Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(9):2431-2441. doi:10.7150/jca.39083 This issue Cite
Research Paper
A novel classification based on B-cell receptor signal gene expression correlates with prognosis in primary breast diffuse large B-cell lymphoma
1. Department of Hematology, First Affiliated Hospital of Harbin Medical University, Harbin Medical University, Harbin 150081, Heilongjiang, China
2. Department of Medical Oncology, Harbin Medical University Cancer Hospital, Harbin Medical University, Harbin 150081, Heilongjiang, China
3. Institute of Cancer Prevention and Treatment, Harbin Medical University, Harbin 150081, Heilongjiang, China
4. Heilongjiang Academy of Medical Sciences, Harbin 150081, Heilongjiang, China
5. Department of General Surgery, Second Affiliated Hospital of Harbin Medical University, Harbin Medical University, Harbin 150081, Heilongjiang, China
* Wenjia Su and Xingjian Niu contributed equally to this work
Received 2019-8-6; Accepted 2020-1-3; Published 2020-2-10
Abstract
Primary breast diffuse large B-cell lymphoma (PB-DLBCL), the most common histologic subtype of lymphoid malignancy in the breast, is a clinically and genetically heterogeneous disease that has insufficient systematic studies on the pathological and molecular features, optimal treatment scheme, as well as the prognostic factors. The aim of our study was to identify biomarkers and distinct subtypes of PB-DLBCLs and then evaluate the prognosis of this rare malignant lymphoma. We carried out hierarchical clustering analysis to evaluate protein expressions of potential biomarkers detected by immunohistochemistry staining of samples from 68 PB-DLBCL patients. The gene expression data from TCGA database was obtained to validate the identified clusters. We identified three robust clusters based on the B-cell receptor (BCR) signaling pathway, including two recognized NF-κB-dependent and PI3K-dependent clusters, and a distinct subset of PB-DLBCL with NF-κB-independent anti-apoptotic overexpression plus PI3K signaling, which exhibited an evolving definition and distinctive characters of a cluster group. Furthermore, survival analysis results showed an inferior outcome in NF-κB-dependent cluster patients and favorable survival in the PI3K-dependent cluster patients, suggesting an important predictive value of the three clusters. Our study provided a new perspective for understanding clinical complexity of PB-DLBCLs, and gave evidence for finding targeted biomarkers and strategies.
Keywords: Primary breast diffuse large B-cell lymphoma, B-cell receptor signaling pathway, Cluster analysis, Prognosis
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common aggressive lymphoma subtype with distinct genetic backgrounds and clinical characteristics [1-3]. Due to the biological heterogeneity of DLBCL, the responses to therapy and the prognostic survivals of patients are also different [1, 3, 4]. Currently, the cell-of-origin (COO) classification [activated B-cell like (ABC) and germinal center B-cell like (GCB)] and the International Prognostic Index (IPI) score are two most commonly used prognostic factors for DLBCL patients [4-6]. However, these predictors do not completely elucidate the risk stratification, variable outcomes, as well as the complex mechanisms underlying tumor biology of DLBCL [7]. The latest findings based on whole-exome sequencing have identified five robust DLBCL subsets which linked genetic signatures with pathogenetic mechanisms [8]. More importantly, these newly defined DLBCL subsets provided new insights into assessment of clinical outcomes and more rational therapies [8].
Up to a third of DLBCLs arise from extranodal organs, which are usually characterized by poor survival [9]. DLBCL primary of the breast (PB-DLBCL) are a rare presentation of extranodal DLBCL, representing less than 2.0% of all cases [10-12]. Because of the rare incidence of PB-DLBCL, there exists limited data on the natural history of this lymphoma entity [13-15]. Generally, PB-DLBCL mainly shows a phenotype of ABC (60-90%) according to the COO classification [13, 16]. However, as an uncommon site of involvement in extranodal DLBCL, PB-DLBCL has its own particularity with heterogenic biological and clinical characters [16]. In addition, PB-DLBCL has been reported to exhibit a worse prognosis compared with other extranodal DLBCLs, and the 5-year overall survival (OS) rates are nearly 50% [17]. Therefore, valuable biomarkers based on profound understanding the distinct subtypes are warranted to guide prognostic factors and therapeutic approaches for PB-DLBCL.
The genetic heterogeneity is usually reflected by gene-expression profiling, in which B cell receptor (BCR) signaling pathway plays the key role in DLBCL [18-20]. BCR signaling pathway mediates the survival signals in almost all DLBCL cells, including “chronic active” and “tonic” BCR signaling [18]. Previous studies have shown that extranodal DLBCL cells were mainly derived by chronic active BCR signaling with selectively acquiring mutations that target the BCR [21]. Active BCR signaling engages many complex transcriptional networks and pathways. After ligand binding, BCRs cluster, BCR pathway adaptor caspase recruitment domain family member 11 (CARD11), and resultant protein tyrosine kinases (PTK) will recruit and activate, thus initiate downstream NF-κB signaling pathway mainly [19, 22, 23]. BCR-pathway components and networks are complex and variable, which may provide the basis of underlying the biological diversity of the PB-DLBCL. More importantly, comprehensive establishment of the expression profile of PB-DLBCL will be helpful for classifying the distinct subsets and determining the subtype-specific signaling targets, treatments as well as the outcomes of PB-DLBCL. Herein, we identified potential subtypes by performing clustering analysis of the downstream components of BCR signaling pathways in PB-DLBCL patients, and then characterized each cluster in order to predict therapeutic effects as well as the prognostic survival of PB-DLBCL patients.
