3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(12):3512-3518. doi:10.7150/jca.42798 This issue Cite
Research Paper
LIN28B gene polymorphisms modify hepatoblastoma susceptibility in Chinese children
1. Department of Pediatric Surgery, Shengjing Hospital of China Medical University, Shenyang 110004, Liaoning, China.
2. Department of Clinical Medicine, The Fourth Affiliated Hospital of China Medical University, Shenyang 110001, Liaoning, China.
3. Department of Clinical Laboratory, Biobank, Harbin Medical University Cancer Hospital, Harbin 150040, Heilongjiang, China.
4. Department of Pediatric Surgery, the First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan, China.
5. Clinical Laboratory Medicine Center of PLA, Xijing Hospital, Air Force Medical University, Xi'an 710032, Shaanxi, China.
6. Kunming Key Laboratory of Children Infection and Immunity, Yunnan Key Laboratory of Children's Major Disease Research, Yunnan Institute of Pediatrics Research, Yunnan Medical Center for Pediatric Diseases, Kunming Children's Hospital, Kunming 650228, Yunnan, China.
7. Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou 510623, Guangdong, China.
#These authors contributed equally to this work.
Received 2019-12-6; Accepted 2020-3-2; Published 2020-3-15
Abstract
Hepatoblastoma is one of the malignant liver tumors in children. However, genetic mechanisms underpinning the initiation of hepatoblastoma remain largely unclear. The previous study showed that lin-28 homolog B (LIN28B) might play a role in the development of hepatoblastoma. To detect the association between LIN28B gene polymorphisms and hepatoblastoma risk in Chinese children, we conducted a five-center case-control study of 275 hepatoblastoma patients and 1018 cancer-free controls. Four potentially functional polymorphisms were genotyped using the Taqman method. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to evaluate the strength of the associations. We found that the rs314276 C>A polymorphism (AA vs. CC: adjusted OR=2.05, 95% CI=1.36-3.10, P=0.0006; AA vs. CA/CC: adjusted OR=2.11, 95% CI=1.43-3.12, P=0.0002) and rs9404590 T>G (GG vs. TT: adjusted OR=1.89, 95% CI=1.20-3.00, P=0.007; GG vs. TT/TG: adjusted OR=1.87, 95% CI=1.20-2.92, P=0.006) were associated with increased hepatoblastoma risk. Combination analysis of risk genotypes showed that patients with four risk genotypes had a higher chance of developing hepatoblastoma than carriers of 1 to 3 risk genotypes. Stratification analysis showed the significant association between the rs314276 AA genotype and hepatoblastoma risk in both age and sex groups, as well as clinical stages III+IV cases. The rs9404590 GG genotype was associated with hepatoblastoma risk in participants' ≥17 months, in females, and for those with clinical stages III+IV disease. Furthermore, four risk genotypes confer higher hepatoblastoma susceptibility in both age and sex groups, as well as groups with clinical stages III+IV disease. Genotype-based gene expression analysis confirmed that the rs9404590 T>G polymorphism was significantly associated with altered LIN28B gene expression. We further validated our findings using false-positive probability analysis. This finding suggested that LIN28B gene polymorphisms may be associated with an increased predisposition to hepatoblastoma.
Keywords: LIN28B, polymorphism, hepatoblastoma, susceptibility, case-control study
Introduction
Hepatoblastoma is one of the major malignant tumors in children and originates from the progenitor cell during embryogenesis, accounting for two-thirds of the pediatric malignant tumors [1]. The incidence rate is approximately 1.5 cases/million population per year (around 1% of cancers for young children) [2] and has been rising 4% per year continually [3]. This phenomenon is partly due to increased prematurity and very low birth weight [4,5]. The development of hepatoblastoma is also associated with some environmental factors, such as parental exposure to tobacco, metal and petroleum products [6-9]. Moreover, it is strongly associated with familial adenomatous polyposis [10], Beckwith-Wiedemann syndrome [11], and Glycogen storage disease [12], indicating some genetic underpinnings. Although progressive surgical techniques and chemotherapy regimens have increased survival rates, the prognosis of high-risk hepatoblastoma remains very poor [13]. As for the molecular mechanism, although Wnt/β-catenin, MYC, and Hippo pathways have been reported to be involved in the pathogenesis of hepatoblastoma [14-16], little is known about the molecular basis of this disease, and there are no validated prognostic or therapeutic biomarkers for hepatoblastoma patients.
