3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(8):2181-2191. doi:10.7150/jca.37150 This issue Cite
Review
Association of microRNA biosynthesis genes XPO5 and RAN polymorphisms with cancer susceptibility: Bayesian hierarchical meta-analysis
1. Department of Epidemiology and Health Statistics, School of Public Health, Beijing Municipal Key Laboratory of Clinical Epidemiology, Capital Medical University, Beijing, China.
2. Department of Molecular Physiology and Biophysics, Holden Comprehensive Cancer Center, University of Iowa Carver College of Medicine, Iowa City, IA, United States.
Received 2019-5-30; Accepted 2019-12-7; Published 2020-2-3
Abstract
XPO5/RAN-GTP complex mediates the nuclear transport of pre-miRNAs in the miRNA processing system, its altered expression is indicated to be correlated with cancer risk. Several studies have inspected the association between XPO5 or RAN polymorphisms and the risk of various cancers, but the findings remain controversial. A Bayesian hierarchical meta-analysis was carried out to review and analyze the effect of XPO5 and RAN polymorphisms on cancer risk. The association was estimated by calculating the logarithm of odds ratio (Log OR) and 95% credible interval (95% CrI). The expression quantitative trait loci (eQTL) analysis was used for in silico functional validation of the identified significant susceptibility loci. Consequently, 38 case-control studies (from 27 citations) with 27,459 cancer cases and 25,151controls were included in the meta-analysis of the five most prevalent SNPs (rs11077 A/C, rs2257082 G/A, rs3803012 A/G, rs14035 C/T, rs3809142 C/T). In the XPO5 gene rs11077 SNP, the minor C allele significantly increased the risk of cancer (Log OR = 0.120, 95% CrI = 0.013, 0.241), and a strong association between rs11077 SNP and cancer risk was also found in the dominant model (CC + AC vs. AA: Log OR = 0.132, 95% CrI = 0.009, 0.275). In addition, the minor GG genotype allele of the RAN gene rs3803012 SNP significantly increased the cancer risk (Log OR = 0.707, 95% CrI = 0.059, 1.385). Statistically significant associations between rs3803012 SNP and cancer risk were also observed in the recessive model (GG vs. AG + AA: Log OR = 0.708, 95% CrI = 0.059, 1.359). Furthermore, the eQTL analysis revealed that rs11077 SNP was significantly correlated with XPO5 mRNA expression, which provided additional biological basis for the observed positive association. Our results suggest that XPO5 rs11077 may be a possible functional susceptibility locus for cancer risk.
Keywords: Bayesian hierarchical meta-analysis, XPO5, RAN, polymorphism, cancer
Introduction
MicroRNAs (miRNAs) are a highly conserved class of small, noncoding RNAs, which mediate post-transcriptional gene silencing [1]. Over the past decade, they have increasingly been recognized to be involved in the initiation and progression of human carcinogenesis [2]. The biosynthesis of miRNAs involves a multiple-step process that starts in the nucleus of the cell, where miRNA genes are initially transcribed as primary miRNAs (pri-miRNAs), and then converted into precursor miRNAs (pre-miRNAs). Secondly, with the assistance of GTP-binding nuclear protein Ran/exportin-5 (XPO5) complex, the pre-miRNAs are exported from the nucleus to the cytoplasm, where the mature miRNA molecule exerts its main function [3, 4].
During the miRNA maturing processing, the XPO5/RAN-GTP complex mediates the nuclear transport of pre-miRNAs, which are crucial components. XPO5, a member of the nucleo-cytoplasmic exportins, is related to the human export receptor that uses the Ran-GTPase to control cargo association [5, 6]. Studies have shown that the overexpression of XPO5 is found to improve transport efficiency and further enhance miRNA activity, while the downregulation of XPO5 leads to a loss of pre-miRNA function [7, 8]. RAN encodes a small G protein that is crucial for the translocation of RNA and proteins through the nuclear pore complex. If the RAN is depleted, the output of the pre-miRNA will be greatly reduced [9, 10]. Therefore, impaired miRNA processing caused by the dysregulation expression of miRNA biosynthesis genes XPO5 or RAN can noticeably promote the tumorigenesis [11].
Increasing evidence proposed shows that single-nucleotide polymorphisms (SNPs) in core components of miRNA biogenesis may impair or enhance miRNA processing efficiency or function, which can function as an oncogene or tumor suppressor [12]. Formerly, several studies have been conducted to assess the association between XPO5 and RAN SNPs and cancer susceptibility in diverse populations. However, the conclusions of the findings remain inconsistent. Hence, a meta-analysis is required to combine data from all the individual studies to obtain a more comprehensive and effective estimation. Previous studies have reviewed the relationship between polymorphisms of miRNA processing genes and cancer risk through classical meta-analysis approach [13]. However, the results did not indicate a correlation between SNPs in XPO5 and RAN genes with cancer risk. This might be as a result of the small number of articles included. Indeed, classical meta-analysis requires the initial sample to be large enough to ensure the asymptotic normality of the effect size and, to further obtain accurate and realistic results [14]. Bayesian hierarchical meta-analysis, however, provides a more accurate pooled effect size compared to classical meta-analysis approaches, especially in situations with a small number of studies [15]. Therefore, in the present study, we carried out a Bayesian hierarchical meta-analysis including newly published articles to find a vivid and precise association between SNPs in XPO5 and RAN genes with cancer risk based on all available eligible studies. We also used expression quantitative trait loci analysis (eQTL) to validate the potential function of the identified significant susceptibility loci.
Materials and methods
Retrieval strategy
To identify all potentially eligible publications, PubMed, PubMed Central (PMC), Web of science, Embase, China National Knowledge Infrastructure (CNKI), Chinese Wanfang databases, Wiley, Google Scholar, Cochrane, the Cochrane Central Register of Controlled Trials were searched, using a combination of the following keywords: 'XPO5/ exportin 5/ exp5/ RAN/ ARA24/ Gsp1/ TC4'; 'SNP/ polymorphism/ variation/ variant'; and 'tumor/ cancer/ carcinoma/ neoplasm'. The search was limited to articles published in English or Chinese through April 9, 2019. References of the relevant literature and review articles were also evaluated to identify all potentially eligible articles. This meta-analysis was carried out in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (Supplementary PRISMA 2009 Checklist) [16].