Methods
Patients
We retrospectively studied 68 cases of female patients with PB-DLBCLs, of whom 50 cases were treated in Harbin Medical University Cancer Hospital, 6 cases in the First Affiliated Hospital of Harbin Medical University and 12 cases in the Second Affiliated Hospital of Harbin Medical University from June 1976 through December 2016. Patients who fulfilled the following criteria were included in our current study: (1) diagnosed with primary breast lymphomas (PBLs) according to the criteria of Wiseman and Liao [24]: adequate pathologic specimen technically and close association of lymphomatous infiltration and breast tissue; (2) histologic classification of DLBCL according to the 2016 World Health Organization (WHO) criteria [25]. Patients were excluded if they were in the following cases: (1) a previous history of extramammary lymphoma or indolent B-cell lymphoma; (2) post-transplant lymphoproliferative disorders; (3) receiving major surgery within 4 weeks; (4) uncontrolled systemic infection; (5) EBV-positive DLBCL; (6) serological positivity for Hepatitis B, C virus or HIV infection. Ethical protocol was approved by the Institutional Review Board of Harbin Medical University and written informed consents were obtained from the patients or guardians. All methods were performed in accordance with the relevant guidelines and regulations. The pretreatment workups included a complete history and physical examination. We collected the histological types and various clinical or laboratory parameters.
Immunohistochemistry (IHC) staining and data analysis
Formalin-fixed, paraffin-embedded tissue specimens were collected from 68 patients diagnosed with PB-DLBCL patients for IHC analysis. Briefly, tissue sections were de-paraffinized in xylene and rehydrated in grade ethanol. High-pressure antigen retrieval was conducted using citrate buffer (pH 6.0). Next, endogenous peroxidase activity was blocked using 3% H2O2. Subsequently, the slides were incubated with single primary antibody respectively [anti-CD10 antibody (dilution 1:200); anti-BCL6 antibody (dilution 1:200); anti-MUM1 antibody (dilution 1:200); anti-PI3K antibody (dilution 1:300); anti-AKT2 antibody (dilution 1:200); anti-JAK2 antibody (dilution 1:300); anti-STAT3 antibody (dilution 1:300); anti-MAPK antibody (dilution 1:300); anti-BCL10 antibody (dilution 1:500); anti-NF-κB (p50) antibody (dilution 1:200); anti-Myc antibody (dilution 1:200); anti-BCL2 antibody (dilution 1:300); anti-MCL1 antibody (dilution 1:200); anti-BCL-xL antibody (dilution 1:200); anti-Ki67 antibody (dilution 1:300); and anti-P53 antibody (dilution 1:100), Abcam, Cambridge, USA)] at 4°C overnight. Then the slides were further incubated with HRP-labelled secondary antibody (Abcam, Cambridge, USA) for 30 min at room temperature. Then the slides were counterstained with instant hematoxylin, and then dehydrated, cleared, and mounted. All tissue specimens were examined by three independent well-experienced pathologists in a blinded manner (200 × magnification) without any prior information of the patient samples. According to a commonly used standard for IHC staining in DLBCL defined by Hans et al [26], the highest percentage of stained tumor cells was calculated to decide the positive cells in each case, and positive expression result was based on the cut-off value. The cut-off values for the proteins were shown as reported previously [7, 26-32]: CD10 (30%), BCL6 (30%), MUM1 (30%), PI3K (30%), AKT2 (20%), JAK2 (30%), STAT3 (30%), MAPK (20%), BCL10 (20%), NF-κB p50 (nuclear 20%), Myc (40%), BCL2 (50%), MCL1 (50%), BCL-xL (50%), Ki67 (70%) and P53 (30%) respectively. Staining intensity of each tissue sections were also evaluated, but it was not applied for determining the positivity due to the variability in tissue fixation and processing according to the previous study [26]. Phosphate buffered saline was used as negative control. The expression data of these proteins were evaluated by hierarchical clustering analysis using Manhattan Distance Method in Multi Experiment View (MEV) cluster software.
Gene expression data from TCGA database
The gene expression data and detailed clinical information from TCGA database (https://cancergenome.nih.gov/) were obtained and enrolled. The values of gene expressions from the DLBCL data sets were standardized by log 2. Hierarchical clustering analysis using Euclidean Distance Method was performed to assess TCGA data by Multi Experiment View (MEV) cluster software.
Statistical analysis
The last follow-up was in June 2017. The Kaplan-Meier methods was used to evaluate the OS, defined as the period of time from the date of diagnosis to the date of death from any causes or last follow-up. Different groups were compared using the log-rank test. Chi-squared and Fisher's Exact tests were carried out to evaluate the associations between the clusters and clinicopathological parameters. Multivariate analysis to evaluate the variables was performed using Cox proportional hazards models, and the results were presented as Hazard ratios (HRs) and 95% confidence intervals (CIs). SPSS 20.0 was used for statistical analysis. P-values < 0.05 were reckoned to be statistically significant.
Results
Protein expression signature based on IHC distinguished subgroups in PB-DLBCL
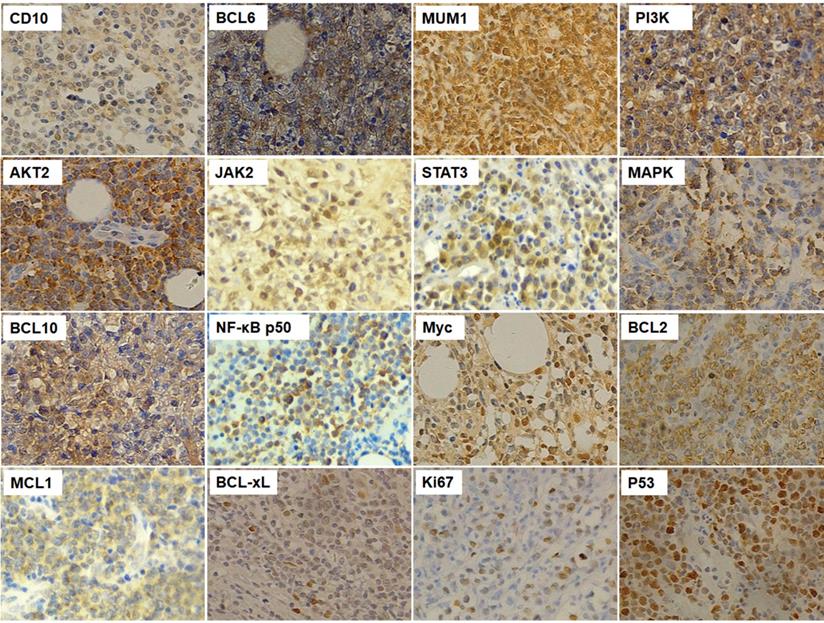
In order to comprehensively understand the distinct pathological and molecular characteristics of PB-DLBCL, we firstly screened several general clinical and biological markers of lymphoma cells, and then detected the protein expressions of these markers. First of all, we detected the expressions of CD10 (positive rate: 60.3%), BCL6 (45.6%) and MUM1 (85.3%) to demonstrate the COO classification PB-DLBCL patients according to the Hans method [26]. BCR signaling pathway was associated with survival and development of DLBCL cells, and we determined several BCR signaling components, including PI3K (47.1%), AKT2 (41.2%), JAK2 (42.7%), STAT3 (50.0%), MAPK (48.5%), BCL10 (60.3%), and NF-κB p50 (58.8%). Myc together with BCL2 led to a rapidly clinical progression and short survival [33], and the positive rates of expressions of Myc and BCL2 were 66.2% and 67.7% respectively. In addition, we detected the expressions of apoptotic factors, such as MCL1 (22.1%) and BCL-xL (35.3%). the positive expressions of IHC results were shown in Fig. 1.