LIN28B gene encodes an RNA-binding protein, which is featured by the conservative N-terminal cold-shock domain and two C-terminal CysCysHisCys zinc fingers [17]. LIN28B functions by blocking the maturity of tumor-suppressing microRNA (miRNA) let-7 family, which subsequently causes overexpression of numerous oncogenes, such as C-MYC and K-RAS, thereby supporting tumorigenesis and tumor growth [18,19]. Meanwhile, LIN28B itself is downregulated by let-7, leading to the formation of double-negative feedback [20]. In this way, LIN28B is aberrantly expressed in a broad spectrum of tumors and engaged in the regulation of miRNAs [18,20,21]. A study showed that LIN28B is sufficient to drive liver tumors in the let-7 miRNA dependent and independent ways in endogenous tumor models and is over-activated in mouse models of MYC-driven hepatoblastoma [19], which indicates its essential role in the development of hepatoblastoma.
The elucidation of genetic mechanisms of hepatoblastoma may accelerate the development of preventive oncology. Genome-wide association study (GWAS) is one of the emerging and promising techniques to associate genetic variations with disease risk. Most of the genetic variants are single nucleotide polymorphisms (SNPs). LIN28B polymorphisms were associated with Wilms tumor [22] and neuroblastoma [23] susceptibility in Chinese children. However, the relationship of LIN28B polymorphisms with hepatoblastoma susceptibility has not been investigated. In this study, we did a five-center case-control study to investigate the association between LIN28B gene polymorphisms and hepatoblastoma susceptibility in Chinese Han children.
Material and methods
Patients and controls
We enrolled 275 histopathologically diagnosed hepatoblastoma patients and 1018 cancer-free controls from Guangdong, Henan, Shaanxi, Yunnan and Liaoning provinces (Supplemental Table 1). All controls are unrelated to patients genetically. Moreover, controls were matched to patients by age, gender, and ethnicity. Our study was approved by the Ethics Committee of Guangzhou Women and Children's Medical Center. Written informed consent was obtained from each patient or his/her guardian. The study protocol was compliant with ethical guidelines.
SNP selection and genotyping
Four LIN28B polymorphisms (rs314276 C>A, rs221634 A>T, rs221635 T>C and rs9404590 T>G) were chosen and genotyped using the TaqMan real-time PCR method as we reported previously [22, 23]. Briefly, the selected polymorphisms were all potentially functional SNPs according to SNPinfo online software (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html), which can affect the binding capacity of transcription factor binding sites (rs314276) or microRNA binding sites (rs221634 and rs221635), or leading to amino acids alterations (rs9404590). To validate the accuracy of genotyping results and for quality control, approximately 10% of the samples were randomly selected and re-genotyped. The concordance for the quality control samples was 100%.
Genotype and gene expression correlation analysis
GTEx Portal database (https://www.gtexportal.org/home/) was used to evaluate the correlation between genotypes of the selected polymorphisms and LIN28B mRNA expression levels [24].
Statistical analysis
The χ2 test was used to evaluate the demographic variables distribution, risk factors distribution, and LIN28B genotype distributions between case and control groups. The χ2 test was also performed to assess whether or not the LIN28B genotypes were consistent with Hardy-Weinberg equilibrium (HWE). Unconditional univariate and multivariate logistic regression analyses were used to estimate the strength of association between the selected polymorphisms and hepatoblastoma risk, using odds ratio (ORs) and 95% confidence intervals (CIs). Age and gender were adjusted for in the multivariate analysis. Further stratification analysis was performed based on the age, sex, and clinical stages. Moreover, we also performed false-positive probability analysis (FPRP) analysis to verify the significant results from the combined subjects [25]. Differences with P values <0.05 were counted as statistically significant. All two-sided statistical analyses were performed using SAS software (version 9.1; SAS Institute, Cary, NC, United States).
Results
General characteristics of the subjects
As shown in Supplemental Table 1, there is no significant difference in both cases and controls in terms of age (P=0.365) and gender (P=0.589). The majority of the subjects in both cases and controls are male, accounting for 58.91% (162/275) and 60.71% (618/1018), respectively.
Association of LIN28B SNPs with hepatoblastoma susceptibility
Of the included subjects, 275 cases and 1017 controls were successfully genotyped. The LIN28B genotypes are in accordance with HWE in the controls (P=0.209 for rs314276 C>A, P=0.969 for rs221634 A>T, P=0.139 for rs221635 T>C and P=0.868 for rs9404590 T>G). The genotype frequencies of four SNPs in cases and controls were listed in Table 1. Our results indicated that patients with the rs314276 A allele had higher cancer risk (AA vs. CC: adjusted OR=2.05, 95% CI=1.36-3.10, P=0.0006; AA vs. CA/CC: adjusted OR=2.11, 95% CI=1.43-3.12, P=0.0002). Carriers of the rs9404590 G allele also showed significantly increased risk (GG vs. TT: adjusted OR=1.89, 95% CI=1.20-3.00, P=0.007; GG vs. TT/TG: adjusted OR=1.87, 95% CI=1.20-2.92, P=0.006), compared with the reference group. According to the ORs, risk genotypes were carriers with rs314276 AA, rs221634 AA/AT, rs221635 TC/TT, rs9404590 TG/GG. Meanwhile, patients carrying four risk genotypes had a significantly elevated risk (adjusted OR=1.95, 95% CI=1.31-2.90, P=0.0009) when compared with those with 1-3 risk genotypes. Unfortunately, no significant association was detected for either rs221634 A>T or rs221635 T>C in any comparison.