Inclusion and exclusion criteria
Eligible publications were selected based on the following inclusion criteria: (i) evaluation of genetic association between XPO5 or RAN and susceptibility to cancer; (ii) a case-control designed study; Articles meeting the following criteria were excluded: (i) reviews, meta-analyses, conference reports, or editorial articles; (ii) duplicate records; (iii) no available data to extract; (iv) the control subjects exhibited a departure from Hardy-Weinberg equilibrium (HWE).
Data extraction and Quality assessment
The following information was extracted by two reviewers independently and disagreement was solved through discussion: first author's name, publication year, country, ethnicity, cancer type, polymorphisms, sample size of cases and controls, genotype distribution, source of control groups (population-based (PB) or hospital-based (HB)), genotyping method, HWE in controls. If more than one type of cancer or multistage research was involved in a single article, data for each type of cancer was extracted independently. When the data in eligible articles was unavailable, we tried our best to contact the corresponding authors for original data. Quality assessment of articles was conducted using the Newcastle-Ottawa Quality Assessment Scale (NOS) [17]. NOS scores range from 0 to 9. To the best of our knowledge, there is no established cut-offs for low, moderate and high quality. Hence, we have relied on previous literature [18] to define low quality as a score ≤ 5, moderate quality as a score between 6 and 7, and high quality as a score between 8 and 9.
Statistical methods
In this meta-analysis, the following comparisons for XPO5 and RAN polymorphisms were evaluated in five common genetic models including allele model (V vs. W) (W for wild allele, V for variation allele), heterozygote model (WV vs. WW), homozygote model (VV vs. WW), dominant model (WV+VV vs. WW), and recessive model (VV vs. WW+WV).
Bayesian meta-analysis method
The Bayesian meta-analysis is a Bayesian modeling method which determines the prior distribution with hierarchical prior distribution, and then does the statistical inference [19]. Compared with the classical meta-analysis, the Bayesian hierarchical random-effect model can obtain accurate pooling effects, especially in situations with a small number of studies [14, 20-22]. A Bayesian approach allows one to coherently process the uncertainty in the heterogeneity parameter while focusing on inference for the effect parameters, and interprets the results more intuitively [23]. In order to compare the effect magnitude between different studies, the pooled logarithmic odds ratio (Log OR), between-study standard deviation (τ2) and their respective 95% credible intervals (CrIs) are estimated. The 95% CrI is the Bayesian equivalent for standard confidence intervals. In particular, the model supposes that the mean of the Log ORs has a low-informative normal distribution (mean = 0, variance = 100) and the variance of the Log ORs has a low-informative inverse-gamma distribution (0.01, 0.01) [24]. Sensitivity analyses with different choices of low-information prior distributions showed the robustness of this choice [20]. In addition, we estimated the I2 statistic, which is used to measure the total variation [25]. Forest plots, which illustrate Log ORs and 95% CrIs for both the individual trials and the pooled results, were included in our meta-analysis. Moreover, the heterogeneity plot displayed the joint posterior density of the two parameters, Log OR and τ parameters, with darker shading corresponding to higher probability density. All the statistical analyses were calculated using “bayesmeta” R package (https://cran.r-project.org/web/packages/bayesmeta/index.html).
In silico functional validation
To validate the potential impact of the cancer risk SNP, we examined its association with the expression of corresponding genes using eQTL databases. The eQTL analysis was performed by using the genotyping and expression data of lymphoblastoid cells from 373 European individuals available in the 1000 Genomes Project [26]. Considering that many eQTLs are population-specific, we also extracted eQTL data of East Asian individuals from a study by Stranger et al. [27], in which genome-wide mRNA expression in lymphoblastoid cell lines of 726 individuals from eight global populations in the HapMap3 project was analyzed. Seventeen cases of hepatocellular carcinoma genotype and gene expression data were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo) (GSE65373). Choosing hepatocellular carcinoma and breast cancer as representatives, we also downloaded mRNA sequencing datasets of 154 paired cancer tissue samples and normal adjacent tissue samples from The Cancer Genome Atlas (TCGA-LIHC and TCGA-BRCA) (https://tcga-data.nci.nih.gov/tcga/). A linear regression model was performed to evaluate the correlation between SNPs and specific mRNA expression levels. A paired t-test was used to test for the differences in gene mRNA expression levels between cancer tissue and adjacent normal tissue from the TCGA database. All analyses were performed using R (version 3.5.1).
Results
Study characteristics
The process of selecting eligible studies is depicted in Figure 1. A total of 194 articles were identified based on our search strategy, 118 of the articles were duplicates. After a screening of the titles and abstracts, 24 articles were excluded for irrelevant information (5 were reviews, 19 were not related to our topic). We eliminated 25 records after browsing the full text of the remaining 52 articles (16 were related to prognosis; 5 had overlapping study populations; 1 for unavailable data; 3 were departure from HWE). Finally, 38 studies from 27 articles with 27,459 cases and 25,151 controls were included in our meta-analysis [28-54]. Table 1 shows the characteristics and relevant data of the included studies. The detail of NOS scores for every included study was shown in Table S1. In summary, 38 eligible case-control studies, five SNPs of XPO5 or RAN genes were investigated in the eventual analysis. In XPO5, the analyzed SNPs were rs11077 A/C, rs2257082 G/A; while in RAN, the analyzed SNPs were rs3803012 A/G, rs14035 C/T, rs3809142 C/T.
The flow chart of identification for studies included in the meta-analysis
Quantitative synthesis
The main results of Bayesian hierarchical meta-analysis were calculated as the median of the marginal posterior distribution of the Log ORs and τ parameters. On the basis of the Bayesian hierarchical meta-analysis, XPO5 rs11077 and RAN rs3803012 SNPs were significantly associated with the risk of cancer (Table 2).