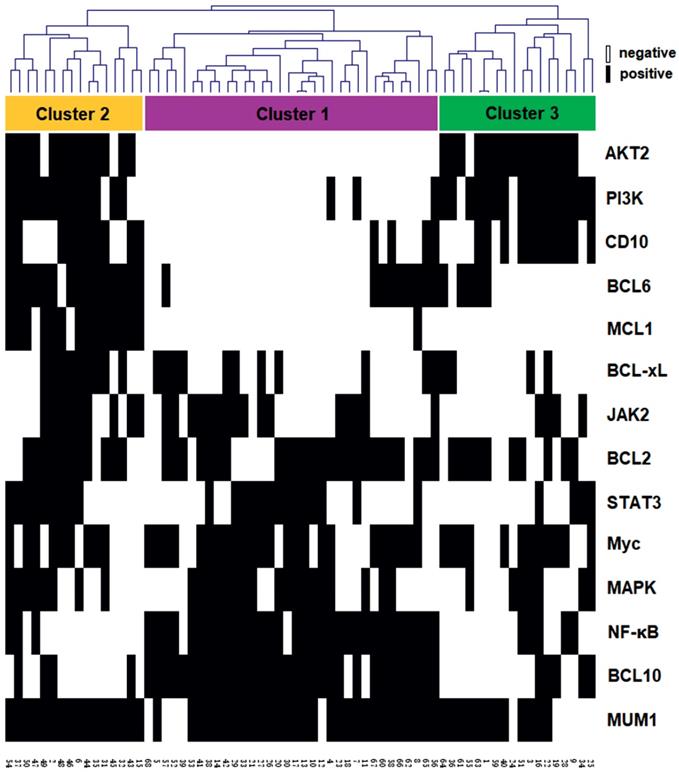
It has been reported that the diversity and complexity of BCR signaling pathway contributed to the biological heterogeneity of DLBCL. To examine whether significant expression patterns of BCR signaling components were informative that they were able to distinguish characteristic subtypes of PB-DLBCL, the protein expression data was hierarchically clustered, as shown in Fig. 2. The vertical columns represented the associated biomarkers whereas the horizontal rows represented 68 samples. According to our clustering result, we identified three robust subsets that differentially expressed protein profiling in PB-DLBCL patients, named Cluster 1, 2 and 3 (Fig. 2).
Cluster 1. In a total of 68 cases, half of the PB-DLBCL patients were included into this cluster, with high NF-κB expression predominantly. These samples also had increasing expression of BCL10, which was an important component of the CBM adapter complex combined with CARD11 and MALT1 that recruited and activated IκB kinase, a key activator of the NF-κB signaling pathway [34]. Therefore, we defined this cluster as NF-κB-dependent cluster. Other candidate proteins including Myc and BCL2 also were also significantly higher in this cluster, and tumors with co-occurring Myc and BCL2 were significantly more frequent, which might be associated with poor survival in DLBCL (called double-expression lymphoma) [7, 35, 36]. In addition, sporadic cases in this cluster had increasing expressions of STAT3 and MAPK in Cluster 1, which might appear as the upstream signaling of the NF-κB.
Cluster 3. The 18 cases of PB-DLBCLs were classified into this cluster, which exhibited significantly strong expressions of PI3K and AKT2 predominantly. Therefore, this cluster was defined as PI3K-dependent cluster. These samples also had increasing expression of CD10, which was the most major determinant of GCB-type of DLBCLs according to the Hans method [26]. We thus hypothesized this cluster be in GCB subtype predominantly, probably associated with more favorable survival.
Cluster 2. The remaining 16 PB-DLBCLs were grouped into this cluster, characterized by low or absent NF-κB expressions, but increasing expressions of MCL1 and BCL-xL, indicating a possible anti-apoptotic effect of this cluster. The cases also had enriched expressions of PI3K and AKT2, which might be associated with the anti-apoptotic protein expressions [37]. Additionally, several NF-κB-independent BCR-associated signaling pathway members, such as JAK2, STAT3 and MAPK were also enriched in this cluster, Therefore, we considered this cluster as NF-κB-independent cluster with high anti-apoptotic potential plus PI3K signaling, suggesting several specific targeted therapies for this cluster.
The immunohistochemistry results of CD10, BCL6, MUM1, PI3K, AKT2, JAK2, STAT3, MAPK, BCL10, NF-κB, Myc, BCL2, MCL1, BCL-xL, Ki67 and P53 in PB-DLBCL patients (200×magnification).
Identification of clusters of PB-DLBCL based on protein expression signatures: hierarchical clustering analysis was performed using all IHC results in the 68 PB-DLBCL samples (columns). Clusters 1-3 with their associated protein expressions were visualized (boxed for each cluster: Cluster 1, purple; Cluster 2, yellow; Cluster 3, green). The positive expression of each protein was presented as black, whereas negative expression was white.