Association between LIN28B gene polymorphisms and hepatoblastoma susceptibility
| Genotype | Cases (N=275) | Controls (N=1017) | P a | Crude OR (95% CI) | P | Adjusted OR (95% CI) b | P b |
|---|---|---|---|---|---|---|---|
| rs314276 (HWE=0.209) | |||||||
| CC | 123 (44.73) | 483 (47.49) | 1.00 | 1.00 | |||
| CA | 107 (38.91) | 448 (44.05) | 0.94 (0.70-1.25) | 0.664 | 0.94 (0.70-1.26) | 0.672 | |
| AA | 45 (16.36) | 86 (8.46) | 2.06 (1.36-3.10) | 0.0006 | 2.05 (1.36-3.10) | 0.0006 | |
| Additive | 0.017 | 1.27 (1.04-1.55) | 0.018 | 1.27 (1.04-1.55) | 0.018 | ||
| Dominant | 152 (55.27) | 534 (52.51) | 0.415 | 1.12 (0.86-1.46) | 0.415 | 1.12 (0.86-1.46) | 0.409 |
| Recessive | 230 (83.64) | 931 (91.54) | 0.0001 | 2.12 (1.44-3.12) | 0.0002 | 2.11 (1.43-3.12) | 0.0002 |
| rs221634 (HWE=0.969) | |||||||
| AA | 94 (34.55) | 342 (33.63) | 1.00 | 1.00 | |||
| AT | 144 (52.36) | 495 (48.67) | 1.05 (0.78-1.40) | 0.758 | 1.04 (0.78-1.40) | 0.772 | |
| TT | 36 (13.09) | 180 (17.70) | 0.72 (0.47-1.10) | 0.129 | 0.72 (0.47-1.10) | 0.130 | |
| Additive | 0.239 | 0.89 (0.73-1.08) | 0.239 | 0.89 (0.73-1.08) | 0.239 | ||
| Dominant | 180 (65.45) | 675 (66.37) | 0.776 | 0.96 (0.73-1.27) | 0.775 | 0.96 (0.72-1.27) | 0.765 |
| Recessive | 239 (86.91) | 837 (82.30) | 0.069 | 0.70 (0.48-1.03) | 0.071 | 0.70 (0.48-1.03) | 0.073 |
| rs221635 (HWE=0.139) | |||||||
| TT | 185 (67.27) | 652 (64.11) | 1.00 | 1.00 | |||
| TC | 79 (28.73) | 315 (30.97) | 0.88 (0.66-1.19) | 0.413 | 0.88 (0.66-1.19) | 0.407 | |
| CC | 11 (4.00) | 50 (4.92) | 0.78 (0.40-1.52) | 0.459 | 0.78 (0.40-1.52) | 0.458 | |
| Additive | 0.300 | 0.88 (0.70-1.12) | 0.300 | 0.88 (0.70-1.12) | 0.296 | ||
| Dominant | 90 (32.73) | 365 (35.89) | 0.330 | 0.87 (0.66-1.15) | 0.330 | 0.87 (0.65-1.15) | 0.325 |
| Recessive | 264 (96.00) | 967 (95.08) | 0.525 | 0.81 (0.41-1.57) | 0.526 | 0.81 (0.41-1.57) | 0.527 |
| rs9404590 (HWE=0.868) | |||||||
| TT | 141 (51.27) | 558 (54.87) | 1.00 | 1.00 | |||
| TG | 102 (37.09) | 392 (38.54) | 1.03 (0.77-1.37) | 0.841 | 1.03 (0.78-1.37) | 0.834 | |
| GG | 32 (11.64) | 67 (6.59) | 1.89 (1.19-2.99) | 0.007 | 1.89 (1.20-3.00) | 0.007 | |
| Additive | 0.045 | 1.23 (1.00-1.51) | 0.045 | 1.24 (1.01-1.52) | 0.044 | ||
| Dominant | 134 (48.73) | 459 (45.13) | 0.289 | 1.16 (0.89-1.51) | 0.289 | 1.16 (0.89-1.51) | 0.284 |
| Recessive | 243 (88.36) | 950 (93.41) | 0.005 | 1.87 (1.20-2.91) | 0.006 | 1.87 (1.20-2.92) | 0.006 |
| Combined effect of risk genotypes | |||||||
| 1 | 44 (16.00) | 219 (21.53) | 1.00 | 1.00 | |||
| 2 | 97 (35.27) | 350 (34.41) | 1.38 (0.93-2.05) | 0.100 | 1.37 (0.93-2.04) | 0.115 | |
| 3 | 92 (33.45) | 362 (35.59) | 1.27 (0.85-1.88) | 0.245 | 1.26 (0.85-1.88) | 0.249 | |
| 4 | 42 (15.27) | 86 (8.46) | 2.43 (1.49-3.97) | 0.0004 | 2.43 (1.48-3.96) | 0.0004 | |
| 1-3 | 233 (84.73) | 931 (91.54) | 1.00 | 1.00 | |||
| 4 | 42 (15.27) | 86 (8.46) | 0.0008 | 1.95 (1.31-2.90) | 0.0009 | 1.95 (1.31-2.90) | 0.0009 |