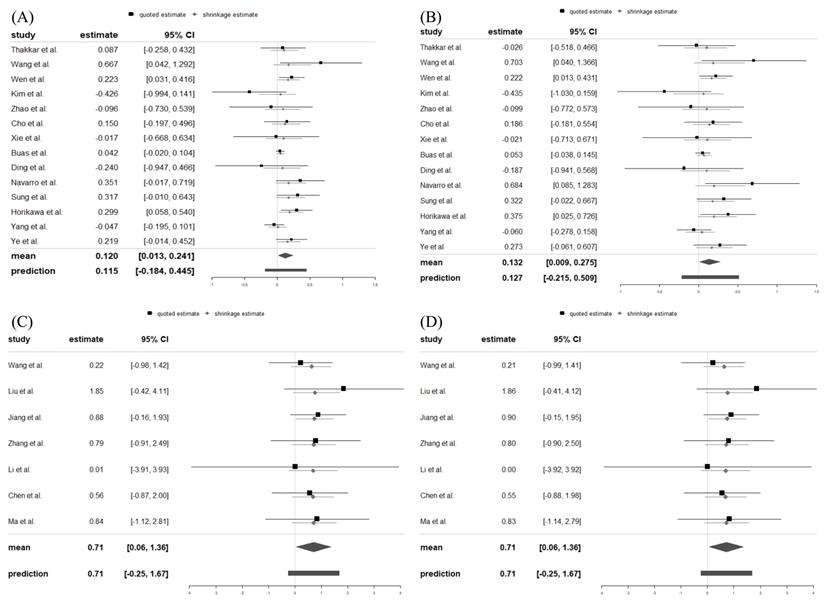
In the XPO5 gene rs11077 SNP (Figure 2A), the minor C allele significantly increased the risk of cancer (Log OR = 0.120, 95% CrI = 0.013, 0.241). A strong association of rs11077 SNP with cancer risk was also found in the dominant model (CC + AC vs. AA: Log OR = 0.132, 95% CrI = 0.009, 0.275) (Figure 2B). In addition, the minor GG genotype allele of the RAN gene rs3803012 SNP (Figure 2C) significantly increased the cancer risk (Log OR = 0.707, 95% CrI = 0.059, 1.385). Statistically significant associations between rs3803012 A/G SNP and cancer risk were also observed in the recessive model (GG vs. AG + AA: Log OR = 0.708, 95% CrI = 0.059, 1.359) (Figure 2D). However, alleles and genotypes in other polymorphisms of XPO5 and RAN genes were not significantly associated with cancer susceptibility (Table 2).
Heterogeneity and publication bias
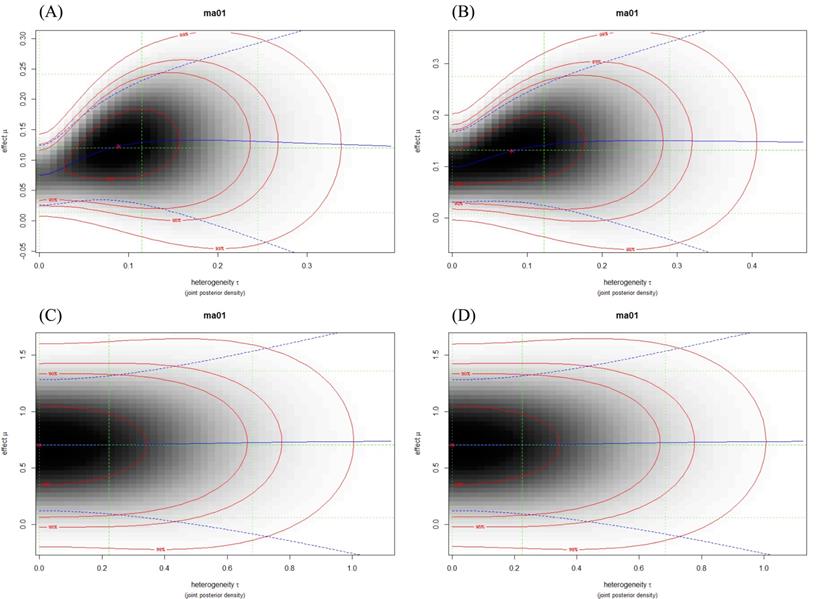
Evaluation of the heterogeneity of the studies was analyzed with the τ2 test. I2 > 0.50 was considered as high value for heterogeneity. On the basis of heterogeneity plots and I2 value, in most of the meta-analyses, the total heterogeneity and between studies heterogeneity were not high, for example, rs11077 and rs3803012 (Figure 3, Table 2). However, these results for rs14035 were significantly high (Table 2).
Begg's and Egger's tests were performed to evaluate the potential publication bias. As shown in Table 2, no publication bias was observed in our meta-analysis.
Functional validation by eQTL analysis
To substantiate the associations between the identified SNPs (XPO5 rs11077 and RAN rs3803012 SNPs) and cancer risk, we performed the eQTL analysis to assess the associations between SNPs and corresponding mRNA expression levels. The eQTL analysis results of lymphoblastoid cell lines from 373 Europeans were visualized through the Geuvadis Data Browser (https://www.ebi.ac.uk/Tools/geuvadis-das), and we found that XPO5 rs11077 was significantly associated with XPO5 mRNA expression levels (P = 5.83E-07).
Characteristics of the studies eligible for the meta-analysis
| Author | Year | Country | Ethnicity | Cancer type | Source ofcontrols | Genotypingmethod | Case/control | Cases | Controls | PHWE | NOS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| XPO5 rs11077 | AA | AC | CC | A | C | AA | AC | CC | A | C | |||||||||
| Thakkar et al. | 2018 | India | Asian | Hodgkin lymphoma | PB | TaqMan | 101/200 | 39 | 41 | 21 | 119 | 83 | 76 | 92 | 32 | 244 | 156 | 0.639 | 8 |
| Wang et al. | 2017 | China | Asian | Breast cancer | HB | PCR-RFLP | 116/120 | 87 | 28 | 1 | 202 | 30 | 103 | 17 | 0 | 223 | 17 | 0.401 | 7 |
| Wen et al. | 2017 | China | Asian | Thyroid cancer | HB | TaqMan | 1134/1228 | 907 | 210 | 17 | 2024 | 244 | 1023 | 194 | 11 | 2240 | 216 | 0.593 | 7 |
| Kim et al. | 2016 | China | Asian | Hepatocellular carcinoma | PB | PCR-RFLP | 147/209 | 128 | 19 | 0 | 275 | 19 | 170 | 38 | 1 | 378 | 40 | 0.465 | 8 |
| Zhao et al. | 2015 | China | Asian | Colorectal cancer | HB | PCR-LDR | 163/142 | 143 | 19 | 1 | 305 | 21 | 123 | 18 | 1 | 264 | 20 | 0.705 | 7 |
| Cho et al. | 2015 | Korea | Asian | Colorectal cancer | HB | PCR-RFLP | 408/400 | 333 | 74 | 1 | 740 | 76 | 337 | 61 | 2 | 735 | 65 | 0.668 | 7 |
| Xie et al. | 2015 | China | Asian | Gastric cancer | HB | PCR-LDR | 137/142 | 119 | 17 | 1 | 255 | 19 | 123 | 18 | 1 | 264 | 20 | 0.705 | 6 |
| Buas et al. | 2015 | Europe | Caucasian | Esophageal cancer | HB | Illumina | 5780/3206 | 1909 | 2826 | 1045 | 6644 | 4916 | 1097 | 1557 | 552 | 3751 | 2661 | 0.991 | 7 |
| Ding et al. | 2013 | China | Asian | Lung cancer | HB | PCR-LDR | 112/80 | 94 | 18 | 0 | 206 | 18 | 65 | 14 | 1 | 144 | 16 | 0.804 | 8 |
| Navarro et al. | 2013 | Spain | Caucasian | Hodgkin lymphoma | HB | TaqMan | 127/104 | 25 | 67 | 35 | 117 | 137 | 34 | 46 | 24 | 114 | 94 | 0.275 | 7 |
| Sung et al. | 2011 | Korea | Asian | Breast cancer | HB | TaqMan | 559/567 | 473 | 82 | 4 | 1028 | 90 | 501 | 64 | 2 | 1066 | 68 | 0.977 | 7 |