The relationship between protein expressions of the three clusters of the PB-DLBCL patients and the gene-expression profile from TCGA DLBCL database
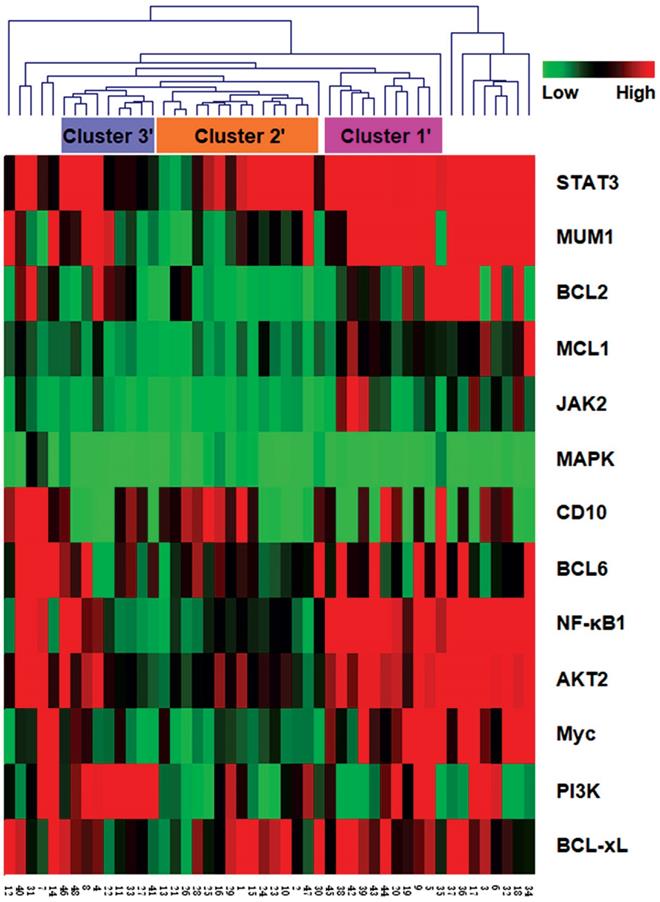
To validate our cluster classification based on the BCR signaling pathway, we collected the DLBCL datasets from the TCGA databases, and examined the associated differential expression data of the BCR signaling pathway members from 48 DLBCL patients accordingly (Fig. 3). The results also demonstrated three clusters (termed Cluster 1', 2' and 3') similar to the above identified clusters, which covered more than 70% of the DLBCL patients. Other cases that were not classified into the clusters might be due to the differences between protein expression and mRNA expression levels.
Identification of the clusters was associated with different COO subtypes in PB-DLBCL
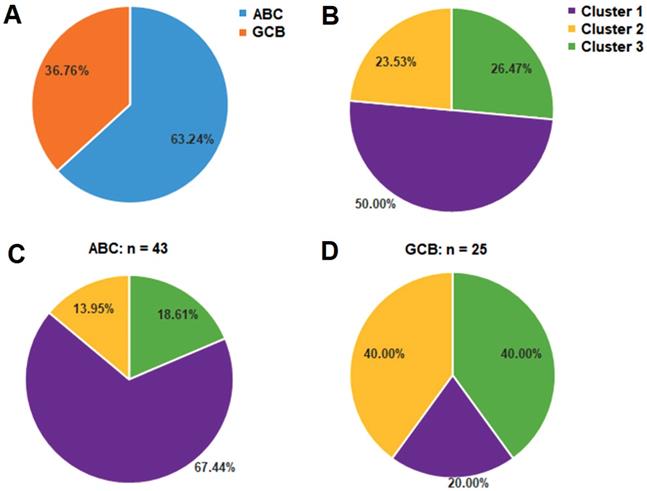
According to Hans method, CD10, BCL6 and MUM1 expression by IHC analysis has defined two major DLBCL subtypes as ABC and GCB based on the COO classification as mentioned above [26]. The majority of PB-DLBCL patients (43 of 68 cases, 63.24%) were classified into ABC subgroup, and 25 cases (36.76%) were GCB (Fig. 4A). To demonstrate whether our identification of the three clusters based on the BCR signaling components (Fig. 4B) was associated with ABC or GCB subgroups, we compared the percentages of the three clusters in the ABC and GCB subgroups respectively. The Cluster 1 was dominated by ABC cases, accounting for 67.44% of the ABC PB-DLBCL patients (Fig. 4C). However, Cluster 3 included mostly GCB cases (10 of 18 cases, 55.56%), although the percentage of Cluster 3 to GCB cases was 40.00% (Fig. 4D). Additionally, the Cluster 2 was composed by both the ABC and GCB gene expression subgroups (Fig. 4C and 4D). These data demonstrated that our classification based on BCR signaling components might be independent from the traditional COO classification.
The hierarchical clustering analysis was also carried out to classify the gene expression data of 48 DLBCL patients from TCGA. Cluster 1'-3' were labeled according to the clustering results.
Classification of the three clusters corresponds to different ABC or GCB subtypes of PB-DLBCL: (A) the distribution of ABC and GCB subgroups within 68 PB-DLBCL patients; (B, C, D) the distribution of Cluster 1-3 within 68 PB-DLBCL patients (B), 43 ABC subtype of PB-DLBCL patients (C), and 25 GCB subtype of PB-DLBCL patients (D).