OR, odds ratio; CI, confidence interval; HWE, Hardy-Weinberg equilibrium. a χ2 test for genotype distributions between hepatoblastoma patients and controls, b Adjusted for age and gender, c Risk genotypes were carriers with rs314276 AA, rs221634 AA/AT, rs221635 TC/TT, rs9404590 TG/GG.
Stratification analysis of LIN28B risk genotypes with hepatoblastoma susceptibility
| Variables | rs314276 (cases/controls) | AOR (95% CI) a | P a | rs9404590 (cases/controls) | AOR (95% CI) a | P a | Combined (cases/controls) | AOR (95% CI) a | P a | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC/CA | AA | TT/TG | GG | 1-3 | 4 | |||||||
| Age, month | ||||||||||||
| <17 | 123/420 | 25/39 | 2.19 (1.28-3.76) | 0.005 | 131/427 | 17/32 | 1.73 (0.93-3.21) | 0.084 | 126/420 | 22/39 | 1.88 (1.08-3.29) | 0.027 |
| ≥17 | 107/511 | 20/47 | 2.04 (1.16-3.58) | 0.013 | 112/523 | 15/35 | 2.02 (1.07-3.83) | 0.031 | 107/511 | 20/47 | 2.04 (1.16-3.58) | 0.013 |
| Gender | ||||||||||||
| Females | 94/366 | 19/33 | 2.21 (1.20-4.06) | 0.011 | 100/376 | 13/23 | 2.09 (1.02-4.28) | 0.044 | 96/366 | 17/33 | 1.94 (1.03-3.63) | 0.039 |
| Males | 136/565 | 26/53 | 2.04 (1.23-3.39) | 0.006 | 143/574 | 19/44 | 1.74 (0.98-3.07) | 0.057 | 137/565 | 25/53 | 1.95 (1.17-3.25) | 0.010 |
| Clinical stages | ||||||||||||
| I+II | 124/931 | 18/86 | 1.57 (0.92-2.70) | 0.101 | 129/950 | 13/67 | 1.44 (0.78-2.69) | 0.247 | 125/931 | 17/86 | 1.48 (0.85-2.56) | 0.168 |
| III+IV | 55/931 | 16/86 | 3.15 (1.73-5.73) | 0.0002 | 59/950 | 12/67 | 2.88 (1.47-5.62) | 0.002 | 55/931 | 16/86 | 3.15 (1.73-5.73) | 0.0002 |
AOR, adjusted odds ratio; CI, confidence interval. a Adjusted for age and gender, omitting the corresponding factor.
Stratification analysis
As shown in Table 2, we found a more prominent association for rs314276 AA genotype in the following subgroups: patients <17 months (adjusted OR=2.19, 95% CI=1.28-3.76, P=0.005) and ≥17 months (adjusted OR=2.04, 95% CI=1.16-3.58, P=0.013), females (adjusted OR=2.21, 95% CI=1.20-4.06, P=0.011) and males (adjusted OR=2.04, 95% CI=1.23-3.39, P=0.006), as well as patients with tumors in clinical stages III+IV (adjusted OR=3.15, 95% CI=1.73-5.73, P=0.0002). The association between the rs9404590 GG genotype and increased cancer risk was more pronounced in the strata of patients ≥17 months (adjusted OR=2.02, 95% CI=1.07-3.83, P=0.031), females (adjusted OR=2.09, 95% CI=1.02-4.28, P=0.044) and patients with tumor in III+IV clinical stages (adjusted OR=2.88, 95% CI=1.47-5.62, P=0.002). As for risk genotypes, the association between 4 risk genotypes and HB risk was statistically significant in patients younger (adjusted OR=1.88, 95% CI=1.08-3.29, P=0.027) and ≥17 months (adjusted OR=2.04, 95% CI=1.16-3.58, P=0.013), males (adjusted OR=1.94, 95% CI=1.03-3.63, P=0.039), and females (adjusted OR=1.95, 95% CI=1.17-3.25, P=0.010) and patients with tumor in III+IV clinical stages (adjusted OR=3.15, 95% CI=1.73-5.73, P=0.0002).