| Horikawa et al. | 2008 | USA | Caucasian | Renal cell carcinoma | PB | SNPlex | 276/277 | 86 | 136 | 54 | 308 | 244 | 110 | 129 | 38 | 349 | 205 | 0.985 | 8 |
| Yang et al. | 2008 | USA | Caucasian | Bladder cancer | HB | SNPlex | 718/726 | 248 | 356 | 114 | 852 | 584 | 241 | 363 | 122 | 845 | 607 | 0.457 | 7 |
| Ye et al. | 2008 | USA | Caucasian | Esophageal cancer | HB | SNPlex | 300/295 | 101 | 146 | 53 | 348 | 252 | 118 | 137 | 40 | 373 | 217 | 0.981 | 7 |
| XPO5 rs2257082 | GG | AG | AA | G | A | GG | AG | AA | G | A | |||||||||
| Bermisheva et al. | 2018 | Russia | Caucasian | Breast cancer | NM | PCR | 417/361 | 182 | 187 | 48 | 551 | 283 | 172 | 154 | 35 | 498 | 224 | 0.951 | 6 |
| Liang et al. | 2010 | USA | Caucasian | Ovarian cancer | HB | Illumina | 339/349 | 192 | 126 | 21 | 510 | 168 | 181 | 141 | 27 | 503 | 195 | 0.949 | 7 |
| Martin-Guerrero et al. | 2015 | Spain | Caucasian | Lymphocytic leukemia | HB | TaqMan | 101/346 | 59 | 37 | 5 | 155 | 47 | 202 | 123 | 21 | 527 | 165 | 0.694 | 7 |
| RAN rs3803012 | AA | AG | GG | A | G | AA | AG | GG | A | G | |||||||||
| Wang et al. | 2016 | China | Asian | Hepatocellular carcinoma | HB | TaqMan | 312/320 | 250 | 56 | 6 | 556 | 68 | 260 | 55 | 5 | 575 | 65 | 0.298 | 7 |
| Liu et al. | 2013 | China | Asian | Hepatocellular carcinoma | PB | TaqMan | 1256/2678 | 1158 | 95 | 3 | 2411 | 101 | 2450 | 227 | 1 | 5127 | 229 | 0.066 | 8 |
| Jiang et al. | 2013 | China | Asian | Breast cancer | PB | TaqMan | 870/884 | 766 | 92 | 12 | 1624 | 116 | 772 | 107 | 5 | 1651 | 117 | 0.539 | 8 |
| Zhang et al. | 2012 | China | Asian | Gastric cancer | PB | TaqMan | 1654/1844 | 1517 | 133 | 4 | 3167 | 141 | 1674 | 168 | 2 | 3516 | 172 | 0.292 | 8 |
| Li et al. | 2012 | China | Asian | Hepatocellular carcinoma | PB | PCR-RFLP | 560/560 | 508 | 52 | 0 | 1068 | 52 | 512 | 48 | 0 | 1072 | 48 | 0.289 | 8 |
| Chen et al. | 2012 | China | Asian | Cervical carcinoma | PB | TaqMan | 1471/1529 | 1325 | 141 | 5 | 2791 | 151 | 1397 | 129 | 3 | 2923 | 135 | 0.990 | 8 |
| Ma et al. | 2012 | China | Asian | Head and neck cancer | PB | TaqMan | 391/892 | 344 | 45 | 2 | 733 | 49 | 799 | 91 | 2 | 1689 | 95 | 0.725 | 8 |
| RAN rs14035 | CC | CT | TT | C | T | CC | CT | TT | C | T | |||||||||
| Kim et al. | 2016 | China | Asian | Hepatocellular carcinoma | PB | PCR-RFLP | 147/229 | 98 | 42 | 7 | 238 | 56 | 137 | 69 | 3 | 343 | 75 | 0.080 | 8 |
| Meng et al. | 2015 | China | Asian | Hepatocellular carcinoma | PB | SNPstream | 324/343 | 208 | 105 | 11 | 521 | 127 | 229 | 107 | 7 | 565 | 121 | 0.172 | 8 |
| Cho et al. | 2015 | Korea | Asian | Colorectal cancer | HB | PCR-RFLP | 408/400 | 267 | 128 | 13 | 662 | 154 | 233 | 150 | 17 | 616 | 184 | 0.240 | 7 |
| Martin-Guerrero et al. | 2015 | Spain | Caucasian | Lymphocytic leukemia | HB | TaqMan | 99/342 | 48 | 41 | 10 | 137 | 61 | 138 | 164 | 40 | 440 | 244 | 0.407 | 7 |
| Xie et al. | 2015 | China | Asian | Gastric cancer | HB | PCR-LDR | 137/142 | 86 | 45 | 6 | 217 | 57 | 35 | 71 | 36 | 141 | 143 | 0.999 | 6 |
| Buas et al. | 2015 | Europe | Caucasian | Esophageal cancer | HB | Illumina | 5783/3202 | 2760 | 2470 | 553 | 7990 | 3576 | 1525 | 1370 | 307 | 4420 | 1984 | 0.978 | 7 |
| Zhao et al. | 2015 | China | Asian | Colorectal cancer | HB | PCR-LDR | 163/142 | 113 | 45 | 5 | 271 | 55 | 107 | 33 | 2 | 247 | 37 | 0.761 | 7 |
| Roy et al. | 2014 | India | Asian | Oral cancer | HB | PCR-RFLP | 439/438 | 281 | 134 | 24 | 696 | 182 | 301 | 124 | 13 | 726 | 150 | 0.958 | 7 |
| Li et al. | 2012 | China | Asian | Hepatocellular carcinoma | PB | PCR-RFLP | 560/560 | 376 | 160 | 24 | 912 | 208 | 390 | 160 | 10 | 940 | 180 | 0.162 | 8 |
| Kim et al. | 2010 | Korea | Asian | Lung cancer | HB | MS | 93/90 | 65 | 23 | 5 | 153 | 33 | 52 | 33 | 5 | 137 | 43 | 0.937 | 7 |
| Horikawa et al. | 2008 | USA | Caucasian | Renal cell carcinoma | PB | SNPlex | 276/278 | 143 | 110 | 23 | 396 | 156 | 129 | 125 | 24 | 383 | 173 | 0.415 | 8 |