Clinicopathologic features of PB-DLBCL patients according to the three clusters
| Characteristics | n=68 (%) | Cluster 1 (n=34) | Cluster 2 (n=16) | Cluster 3 (n=18) | P-value |
|---|---|---|---|---|---|
| Age | |||||
| < 50 years | 45 (66.18) | 21 | 14 | 10 | 0.108 |
| ≥ 50 years | 23 (33.82) | 13 | 2 | 8 | |
| Laterality | |||||
| Right | 29 (42.65) | 13 | 10 | 6 | 0.175 |
| Left | 39 (57.35) | 21 | 6 | 12 | |
| B symptoms | |||||
| Absent | 64 (94.12) | 32 | 15 | 17 | 0.996 |
| Present | 4 (5.88) | 2 | 1 | 1 | |
| ECOG PS | |||||
| 0 | 44 (64.71) | 20 | 10 | 14 | 0.410 |
| 1 | 18 (26.47) | 12 | 4 | 2 | |
| ≥ 2 | 6 (8.82) | 2 | 2 | 2 | |
| Stage | |||||
| IE | 46 (67.65) | 24 | 11 | 11 | 0.781 |
| IIE | 22 (32.35) | 10 | 5 | 7 | |
| LDH | |||||
| Normal | 54 (79.41) | 28 | 9 | 17 | 0.019 |
| Elevated | 14 (20.59) | 6 | 7 | 1 | |
| Adjusted IPI | |||||
| 0 | 35 (51.47) | 16 | 8 | 11 | 0.800 |
| 1 | 28 (41.18) | 14 | 7 | 7 | |
| 2 | 4 (5.88) | 3 | 1 | 0 | |
| 3 | 1 (1.47) | 1 | 0 | 0 | |
| Ki67 expression | |||||
| < 70% | 16 (23.53) | 5 | 8 | 3 | 0.017 |
| ≥ 70% | 52 (76.47) | 29 | 8 | 15 | |
| P53 expression | |||||
| <30% | 30 (44.12) | 14 | 8 | 8 | 0.842 |
| ≥ 30% | 38 (55.88) | 20 | 8 | 10 |
ECOG PS Eastern Cooperative Oncology Group performance status, LDH lactate dehydrogenase, IPI international prognostic index
There existed close relationship between the three clusters and baseline clinical features
Baseline characteristics were analyzed in a whole cohort of 68 patients diagnosed with PB-DLBCL, as shown in Table 1. All patients were females, and the median age at diagnosis was 50 years (range from 28-80 years). The right breast was involved in 29 patients (42.65%), and the B symptoms were absent in 64 patients (94.12%). The Eastern Cooperative Oncology Group (ECOG) performance status was 0 or 1 in 62 patients (91.18%). Most of the patients (67.65%) had Ann Arbor stage IE. Serum lactate dehydrogenase (LDH) was elevated in 14 (20.59%) of 68 patients. Regarding as the IPI score, the majority of the patients (92.65%) were in the low risk group. Ki67 expressions were elevated in most of the patients (76.47%), and P53 were overexpressed in over half of the patients (55.88%). We also analyzed the correlation between our identified three clusters and the clinicopathologic features. As shown in Table 1, the clusters were significantly associated with the LDH level (P = 0.019) and the Ki67 expressions (P = 0.017).
Clinical outcomes of the PB-DLBCL patients in different clusters
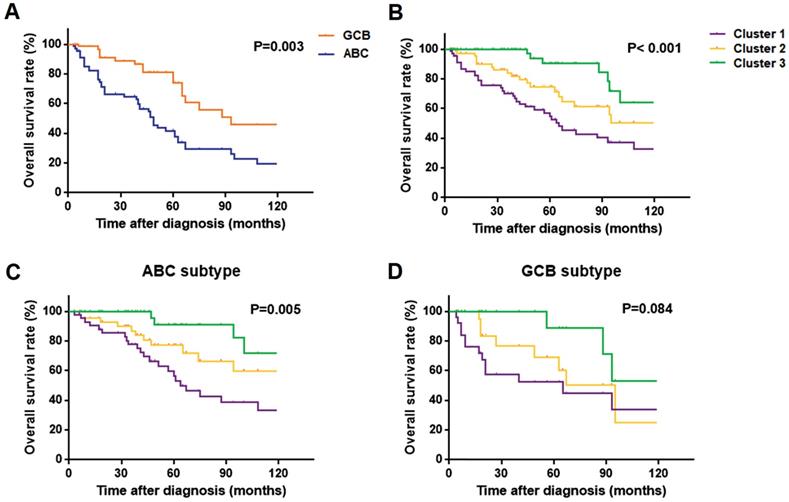
For the survival analysis, we selected all 68 PB-DLBCL patients with outcome data. Fig. 5A showed a better OS rate of GCB subgroup compared with the ABC group (P = 0.003), as previously reported. The three subtypes based on our clustering analysis differed significantly in OS, with the Cluster 3 patients having much more favorable outcomes than the other two groups (P < 0.001), and Cluster 1 patients having the worst survival outcome (Fig. 5B). Within ABC PB-DLBCL patients in Fig. 5C, there existed the similar significant OS rates of the three subgroups, with P-value as 0.005. Regarding as only 25 PB-DLBCL patients in GCB subgroup, although no significant result was obtained, there was a trend toward favorable OS among patients with Cluster 3 as compared with patients with Cluster 1 and 2 subtypes (Fig. 5D). The distinct prognosis regarding our protein expression subsets of BCR components provided the basis of assessing the prognosis of the PB-DLBCL patients. In addition, the identification of the three clusters was considered as an independent prognostic marker for OS (P = 0.013, HR = 0.413; 95%CI: 0.206-0.830), as shown in Table 2. The expression of P53 could also be a prognostic factor in PB-DLBCL patients (Table 2).
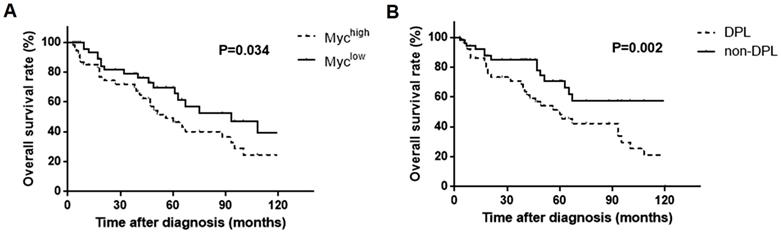
It was noteworthy that Myc expression (66.18%) in our study was significantly higher in PB-DLBCL patients than DLBCL patients (30% of MYC gene expression in TCGA database; 20-30% of Myc protein expression as previously reported). Overexpression of Myc usually resulted in poor prognosis of lymphoma, and this effect can be augmented among tumors that co-expressed BCL2 (DPL) [7, 35, 36]. Therefore, we further performed the survival analysis, and the results showed that patients with higher Myc expression and double expressions of Myc and BCL2 were associated with a significantly worse survival rates (P = 0.034, Fig. 6A; P = 0.002, Fig. 6B respectively), suggesting Myc together with BCL2 probably played a role in PB-DLBCL besides BCR signaling components.