Genotype-based mRNA expression analysis
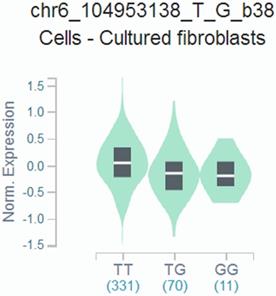
We found that the rs9404590 T>G polymorphism was significantly associated with altered gene expression in transformed fibroblast cells using data from the GTEx Portal (P=2.20*10-4, Figure 1).
False-positive report probability results
We preset 0.2 as the FPRP threshold. As shown in Table 3, at the prior probability of 0.1, the significant findings for the rs314276 C>A polymorphism remained noteworthy on the rs314276 AA genotype (FPRP=0.064), recessive model (FPRP=0.037) and the III+IV stage tumor (FPRP=0.144). All of the significant finding in rs9404590 T>G disappeared. Moreover, in the combined analysis, results with 4 risk genotypes remained noteworthy when compared with results with a risk genotype (FPRP=0.052) and 1-3 genotypes (FPRP=0.078), and in the stratified analysis, results of stage III+IV tumors also reached FPRP threshold (FPRP=0.144).
Genotype-based gene expression in transformed fibroblast cells using data from the GTEx Portal for rs9404590 T>G polymorphism (P=2.20*10-4).
Discussion
In this case-control study, we investigated the association between LIN28B SNPs with the risk of hepatoblastoma in Chinese children. To the best of our knowledge, our team is the first group to assess the association of LIN28B SNPs with hepatoblastoma susceptibility.
The LIN28B gene is located on chromosome 6q21 and encodes a miRNA-binding protein [26]. The heterochronic LIN28 gene the in Caenorhabditis elegans [27], and its mammalian homologue LIN28B gene is complementary to lin-4 homologues miR-125 and let-7 critical by the segment of the unusually long 3' untranslated region (UTR) in diverse mammalian tissues [17, 27-29]. In this way, LIN28B sustains the proliferative and metabolic capacities of pluripotent stem cells and facilitates the transition of them from naive to primed pluripotency [17, 30-32]. Since LIN28B functions as a key regulator in the diverse developing events, SNPs in the 3' UTR may play important roles. SNPs in this gene have been extensively explored concerning the regulation of secondary sexual characteristics and tumorigenesis. For example, LIN28B rs314276 C>A polymorphism has been shown to associate with reproductive timing [33,34], central precocious puberty [35], the linkage between puberty timing and adult disease [36], and the finger-length ratio [37]. Meanwhile, this variant was also found to be associated with the risk and survival of cancers, such as Wilms tumor [22], neuroblastoma [23] and epithelial ovarian cancer [38]. Previous data also showed that LIN28B rs221634 A>T, rs221635 T>C, and rs9404590 T>G were related to the susceptibility to Wilms tumor [22] and neuroblastoma [23].
The LIN28B is upregulated to repress the function of the tumor suppressor miRNA let-7 family in the diverse tumor types and serve as a regulator of miRNA [20,39-41]. The overexpression of LIN28B can lead to the activation of several oncogenes [42]. In the previous study, miR-100, let-7a, and the Caenorhabditis elegans lin-4 orthologue miR-125b are sharply downregulated in the hepatoblastoma [43]. It is noteworthy that two of the miRNAs (miR-125b and let-7a) in the cluster are LIN28B-related miRNA [27], indicating a potential molecular mechanism. Meanwhile, LIN28B and AURKA expressions were upregulated in a transcriptomic and genomic analysis of human hepatoblastoma, and miRNA-26-5p inhibited hepatoblastoma by repressing LIN28B [44]. LIN28B might also increase the risk of hepatoblastoma through LIN28B-RAN-AURKA pathway [44]. The previous study showed that LIN28B is an oncofetal cancer stem cell-like marker in the recurrence of hepatocellular carcinoma [45]. Hepatocellular carcinoma may have similar molecular mechanisms with hepatoblastoma, such as the activation of the Wnt/β-catenin signaling pathway [46-49]. Therefore, it was reasonable to investigate the genetic implication of the LIN28B gene in the risk of hepatoblastoma.