| Ye et al. | 2008 | USA | Caucasian | Esophageal cancer | HB | SNPlex | 304/301 | 127 | 139 | 38 | 393 | 215 | 166 | 115 | 20 | 447 | 155 | 0.989 | 7 |
| RAN rs3809142 | CC | CT | TT | C | T | CC | CT | TT | C | T | |||||||||
| Bermisheva et al. | 2018 | Russia | Caucasian | Breast cancer | NM | PCR | 415/359 | 313 | 89 | 13 | 715 | 115 | 208 | 130 | 21 | 546 | 172 | 0.908 | 6 |
| Jiang et al. | 2013 | China | Asian | Breast cancer | PB | TaqMan | 862/886 | 602 | 232 | 28 | 1436 | 288 | 615 | 239 | 32 | 1469 | 303 | 0.149 | 8 |
Abbreviations: HB: hospital-based; PB: population-based; HWE: Hardy-Weinberg equilibrium; NOS: Newcastle-Ottawa scale; MS: sequenome MS-based genotyping assay; PCR: polymerase chain reaction; PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; PCR-LDR: polymerase chain reaction-ligase detection reaction; NM: not mentioned.
Forest plots displayed Log ORs and 95% credible intervals for both the individual trials and the pooled results. (A) rs11077: C vs. A; (B) rs11077: CC+AC vs. AA; (C) rs3803012: GG vs. AA; (D) rs3803012: GG vs. AG+AA. OR: odds ratio
Bayesian hierarchical meta-analysis of the pooled associations between XPO5 and RAN genes polymorphisms and risk of cancer
| SNPs | Variations | Percentage heterogeneity I2 | Association test | Absolute heterogeneitytest τ2 (95% CrI) | Publication bias (Begg's test, p value; Egger's test, p value) | |
|---|---|---|---|---|---|---|
| Pooled Log OR | 95% CrI | |||||
| XPO5 rs11077 (A>C) | C vs. A | 0.486 | 0.120 | 0.013, 0.241 | 0.115 (0.000, 0.245) | 0.443, 0.166 |
| AC vs. AA | 0.255 | 0.110 | -0.004, 0.243 | 0.094 (0.000, 0.254) | 0.827, 0.253 | |
| CC vs. AA | 0.345 | 0.216 | -0.014, 0.508 | 0.207 (0.000, 0.470) | 0.228, 0.174 | |
| CC+AC vs. AA | 0.385 | 0.132 | 0.009, 0.275 | 0.122 (0.000, 0.289) | 0.827, 0.228 | |
| CC vs. AC+AA | 0.193 | 0.134 | -0.042, 0.367 | 0.126 (0.000, 0.354) | 0.324, 0.194 | |
| XPO5 rs2257082 (G>A) | A vs. G | 0.643 | -0.012 | -0.405, 0.369 | 0.181 (0.000, 0.603) | 1.000, NA |
| AG vs. GG | 0.499 | -0.003 | -0.412, 0.407 | 0.178 (0.000, 0.616) | 1.000, NA | |
| AA vs. GG | 0.384 | -0.022 | -0.640, 0.557 | 0.265 (0.000, 0.767) | 1.000, NA | |
| AA+AG vs. GG | 0.572 | -0.008 | -0.434, 0.414 | 0.197 (0.000, 0.638) | 1.000, NA | |
| AA vs. AG+GG | 0.339 | -0.025 | -0.606, 0.521 | 0.233 (0.000, 0.727) | 1.000, NA | |
| RAN rs3803012 (A>G) | G vs. A | 0.187 | 0.032 | -0.104, 0.169 | 0.070 (0.000, 0.222) | 0.707, 0.981 |
| AG vs. AA | 0.232 | -0.018 | -0.168, 0.136 | 0.084 (0.000, 0.259) | 0.707, 0.919 | |
| GG vs. AA | 0.070 | 0.707 | 0.059, 1.358 | 0.223 (0.000, 0.681) | 0.806, 0.741 | |
| GG+AG vs. AA | 0.208 | 0.007 | -0.137, 0.155 | 0.077 (0.000, 0.243) | 0.707, 0.983 | |
| GG vs. AG+AA | 0.071 | 0.708 | 0.059, 1.359 | 0.224 (0.000, 0.684) | 0.806, 0.774 | |
| RAN rs14035 (C>T) | T vs. C | 0.933 | -0.068 | -0.354, 0.213 | 0.435 (0.251, 0.683) | 0.115, 0.676 |
| CT vs. CC | 0.861 | -0.125 | -0.396, 0.128 | 0.373 (0.166, 0.637) | 0.193, 0.326 | |
| TT vs. CC | 0.859 | 0.082 | -0.457, 0.623 | 0.790 (0.449, 1.195) | 0.150, 0.631 | |
| TT+CT vs. CC | 0.919 | -0.122 | -0.445, 0.190 | 0.482 (0.266, 0.763) | 0.244, 0.465 | |
| TT vs. CT+CC | 0.791 | 0.142 | -0.301, 0.593 | 0.606 (0.267, 1.005) | 0.193, 0.464 | |
| RAN rs3809142 (C>T) | T vs. C | 0.941 | -0.333 | -1.169, 0.487 | 0.453 (0.117, 0.998) | 1.000, NA |
| CT vs. CC | 0.927 | -0.373 | -1.265, 0.497 | 0.494 (0.137, 1.043) | 1.000, NA | |
| TT vs. CC | 0.566 | -0.427 | -1.296, 0.382 | 0.363 (0.000, 0.933) | 1.000, NA | |
| TT+CT vs. CC | 0.934 | -0.388 | -1.280, 0.486 | 0.498 (0.144, 1.043) | 1.000, NA | |
| TT vs. CT+CC | 0.470 | -0.326 | -1.117, 0.423 | 0.297 (0.000, 0.868) | 1.000, NA | |
Abbreviations: Marginal posterior summary, bold pooled Log OR indicated as statistically significant at 0.05 level. I2: relative heterogeneity; CrI: credible interval; OR: odds ratio; SNP: single nucleotide polymorphism; NA: not available.