Multivariate analysis of prognostic factors for survival in PB-DLBCL patients
| Covariate | OS | ||
|---|---|---|---|
| HR | 95%CI | P-valuea | |
| Age, y | 1.022 | 0.391-2.672 | 0.965 |
| Laterality | 0.830 | 0.392-1.759 | 0.627 |
| B symptoms | 0.532 | 0.064-4.388 | 0.558 |
| ECOG PS | 0.838 | 0.435-1.617 | 0.599 |
| Stage | 2.254 | 0.797-6.376 | 0.126 |
| LDH | 0.649 | 0.156-2.709 | 0.553 |
| Adjusted IPI | 1.214 | 0.454-3.242 | 0.699 |
| Ki67 | 2.021 | 0.554-7.376 | 0.287 |
| P53 | 3.599 | 1.465-8.839 | 0.005 |
| Myc | 2.742 | 0.648-11.611 | 0.171 |
| DPL | 0.606 | 0.228-1.611 | 0.315 |
| Three clusters | 0.413 | 0.206-0.830 | 0.013 |
aCox analysis
Kaplan-Meier models of overall survival according to ABC or GCB subgroups as well as the Cluster 1-3: (A) overall survival rates by ABC and GCB subgroups of 68 PB-DLBCL patients; (B, C, D) overall survival rates by Cluster 1-3 of 68 PB-DLBCL patients (B), 43 ABC subtype of PB-DLBCL patients (C), and 25 GCB subtype of PB-DLBCL patients (D).
Kaplan-Meier models of overall survival according to Myc expression (A) and the double expressions of Myc and BCL2 (DPL) (B).
Discussion
The robust protein expression signature for PB-DLBCL that we presented in this study showed a novel landscape of the biological heterogeneity of the PB-DLBCL determined by BCR signaling pathway and the characteristic attributes of our identified three clusters of PB-DLBCLs that might influence the clinical outcomes. Our studies provided the basis of understanding the clinical complexity of PB-DLBCLs, which could also contribute to the development of specific therapeutic targets for PB-DLBCL patients in the future.
Despite that COO classification of ABC and GCB subtypes has been widely used to distinguish DLBCL cells to predict survival outcomes of patients, it may not comprehensively demonstrate the distinct biological feathers of all DLBCLs, especially in several special types of lymphomas [8, 21]. PB-DLBCL is a rare subtype of DLBCL derived from extranodal tissues with poor survival [10, 11, 13]. Although a series of evidences showed its predominance of ABC subtype, the PB-DLBCL patients also exhibited clinical heterogeneity in therapeutic responses and survival outcomes [13, 16]. Furthermore, limited studies have focused on PB-DLBCL's biological diversity, optimal therapeutic regimens, as well as prognosis [10-12, 17]. Therefore, novel classification approaches are required to reclassify PB-DLBCLs to better understand this specific type of extranodal lymphoma.
BCR signaling pathway is central to the pathogenesis and development of B cell malignancies, and is composed by many pathways and transcriptional networks [18]. Generally, the BCR signaling can be divided into the upstream and downstream signaling pathways. The upstream BCR signaling engages the activation signals, whereas the downstream signaling mediates the functional survival signals, which is more important for research and intervention [20]. Furthermore, the complexity and heterogeneity of the BCR signaling pathway components made them possible to distinguish DLBCL cells. In this study, we identified three robust clusters according to BCR signaling components of PB-DLBCL, which were proven to be different from the traditional ABC and GCB catalogue. Notably, our results showed that the ABC and GCB subgroups were both constituted by the three clusters of the PB-DLBCL patients, suggesting a more comprehensive classification intergrating the COO subtypes as well as our clustering analysis based on the BCR signaling pathway members. Moreover, our identified clusters were roughly consistent with the results from gene expression data of TCGA exon, indicating that our results might be implemented clinically.
In our study, the Cluster 1, a genotype that has been characterized by frequent NF-κB expression and activation of the NF-κB-dependent signaling, accounted for nearly a half of the PB-DLBCL patients. In general, NF-κB activation has been considered as an important feature of ABC subgroup of DLBCL, which relies on constitutive NF-κB signaling to reduce apoptosis and sustain viability, called “chronic active” BCR signaling [19, 22]. Constitutive activation of NF-κB mediates several downstream survival signals, and inhibition of NF-κB activity by blocking BCR signaling causes cell cycle arrest and apoptosis [38]. In our results, the Cluster 1 associated with NF-κB activation was dominated by ABC subtype, which was coincidence with the previous studies related to primary extranodal lymphomas. Furthermore, survival analysis in our study showed that patients in Cluster 1 suffered from the worst survival outcome, probably due to their constitutive activation of the NF-κB pathway, which is considered to weaken the effects of cytotoxic agents. These results might account for the blunt therapeutic response of the majority of PB-DLBCL patients in clinic, demonstrating that NF-κB-dependent signaling pathway might be an important intervention target. In addition, the Cluster 1 included cases with both Myc and BCL2 overexpression also provided another explanation of poor survival of patients in this group, due to their important role in malignant proliferation of the lymphoma cells [35].