Despite the findings that LIN28B rs94904590 T>G and rs314276 C>A could increase hepatoblastoma risk, the study suffered from some minor limitations. First, we did not find the exact molecular mechanism underlying the established associations in the study. Secondly, the sample size might be small to conclude, partly due to the low incidence rate of the disease. Validation experiments are needed in the future. Thirdly, we only recruited the Han Chinese people in our study. Fourthly, we only included four potentially functional SNPs in the LIN28B gene, other SNPs including the ones without functional were not included in the current study. Finally, functional experiments should be performed to further explore the role of LIN28B in the carcinogenesis of hepatoblastoma.
False-positive report probability analysis for significant findings
| Genotype | OR (95% CI) | P a | Statistical power b | Prior probability | ||||
|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | ||||
| rs314276 C>A | ||||||||
| AA vs. CC | 2.06 (1.36-3.10) | 0.0006 | 0.079 | 0.022 | 0.064 | 0.429 | 0.883 | 0.987 |
| AA vs. CC/CA | 2.12 (1.44-3.12) | 0.0002 | 0.047 | 0.013 | 0.037 | 0.296 | 0.809 | 0.977 |
| <17 month | 2.19 (1.27-3.76) | 0.005 | 0.085 | 0.137 | 0.324 | 0.840 | 0.982 | 0.998 |
| ≥17 month | 2.03 (1.16-3.57) | 0.014 | 0.147 | 0.217 | 0.454 | 0.902 | 0.989 | 0.999 |
| Females | 2.24 (1.22-4.12) | 0.009 | 0.098 | 0.221 | 0.460 | 0.903 | 0.990 | 0.999 |
| Males | 2.04 (1.23-3.38) | 0.006 | 0.117 | 0.127 | 0.304 | 0.828 | 0.980 | 0.998 |
| Stage III+IV | 3.15 (1.73-5.73) | 0.0002 | 0.011 | 0.053 | 0.144 | 0.649 | 0.949 | 0.995 |
| rs9404590 T>G | ||||||||
| GG vs. TT | 1.89 (1.19-2.99) | 0.007 | 0.177 | 0.102 | 0.254 | 0.789 | 0.974 | 0.997 |
| GG vs. GT/TT | 1.87 (1.20-2.91) | 0.006 | 0.168 | 0.095 | 0.240 | 0.777 | 0.972 | 0.997 |
| ≥17 month | 2.00 (1.06-3.79) | 0.033 | 0.190 | 0.344 | 0.612 | 0.945 | 0.994 | 0.999 |
| Females | 2.13 (1.04-4.34) | 0.039 | 0.170 | 0.406 | 0.673 | 0.958 | 0.996 | 1.000 |
| Stage III+IV | 2.88 (1.48-5.63) | 0.002 | 0.032 | 0.150 | 0.346 | 0.853 | 0.983 | 0.998 |
| Risk genotypes | ||||||||
| 4 vs. 1 | 2.43 (1.49-3.97) | 0.0004 | 0.065 | 0.018 | 0.052 | 0.377 | 0.859 | 0.984 |
| 4 vs. 1-3 | 1.95 (1.31-2.90) | 0.0009 | 0.095 | 0.028 | 0.078 | 0.484 | 0.904 | 0.990 |
| <17 month | 1.88 (1.08-3.29) | 0.027 | 0.213 | 0.274 | 0.532 | 0.926 | 0.992 | 0.999 |
| ≥17 month | 2.03 (1.16-3.57) | 0.014 | 0.147 | 0.217 | 0.454 | 0.902 | 0.989 | 0.999 |
| Females | 1.96 (1.05-3.68) | 0.035 | 0.200 | 0.343 | 0.611 | 0.945 | 0.994 | 0.999 |
| Males | 1.95 (1.17-3.24) | 0.011 | 0.160 | 0.167 | 0.376 | 0.869 | 0.985 | 0.999 |
| Stage III+IV | 3.15 (1.73-5.73) | 0.0002 | 0.011 | 0.053 | 0.144 | 0.649 | 0.949 | 0.995 |
OR, odds ratio; CI, confidence interval.
aChi-square test was used to calculate the genotype frequency distributions, bStatistical power was calculated using the number of observations in each subgroup and the corresponding ORs and P values in this table.