Heterogeneity plots illustrated the joint posterior density of heterogeneity τ and effect μ (Log OR), with darker shading corresponding to higher probability density. The red lines indicate (approximate) 2-dimensional credible regions, and the green lines indicate marginal medians and shortest 95% credible intervals for Log OR and τ. Blue lines show the conditional mean effect (Log OR) as a function of the heterogeneity τ (solid line) along with conditional 95% confidence bounds (dashed lines). (A) rs11077: C vs. A; (B) rs11077: CC+AC vs. AA; (C) rs3803012: GG vs. AA; (D) rs3803012: GG vs. AG+AA
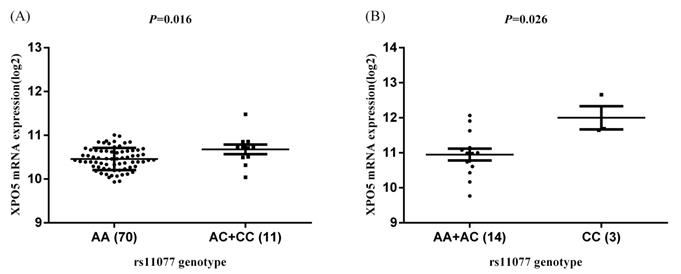
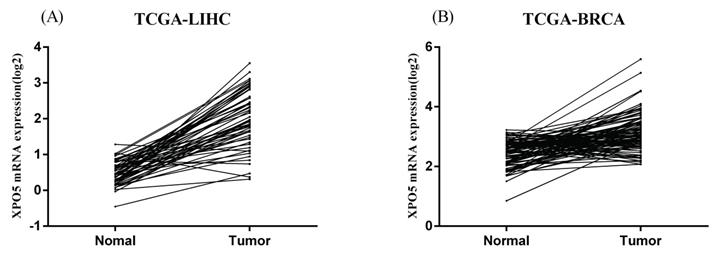
Similarly, in the HapMap3 East Asian samples (81 Japanese samples) from Stranger et al. [27], rs11077 was significantly associated with the expression of XPO5 gene (P = 0.016, Figure 4A), with the risk of C allele predicting higher mRNA levels of XPO5. According to the genotyping and expression data of 17 hepatocellular carcinoma obtained from the GEO database (GSE65373), we also found that rs11077 C allele had a significant association with an increased mRNA expression levels of XPO5 in the recessive model (P = 0.026, Figure 4B). However, no significant associations between rs3803012 and RAN mRNA expression levels were found in the above datasets. In addition, we compared mRNA expression levels of XPO5 in 154 paired cancer tissue samples with normal adjacent tissue samples from two TCGA projects (58 paired samples in TCGA-LIHC and 96 paired samples in TCGA-BRCA). We found that XPO5 mRNA expression levels were significantly increased in the tumor tissues compared to the normal tissues (P = 1.50E-20 and P = 5.27E-11, respectively) (Figure 5).
Discussion
In the present study, a total of five SNPs in XPO5 and RAN genes were comprehensively reviewed and analyzed to estimate their associations with the risk of overall cancer by Bayesian hierarchical meta-analysis. Of these five SNPs, two (rs2257082, rs3809142) were analyzed for the first time. In contrast to the classical meta-analysis already performed with a fewer number of articles included [13], the Bayesian hierarchical meta-analysis applied here indicated that rs11077 SNP of XPO5 and rs3803012 SNP of RAN might facilitate the carcinogenesis. Nonetheless, we also performed a classical meta-analysis of the current data (results were not mentioned) that demonstrated the association of most of the genetic models in rs11077 SNP and the relevance of the rs3803012 SNP in homozygous and recessive models.
The eQTL analysis of rs11077 with the mRNA expression of XPO5. (A) dominant model in Stranger et al. study [27], P = 0.016; (B) recessive model in GEO database (GSE65373), P = 0.026
The mRNA expression of XPO5 in the 154 paired cancer tissue samples and normal adjacent tissue samples from the TCGA database. (A) 58 paired hepatocellular carcinoma samples in TCGA-LIHC, P = 1.50E-20; (B) 96 paired breast cancer samples in TCGA-BRCA, P = 5.27E-11
Since the Bayesian hierarchical meta-analysis is much more sensitive and confers more precise estimation compared with classical meta-analysis, it is powerful suggested that rs11077 SNP of XPO5 and rs3803012 SNP of RAN are associated with cancer risk. Furthermore, eQTL analysis demonstrated that rs11077 SNP may influence the mRNA expression levels of XPO5. However, no associations were revealed amongst other studied SNPs in our meta-analysis, therefore future studies with a larger sample size are needed to determine their relationships.
For the classical meta-analysis, when faced with extreme values or small research quantum, the accuracy of the results cannot be guaranteed and the correctness of its conclusions will be questionable [14]. However, with the development of Markov Chain Monte Carlo (MCMC) methods, Bayesian hierarchical meta-analysis can avoid these defects and address the actual research question more directly [55-57]. Chen et al. [14] compared the difference between fully Bayesian hierarchical meta-analysis and classical meta-analysis, and found that if fixed effect is used to determine the real effect, both types of meta-analysis can be used. When random effect is adopted, if the study quantum is < 20, the Bayesian hierarchical meta-analysis should be the analysis of choice. The number of included articles for each studied SNP was < 20 in our meta-analysis, therefore the Bayesian hierarchical meta-analysis was utilized. From a statistical point of view, the number of included studies was not large enough for a classical meta-analysis, thus the results should be interpreted with caution. In the Bayesian hierarchical meta-analysis, however, the credible interval is slightly wider than that of classical meta-analysis and the results tend to be more consistent [14-15]. Hence, the significant result of Bayesian hierarchical meta-analysis is conservative and more reliable in comparison with the classical meta-analysis.