From our clustering results, Cluster 3 was enriched for PI3K/AKT signaling, which also played an important role in mediating survival signals of BCR signaling pathways. In comparison to “chronic active” BCR signaling characterized by NF-κB, the PI3K was considered as another signaling pathway member in “tonic” BCR signaling, where a constitutively active form of PI3K rescued the B-cell survival while surface BCR expression was genetically ablated [18]. Although the PI3K/AKT also played a role in chronic BCR signaling ABC subtype to indirectly modulate downstream NF-κB signals, most PI3K activation by tonic BCR signaling occurs in GCB tumors, which usually had an improved clinical outcome compared with ABC DLBCLs [38, 39]. As expected, PB-DLBCL patients in Cluster 3 included mostly GCB cases (70%), with the much more favorable outcomes than the other two groups, demonstrating that this cluster driven by PI3K-dependent signaling might have a response to therapy.
Besides the above two clusters with specific characters, we also defined a group of PB-DLBCL patients with shared molecular features using our clustering map, as Cluster 2. The Cluster 2 subtype showed unique features compared with Cluster 1 and 3, and more importantly, exhibited a new and evolving definition of a cluster group of PB-DLBCLs. Firstly, this group lacked the single ABC and GCB characters, but combined some features of both subtypes from our results. Secondly, this cluster shed light on non-NF-κB-mediated BCR signaling pathways, such as JAK/STAT, MAPK, but enriched anti-apoptotic factors MCL1 and BCL-xL, suggesting that there might be other mechanisms regulating the survival signals in this cluster. Last but not least, this cluster combined PI3K/AKT signaling pathways, made this cluster showed some similarities from Cluster 3, and this results might explained the up-regulation of the anti-apoptotic expressions as previously reported [37]. Taken together, this cluster could be characterized by an NF-κB-independent anti-apoptotic subgroup plus PI3K signaling, which displayed distinctive features from both Cluster 1 and 3 classifications in biological behavior. In addition, the survival analysis in our studies showed that PB-DLBCL patients in Cluster 2 had medium survival rates between Cluster 1 and 3, which was consistent with its biological characteristics.
Conclusions
In conclusion, our clustering studies to identify three robust PB-DLBCL clusters distinctively according to BCR signaling components presented a novel signature for assessing previously unrecognized protein expression subsets. These findings in our study provided two important implications in PB-DLBCL patients. First, the distinct protein expression subtypes driven by different BCR signaling pathways might contribute to the subsequent biological behavior of the lymphoma cells. Second, each cluster had distinct survival outcomes after therapy and probably guided to the selection of targeted therapies owing to their distinct signaling abnormalities.
Abbreviations
PB-DLBCL: Primary breast diffuse large B-cell lymphoma; BCR: B-cell receptor; COO: cell-of-origin; ABC: activated B-cell like; GCB: germinal center B-cell like; IPI: International Prognostic Index; OS: overall survival; CARD11: caspase recruitment domain family member 11; PTK: protein tyrosine kinases; WHO: World Health Organization; IHC: Immunohistochemistry; MEV: Multi Experiment View; ECOG: Eastern Cooperative Oncology Group; LDH: lactate dehydrogenase.
Supplementary Material
Supplementary figure and table.
Acknowledgements
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 81730074 & No. 81672599), China Postdoctoral Science Foundation (Grant No. 2018M641858).
Ethics Committee Approval and Patient Consent
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki. It was approved by the institutional review boards or ethics committees of the study sites.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Li S, Young KH, Medeiros LJ. Diffuse large B-cell lymphoma. Pathology. 2018;50:74-87
2. Kubuschok B, Held G, Pfreundschuh M. Management of diffuse large B-cell lymphoma (DLBCL). Cancer treatment and research. 2015;165:271-88
3. Flowers CR, Sinha R, Vose JM. Improving outcomes for patients with diffuse large B-cell lymphoma. CA: a cancer journal for clinicians. 2010;60:393-408
4. Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI. et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. The New England journal of medicine. 2002;346:1937-47
5. Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A. et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503-11
6. Wight JC, Chong G, Grigg AP, Hawkes EA. Prognostication of diffuse large B-cell lymphoma in the molecular era: moving beyond the IPI. Blood reviews. 2018;32:400-15
7. Wang XJ, Medeiros LJ, Bueso-Ramos CE, Tang G, Wang S, Oki Y. et al. P53 expression correlates with poorer survival and augments the negative prognostic effect of MYC rearrangement, expression or concurrent MYC/BCL2 expression in diffuse large B-cell lymphoma. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2017;30:194-203
8. Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA. et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nature medicine. 2018;24:679-90
9. Ollila TA, Olszewski AJ. Extranodal Diffuse Large B Cell Lymphoma: Molecular Features, Prognosis, and Risk of Central Nervous System Recurrence. Current treatment options in oncology. 2018;19:38
10. Ryan G, Martinelli G, Kuper-Hommel M, Tsang R, Pruneri G, Yuen K. et al. Primary diffuse large B-cell lymphoma of the breast: prognostic factors and outcomes of a study by the International Extranodal Lymphoma Study Group. Annals of oncology: official journal of the European Society for Medical Oncology. 2008;19:233-41
11. Aviv A, Tadmor T, Polliack A. Primary diffuse large B-cell lymphoma of the breast: looking at pathogenesis, clinical issues and therapeutic options. Annals of oncology: official journal of the European Society for Medical Oncology. 2013;24:2236-44
12. Yhim HY, Kang HJ, Choi YH, Kim SJ, Kim WS, Chae YS. et al. Clinical outcomes and prognostic factors in patients with breast diffuse large B cell lymphoma; Consortium for Improving Survival of Lymphoma (CISL) study. BMC cancer. 2010;10:321