In all, our study demonstrated that LIN28B rs94904590 T>G and rs314276 C>A might confer increased hepatoblastoma risk in Chinese children. In the future, studies with a larger sample size are called to clarify the impact of LIN28B SNPs on the risk of hepatoblastoma.
Supplementary Material
Supplementary table.
Acknowledgements
This study was funded by grants from the National Natural Science Foundation of China (No: 81800453) and the Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease (No: 2019B030301004). The funders have no role in the manuscript writing, editing, approval, or decision to publish.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Darbari A, Sabin KM, Shapiro CN, Schwarz KB. Epidemiology of primary hepatic malignancies in U.S. children. Hepatology. 2003;38:560-6
2. Lim IIP, Bondoc AJ, Geller JI, Tiao GM. Hepatoblastoma-The Evolution of Biology, Surgery, and Transplantation. Children (Basel). 2018;6:E1
3. Linabery AM, Ross JA. Trends in childhood cancer incidence in the U.S. (1992-2004). Cancer. 2008;112:416-32
4. Tanimura M, Matsui I, Abe J, Ikeda H, Kobayashi N, Ohira M. et al. Increased risk of hepatoblastoma among immature children with a lower birth weight. Cancer Res. 1998;58:3032-5
5. Ikeda H, Matsuyama S, Tanimura M. Association between hepatoblastoma and very low birth weight: a trend or a chance? J Pediatr. 1997;130:557-60
6. McLaughlin CC, Baptiste MS, Schymura MJ, Nasca PC, Zdeb MS. Maternal and infant birth characteristics and hepatoblastoma. Am J Epidemiol. 2006;163:818-28
7. Buckley JD, Sather H, Ruccione K, Rogers PC, Haas JE, Henderson BE. et al. A case-control study of risk factors for hepatoblastoma. A report from the Childrens Cancer Study Group. Cancer. 1989;64:1169-76
8. Janitz AE, Ramachandran G, Tomlinson GE, Krailo M, Richardson M, Spector L. Maternal and paternal occupational exposures and hepatoblastoma: results from the HOPE study through the Children's Oncology Group. J Expo Sci Environ Epidemiol. 2017;27:359-64
9. Secretan B, Straif K, Baan R, Grosse Y, El Ghissassi F, Bouvard V. et al. A review of human carcinogens-Part E: tobacco, areca nut, alcohol, coal smoke, and salted fish. Lancet Oncol. 2009;10:1033-4
10. Hughes LJ, Michels VV. Risk of hepatoblastoma in familial adenomatous polyposis. Am J Med Genet. 1992;43:1023-5
11. Maas SM, Vansenne F, Kadouch DJ, Ibrahim A, Bliek J, Hopman S. et al. Phenotype, cancer risk, and surveillance in Beckwith-Wiedemann syndrome depending on molecular genetic subgroups. Am J Med Genet A. 2016;170:2248-60
12. Ito E, Sato Y, Kawauchi K, Munakata H, Kamata Y, Yodono H. et al. Type 1a glycogen storage disease with hepatoblastoma in siblings. Cancer. 1987;59:1776-80
13. Davies JQ, de la Hall PM, Kaschula RO, Sinclair-Smith CC, Hartley P, Rode H. et al. Hepatoblastoma-evolution of management and outcome and significance of histology of the resected tumor. A 31-year experience with 40 cases. J Pediatr Surg. 2004;39:1321-7
14. Sumazin P, Chen Y, Trevino LR, Sarabia SF, Hampton OA, Patel K. et al. Genomic analysis of hepatoblastoma identifies distinct molecular and prognostic subgroups. Hepatology. 2017;65:104-21
15. Cairo S, Armengol C, Buendia MA. Activation of Wnt and Myc signaling in hepatoblastoma. Front Biosci (Elite Ed). 2012;4:480-6
16. Li N, Xie C, Lu N. Crosstalk between Hippo signalling and miRNAs in tumour progression. FEBS J. 2017;284:1045-55
17. Tsanov KM, Pearson DS, Wu Z, Han A, Triboulet R, Seligson MT. et al. LIN28 phosphorylation by MAPK/ERK couples signalling to the post-transcriptional control of pluripotency. Nat Cell Biol. 2017;19:60-7
18. Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97-100
19. Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M. et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A. 2009;106:3384-9
20. Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R. et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987-93
21. Zhou J, Ng SB, Chng WJ. LIN28/LIN28B: an emerging oncogenic driver in cancer stem cells. Int J Biochem Cell Biol. 2013;45:973-8
22. Fu W, Liu GC, Zhao Z, Zhu J, Jia W, Zhu SB. et al. The correlation between LIN28B gene potentially functional variants and Wilms tumor susceptibility in Chinese children. J Clin Lab Anal. 2018;32:e22200
23. He J, Yang T, Zhang R, Zhu J, Wang F, Zou Y. et al. Potentially functional polymorphisms in the LIN28B gene contribute to neuroblastoma susceptibility in Chinese children. J Cell Mol Med. 2016;20:1534-41