XPO5 is a member of karyopherin β family related to human export receptor CRMI, and is responsible for nuclear export and stabilization to form mature miRNA to produce physiological effects [58-59]. As XPO5 is a key factor for the transportation of miRNA from the nucleolus, it has been postulated as a rate-limiting step in the development of miRNAs, so its impairment could lead to pre-miRNA trapping in the nucleolus, influencing the risk of cancer [60-61]. Current studies have indicated the role of XPO5 in the development of several sorts of cancers such as hepatocellular carcinoma, thyroid cancer, lung cancer, and so on [62-64]. These studies are consistent with the results of the present study in which XPO5 mRNA expression levels were significantly increased in tumor tissues compared to normal tissues in 154 paired cancer tissue samples from the TCGA database. In addition, an increasing number of studies have focused on the correlations of XPO5 polymorphisms with cancer risk. The previous classical meta-analysis of XPO5 gene rs11077 SNP performed by He et al. [13], which included 7 case-control studies, showed no significant correlation with cancer risk. However, our analysis which included 14 case-control studies indicated that the minor C allele of rs11077 SNP significantly increased the risk of cancer and a strong association with cancer risk was also found in the dominant model. This association was further supported by the significant correlation between rs11077 C allele and an increased XPO5 mRNA expression level in the eQTL analysis. These findings suggest that rs11077 was significantly associated with cancer risk possibly by decreasing the mRNA expression levels of XPO5. Located in the 3'-UTR of XPO5, the A to C substitution of rs11077 may affect mRNA stability, alter the expression of XPO5 and, consequently, affect the expression of miRNAs, resulting in an aberrant expression of miRNA target gene at the post-transcriptional level [12,65].
RAN is a key member of the Ras superfamily of GTPases and is essential for translocation of pre-miRNAs from the nucleus to the cytoplasm through the nuclear pore complex in a GTP-dependent manner [66]. Studies have revealed that the up-regulation of RAN expression in various malignancies supports its role in cancer development [67-69]. No significant association was observed in the RAN gene rs3803012 SNP according to the previous classical meta-analysis [13], which included 5 case-control studies. In contrast, our Bayesian hierarchical meta-analysis which included 6,514 cases and 8,707 healthy subjects for the RAN gene rs3803012 SNP from 7 studies, demonstrated a significant association between rs3803012 SNP (homozygote or recessive model) and overall cancer risk. Our classical meta-analysis (the results of this analysis were not included in this study) also demonstrated a significant increased association risk of RAN gene rs3803012 SNP in cancer. Studies have hypothesized that the RAN rs3803012 G allele might affect the targeting of hsa-miR-199a-3p and result in decreased expression of RAN mRNA in tumor cells, which may affect various miRNA biosynthesis [43]. Unfortunately, we failed to obtain a significant eQTL results for SNP rs3803012 because the minor allele frequency (MAF) of rs3803012 was low in the included datasets (MAF ≤ 0.05). Population-specific eQTL analysis are warranted to validate our findings. As for the RAN gene rs14035 and rs3809142 polymorphisms, both types of meta-analysis did not support the significant association with cancer risk. The total heterogeneity as well as between studies heterogeneity was relatively high. Thus, further investigations are required to identify these potential cancer susceptibility loci.
Despite these results, we encountered some limitations during our meta-analysis. Firstly, since we had a limited number of studies, we could not perform a subgroup analysis with respect to the ethnicity, source of control groups (population-based or hospital-based) and cancer type. Heterogeneity among different cancers may cause the real effects to be hidden when pooling all cancer types. Secondly, gene-environmental interactions which may alter cancer risk were not evaluated due to the lack of relevant data across the included studies. Thirdly, studies of XPO5 and RAN SNPs in the cancer predisposition field continue to emerge, which resulted in limited number of the relevant investigations.
In view of all this, Bayesian hierarchical meta-analysis suggests a potential role of the miRNA biogenesis genes XPO5 (rs11077 A/C) and RAN (rs3803012 A/G) SNPs in cancer risk, supplying novel clues to identifying new biomarkers with cancer-forewarning function. Although we used publicly available genotyping and expression data to confirm the biological significance of the variant and suggest that XPO5 rs11077 may be a possible functional susceptibility locus for cancer risk, further high-quality research and functional evaluations are still warranted to validate our findings due to the limitations mentioned above.
Supplementary Material
Supplementary tables.
Acknowledgements
Funding
This work was supported by grants from the National Natural Science Foundation of China (81874277 and 81473056), Beijing Natural Science Foundation (7172025) and Scientific Research Project of Beijing Educational Committee (SQKM201710025006).
Author Contributions
Fen Liu and Yi Shao designed research; Yi Shao, Yi Shen, Xudong Guo and Chen Niu collected and analyzed the data; Yi Shao and Lei Zhao drafted the manuscript; Fen Liu revised and made the decision to submit for publication. All authors contributed to manuscript revision, read and approved the submitted version.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509-524
2. Mishra PJ, Bertino JR. MicroRNA polymorphisms: The future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009;10:399-416
3. Yi R, Qin Y, Macara IG. et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes and Development. 2003;17:3011-3016
4. Winter J, Jung S, Keller S. et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228-234
5. Smith AE, Slepchenko BM, Schaff JC. et al. Systems analysis of Ran transport. Science. 2002;295:488-491
6. Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185-191
7. Yi R, Doehle BP, Qin Y. et al. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11:220-226
8. Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776-4785
9. Izaurralde E, Jarmolowski A, Beisel C. et al. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997;137:27-35
10. Sazer S, Dasso M. The ran decathlon: multiple roles of Ran. J Cell Sci. 2000;113:1111-1118
