensi M, Cuzick J, DeCensi A. PIK3CA Mutation, Aspirin Use after Diagnosis and Survival of Colorectal Cancer. A Systematic Review and Meta-analysis of Epidemiological Studies. Clin Oncol (R Coll Radiol). 2016;28:317-26
13. Viudez A, Hernandez I, Vera R. Aspirin, PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2013;368:289
14. Polom K, Marrelli D, Roviello G, Pascale V, Voglino C, Vindigni C. et al. PIK3CA mutation in gastric cancer and the role of microsatellite instability status in mutations of exons 9 and 20 of the PIK3CA gene. Adv Clin Exp Med. 2018;7:963-9
15. Marmol I, Sanchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int J Mol Sci. 2017;18:197
16. Mei ZB, Duan CY, Li CB, Cui L, Ogino S. Prognostic role of tumor PIK3CA mutation in colorectal cancer: a systematic review and meta-analysis. Ann Oncol. 2016;27:1836-48
17. Day FL, Jorissen RN, Lipton L, Mouradov D, Sakthianandeswaren A, Christie M. et al. PIK3CA and PTEN gene and exon mutation-specific clinicopathologic and molecular associations in colorectal cancer. Clin Cancer Res. 2013;19:3285-96
18. Tsilimigras DI, Ntanasis-Stathopoulos I, Bagante F, Moris D, Cloyd J, Spartalis E. et al. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP53 genetic mutation analysis for resectable and unresectable colorectal liver metastases: A systematic review of the current evidence. Surg Oncol. 2018;27:280-8
19. Li W, Qiu T, Guo L, Ying J. Major challenges related to tumor biological characteristics in accurate mutation detection of colorectal cancer by next-generation sequencing. Cancer Lett. 2017;410:92-9
20. Zhao Y, Ando K, Oki E, Ikawa-Yoshida A, Ida S, Kimura Y. et al. Aberrations of BUBR1 and TP53 gene mutually associated with chromosomal instability in human colorectal cancer. Anticancer Res. 2014;34:5421-7
21. Velho S, Oliveira C, Ferreira A, Ferreira AC, Suriano G, Schwartz S Jr. et al. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur J Cancer. 2005;41:1649-54
22. Barault L, Veyrie N, Jooste V, Lecorre D, Chapusot C, Ferraz JM. et al. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer. 2008;122:2255-9
23. Kim JS, Kim JE, Kim K, Lee J, Park JO, Lim HY. et al. The Impact of Cetuximab Plus AKT- or mTOR- Inhibitor in a Patient-Derived Colon Cancer Cell Model with Wild-Type RAS and PIK3CA Mutation. J Cancer. 2017;8:2713-9
24. Hechtman JF, Sadowska J, Huse JT, Borsu L, Yaeger R, Shia J. et al. AKT1 E17K in Colorectal Carcinoma Is Associated with BRAF V600E but Not MSI-H Status: A Clinicopathologic Comparison to PIK3CA Helical and Kinase Domain Mutants. Mol Cancer Res. 2015;13:1003-8
Author contact
![]() Corresponding authors: Prof. Lei Guo or Dr. Jianming Ying, Department of Pathology, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, No.17 Panjiayuan Nanli, Beijing 100021, China. Tel: +86-10-87788498; Fax: +86-10-87787515; E-mail: guolei0520com (G. L.); jmyingac.cn (Y.J.)
Corresponding authors: Prof. Lei Guo or Dr. Jianming Ying, Department of Pathology, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, No.17 Panjiayuan Nanli, Beijing 100021, China. Tel: +86-10-87788498; Fax: +86-10-87787515; E-mail: guolei0520com (G. L.); jmyingac.cn (Y.J.)

 Global reach, higher impact
Global reach, higher impact