24. Zhu J, Fu W, Jia W, Xia H, Liu GC, He J. Association between NER Pathway Gene Polymorphisms and Wilms Tumor Risk. Mol Ther Nucleic Acids. 2018;12:854-60
25. Zhou L, Zheng Y, Tian T, Liu K, Wang M, Lin S. et al. Associations of interleukin-6 gene polymorphisms with cancer risk: Evidence based on 49,408 cancer cases and 61,790 controls. Gene. 2018;670:136-47
26. Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 2003;258:432-42
27. Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637-46
28. Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE. et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901-6
29. Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659-69
30. Zhang J, Ratanasirintrawoot S, Chandrasekaran S, Wu Z, Ficarro SB, Yu C. et al. LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell Stem Cell. 2016;19:66-80
31. Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12:395-406
32. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917-20
33. Spencer KL, Malinowski J, Carty CL, Franceschini N, Fernandez-Rhodes L, Young A. et al. Genetic variation and reproductive timing: African American women from the Population Architecture using Genomics and Epidemiology (PAGE) Study. PLoS One. 2013;8:e55258
34. Ong KK, Elks CE, Li S, Zhao JH, Luan J, Andersen LB. et al. Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet. 2009;41:729-33
35. Hu Z, Chen R, Cai C. Association of genetic polymorphisms around the LIN28B gene and idiopathic central precocious puberty risks among Chinese girls. Pediatr Res. 2016;80:521-5
36. Leinonen JT, Surakka I, Havulinna AS, Kettunen J, Luoto R, Salomaa V. et al. Association of LIN28B with adult adiposity-related traits in females. PLoS One. 2012;7:e48785
37. Medland SE, Zayats T, Glaser B, Nyholt DR, Gordon SD, Wright MJ. et al. A variant in LIN28B is associated with 2D:4D finger-length ratio, a putative retrospective biomarker of prenatal testosterone exposure. Am J Hum Genet. 2010;86:519-25
38. Permuth-Wey J, Kim D, Tsai YY, Lin HY, Chen YA, Barnholtz-Sloan J. et al. LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res. 2011;71:3896-903
39. Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J. et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696-708
40. Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276-84
41. Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP. et al. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066-79
42. Balzeau J, Menezes MR, Cao S, Hagan JP. The LIN28/let-7 Pathway in Cancer. Front Genet. 2017;8:31
43. Luo JH, Ren B, Keryanov S, Tseng GC, Rao UN, Monga SP. et al. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology. 2006;44:1012-24
44. Zhang Y, Zhao Y, Wu J, Liangpunsakul S, Niu J, Wang L. MicroRNA-26-5p functions as a new inhibitor of hepatoblastoma by repressing lin-28 homolog B and aurora kinase a expression. Hepatol Commun. 2018;2:861-71
45. Cheng SW, Tsai HW, Lin YJ, Cheng PN, Chang YC, Yen CJ. et al. Lin28B is an oncofetal circulating cancer stem cell-like marker associated with recurrence of hepatocellular carcinoma. PLoS One. 2013;8:e80053
46. Ng K, Mogul DB. Pediatric Liver Tumors. Clinics in Liver Disease. 2018;22:753-72
47. Sharma D, Subbarao G, Saxena R. Hepatoblastoma. Semin Diagn Pathol. 2017;34:192-200
48. Kim W, Khan SK, Gvozdenovic-Jeremic J, Kim Y, Dahlman J, Kim H. et al. Hippo signaling interactions with Wnt/beta-catenin and Notch signaling repress liver tumorigenesis. J Clin Invest. 2017;127:137-52
49. Cairo S, Armengol C, De Reynies A, Wei Y, Thomas E, Renard CA. et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471-84
Author contact
![]() Corresponding authors: Weilin Wang, Department of Pediatric Surgery, Shengjing Hospital of China Medical University, 36 Sanhao Street, Heping District, Shenyang 110004, Liaoning, China. E-mail: wangwlorg; or Jing He, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China. E-mail: hejing198374com or hejingorg.
Corresponding authors: Weilin Wang, Department of Pediatric Surgery, Shengjing Hospital of China Medical University, 36 Sanhao Street, Heping District, Shenyang 110004, Liaoning, China. E-mail: wangwlorg; or Jing He, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China. E-mail: hejing198374com or hejingorg.

 Global reach, higher impact
Global reach, higher impact