13. Yhim HY, Kim JS, Kang HJ, Kim SJ, Kim WS, Choi CW. et al. Matched-pair analysis comparing the outcomes of primary breast and nodal diffuse large B-cell lymphoma in patients treated with rituximab plus chemotherapy. International journal of cancer. 2012;131:235-43
14. Lin YC, Tsai CH, Wu JS, Huang CS, Kuo SH, Lin CW. et al. Clinicopathologic features and treatment outcome of non-Hodgkin lymphoma of the breast-a review of 42 primary and secondary cases in Taiwanese patients. Leukemia & lymphoma. 2009;50:918-24
15. Wong WW, Schild SE, Halyard MY, Schomberg PJ. Primary non-Hodgkin lymphoma of the breast: The Mayo Clinic Experience. Journal of surgical oncology. 2002;80:19-25 discussion 6
16. Aviles A, Neri N, Nambo MJ. The role of genotype in 104 cases of diffuse large B-cell lymphoma primary of breast. American journal of clinical oncology. 2012;35:126-9
17. Shao YB, Sun XF, He YN, Liu CJ, Liu H. Clinicopathological features of thirty patients with primary breast lymphoma and review of the literature. Medical oncology. 2015;32:448
18. Young RM, Shaffer AL 3rd, Phelan JD, Staudt LM. B-cell receptor signaling in diffuse large B-cell lymphoma. Seminars in hematology. 2015;52:77-85
19. Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB. et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88-92
20. Wilson WH, Young RM, Schmitz R, Yang Y, Pittaluga S, Wright G. et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nature medicine. 2015;21:922-6
21. Wright GW, Wilson WH, Staudt LM. Genetics of Diffuse Large B-Cell Lymphoma. The New England journal of medicine. 2018;379:493-4
22. Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. The Journal of experimental medicine. 2001;194:1861-74
23. Dal Porto JM, Gauld SB, Merrell KT, Mills D, Pugh-Bernard AE, Cambier J. B cell antigen receptor signaling 101. Molecular immunology. 2004;41:599-613
24. Wiseman C, Liao KT. Primary lymphoma of the breast. Cancer. 1972;29:1705-12
25. Beham-Schmid C. Aggressive lymphoma 2016: revision of the WHO classification. Memo. 2017;10:248-54
26. Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G. et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275-82
27. Merzianu M, Jiang L, Lin P, Wang X, Weber DM, Vadhan-Raj S. et al. Nuclear BCL-10 expression is common in lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia and does not correlate with p65 NF-kappaB activation. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2006;19:891-8
28. Cai Q, Tu M, Xu-Monette ZY, Sun R, Manyam GC, Xu X. et al. NF-kappaB p50 activation associated with immune dysregulation confers poorer survival for diffuse large B-cell lymphoma patients with wild-type p53. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2017;30:854-76
29. Zhuang L, Lee CS, Scolyer RA, McCarthy SW, Zhang XD, Thompson JF. et al. Mcl-1, Bcl-XL and Stat3 expression are associated with progression of melanoma whereas Bcl-2, AP-2 and MITF levels decrease during progression of melanoma. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2007;20:416-26
30. Cui W, Cai Y, Wang W, Liu Z, Wei P, Bi R. et al. Frequent copy number variations of PI3K/AKT pathway and aberrant protein expressions of PI3K subunits are associated with inferior survival in diffuse large B cell lymphoma. Journal of translational medicine. 2014;12:10
31. Kuboki Y, Shiratori K, Hatori T, Fujita I, Kimijima A, Yamamoto M. et al. Association of epidermal growth factor receptor and mitogen-activated protein kinase with cystic neoplasms of the pancreas. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2010;23:1127-35
32. Clay TD, Russell PA, Do H, Sundararajan V, Conron M, Wright GM. et al. EGFR and KRAS mutations do not enrich for the activation of IL-6, JAK1 or phosphorylated STAT3 in resected lung adenocarcinoma. Medical oncology. 2017;34:175
33. Sarkozy C, Traverse-Glehen A, Coiffier B. Double-hit and double-protein-expression lymphomas: aggressive and refractory lymphomas. The Lancet Oncology. 2015;16:e555-e67
34. Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW. et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676-9
35. Friedberg JW. Double hit diffuse large B-cell lymphomas: diagnostic and therapeutic challenges. Chinese clinical oncology. 2015;4:9
36. Teoh CS, Lee SY, Chiang SK, Chew TK, Goh AS. Impact of Double Expression of C-MYC/BCL2 Protein and Cell of Origin Subtypes on the Outcome among Patients with Diffuse Large B-Cell Lymphoma: a Single Asian Center Experience. Asian Pacific journal of cancer prevention: APJCP. 2018;19:1229-36
37. Jiao Y, Ding H, Huang S, Liu Y, Sun X, Wei W. et al. Bcl-XL and Mcl-1 upregulation by calreticulin promotes apoptosis resistance of fibroblast-like synoviocytes via activation of PI3K/Akt and STAT3 pathways in rheumatoid arthritis. Clinical and experimental rheumatology. 2018
38. Chen L, Monti S, Juszczynski P, Ouyang J, Chapuy B, Neuberg D. et al. SYK inhibition modulates distinct PI3K/AKT- dependent survival pathways and cholesterol biosynthesis in diffuse large B cell lymphomas. Cancer cell. 2013;23:826-38
39. Kloo B, Nagel D, Pfeifer M, Grau M, Duwel M, Vincendeau M. et al. Critical role of PI3K signaling for NF-kappaB-dependent survival in a subset of activated B-cell-like diffuse large B-cell lymphoma cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:272-7
Author contact
![]() Corresponding authors: Jin Zhou, Department of Hematology, First Affiliated Hospital of Harbin Medical University, Harbin Medical University, Harbin, Heilongjiang, P. R. China, 150081. Tel.: +86045186298333; Fax: +86045186298333; E-mail: niuxingjianedu.cn; Qingyuan Zhang, Department of Medical Oncology, Harbin Medical University Cancer Hospital, Harbin Medical University, Harbin, Heilongjiang, P. R. China,150081.
Corresponding authors: Jin Zhou, Department of Hematology, First Affiliated Hospital of Harbin Medical University, Harbin Medical University, Harbin, Heilongjiang, P. R. China, 150081. Tel.: +86045186298333; Fax: +86045186298333; E-mail: niuxingjianedu.cn; Qingyuan Zhang, Department of Medical Oncology, Harbin Medical University Cancer Hospital, Harbin Medical University, Harbin, Heilongjiang, P. R. China,150081.

 Global reach, higher impact
Global reach, higher impact