11. Catto JW, Miah S, Owen HC. et al. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Research. 2009;69:8472-8481
12. Guo Z, Shu Y, Zhou H. et al. Identification of diagnostic and prognostic biomarkers for cancer: Focusing on genetic variations in microRNA regulatory pathways (Review). Mol Med Rep. 2016;13:1943-52
13. He J, Zhao J, Zhu W. et al. MicroRNA biogenesis pathway genes polymorphisms and cancer risk: a systematic review and meta-analysis. PeerJ. 2016;4:e2706
14. Chen W, Zhao SY, Ge Y. et al. Comparison between fully Bayesian hierarchical meta-analysis and classical meta-analysis: A Monte Carlo Study based on correlation coefficient. Metallurgical & Mining Industry. 2005;9:194-202
15. Lunn D, Barrett J, Sweeting M. et al. Fully Bayesian hierarchical modelling in two stages, with application to meta-analysis. Journal of the Royal Statistical Society. Series C, Applied Statistics. 2013;62:551-572
16. Liberati A, Altman DG, Tetzlaff J. et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700
17. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta analyses. European Journal of Epidemiology. 2010;25:603-605
18. Fung SM, Wong XY, Lee SX. et al. Performance of Single-Nucleotide Polymorphisms in Breast Cancer Risk Prediction Models: A Systematic Review and Meta-analysis. Cancer Epidemiol Biomarkers Prev. 2019;28:506-521
19. Raudenbush SW, Bryk AS. Hierarchical linear models: Applications and data analysis methods. NewYork: Sage. 2003;98:133-155
20. Babapulle MN, Joseph L, Bélisle P. et al. A hierarchical Bayesian meta-analysis of randomized clinical trials of drug-eluting stents. The Lancet. 2004;364:583-591
21. Huynh T, Perron S, O'Loughlin J. et al. Comparison of primary percutaneous coronary intervention and fibrinolytic therapy in ST-segment-elevation myocardial infarction: Bayesian hierarchical meta-analyses of randomized controlled trials and observational studies. Circulation. 2009;119:3101-3109
22. Rhodes KM, Turner RM, Higgins JP. Empirical evidence about inconsistency among studies in a pair-wise meta-analysis. Research Synthesis Methods. 2016;7:346-370
23. Röver C. Bayesian random-effects meta-analysis using the bayesmeta R package. Journal of Statistical Software. 2018. <. https://cran.r-project.org/web/packages/bayesmeta/vignettes/Roever2018-bayesmeta.pdf.>
24. Mohammadi FS, Aslani S, Mostafaei S. et al. Are genetic variations in IL-21-IL-23R-IL-17A cytokine axis involved in a pathogenic pathway of rheumatoid arthritis? Bayesian hierarchical meta-analysis. J Cell Physiol. 2019;234:17159-17171
25. Huedo-Medina TB, Sánchez-Meca J, Marín-Martínez F. et al. Assessing heterogeneity in meta-analysis: Q statistic or I² index? Psychological Methods. 2006;11:193-206
26. Lappalainen T, Sammeth M, Friedlander MR. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506-511
27. Stranger BE, Montgomery SB, Dimas AS. et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639
28. Thakkar DN, Palugulla S, Selvarajan S. et al. Frequency distribution of BLMH, XPO5 and HFE gene polymorphisms in the South Indian population and their association with Hodgkin Lymphoma. Int J Biol Markers. 2018;33:514-519
29. Bermisheva MA, Takhirova ZR, Gilyazova IR. et al. MicroRNA Biogenesis Pathway Gene Polymorphisms Are Associated with Breast Cancer Risk. Russian Journal of Genetics. 2018;54:568-575
30. Wen J, Gao Q, Wang N. et al. Association of microRNA-related gene XPO5 rs11077 polymorphism and breast cancer. International Journal of Surgery. 2017;96:e6351
31. Wen J, Gao Q, Wang N. et al. Association of microRNA-related gene XPO5 rs11077 polymorphism with susceptibility to thyroid cancer. Medicine (Baltimore). 2017;96:e6351
32. Kim MN, Kim JO, Lee SM. et al. Variation in the Dicer and RAN Genes Are Associated with Survival in Patients with Hepatocellular Carcinoma. PLoS One. 2016;11:e0162279
33. Wang Z, Wang J, Wang Q. et al. Genetic variant in DICER gene is associated with prognosis of hepatocellular carcinoma in a Chinese cohort. Hepatol Res. 2017;47:845-853
34. Zhao Y, Du Y, Zhao S. et al. Single-nucleotide polymorphisms of microRNA processing machinery genes and risk of colorectal cancer. OncoTargets and Therapy. 2015;8:421-425
35. Cho SH, Ko JJ, Kim JO. et al. 3'-UTR Polymorphisms in the MiRNA Machinery Genes DROSHA, DICER1, RAN, and XPO5 Are Associated with Colorectal Cancer Risk in a Korean Population. PLoS One. 2015;6-10:e0131125
36. Xie Y, Wang Y, Zhao Y. et al. Single-nucleotide polymorphisms of microRNA processing machinery genes are associated with risk for gastric cancer. OncoTargets and Therapy. 2015;8:567-571
37. Buas MF, Onstad L, Levine DM. et al. MiRNA-Related SNPs and Risk of Esophageal Adenocarcinoma and Barrett's Esophagus: Post Genome-Wide Association Analysis in the BEACON Consortium. PLoS One. 2015;10:e0128617
38. Meng JF, Li PY, Zhang HX. et al. Association between RAN polymorphisms and susceptibility to hepatitis B related hepatocellular carcinoma in Guangxi. Acta Universitatis Medicinalis Anhui. 2012;47:1070-1074
39. Martin-Guerrero I, Gutierrez-Camino A, Lopez-Lopez E. et al. Genetic variants in miRNA processing genes and pre-miRNAs are associated with the risk of chronic lymphocytic leukemia. PLoS One. 2015;20-10:e0118905
40. Roy R, De Sarkar N, Ghose S. et al. Genetic variations at microRNA and processing genes and risk of oral cancer. Tumour Biol. 2014;35:3409-3414
41. Ding C, Li C, Wang H. et al. A miR-SNP of the XPO5 gene is associated with advanced non-small-cell lung cancer. Onco Targets Ther. 2013;6:877-881
42. Navarro A, Muñoz C, Gaya A. et al. MiR-SNPs as markers of toxicity and clinical outcome in Hodgkin lymphoma patients. PLoS One. 2013;8:e64716
43. Liu L, An J, Liu J. et al. Potentially functional genetic variants in microRNA processing genes and risk of HBV-related hepatocellular carcinoma. Mol Carcinog. 2013;52:148-154
44. Jiang Y, Chen J, Wu J. et al. Evaluation of genetic variants in microRNA biosynthesis genes and risk of breast cancer in Chinese women. Int J Cancer. 2013;133:2216-24
45. Zhang Q. Association studies of polymorphisms in miRNA and it's biological processing genes with the susceptibility of gastric cancer. Master Thesis, Nanjing Medical University, China. 2014
46. Li YC, Song CH, Yang WJ. et al. Correlation between tag single nucleotide polymorphisms of microRNA regulatory genes and the gentic susceptibility of primary liver cancer. Chinese Journal of Preventive Medicine. 2012;46:533-537
47. Chen J, Qin Z, Pan S. et al. Genetic variants in RAN, DICER and HIWI of microRNA biogenesis genes and risk of cervical carcinoma in a Chinese population. Chin J Cancer Res. 2013;25:565-571
48. Ma H, Yuan H, Yuan Z. et al. Genetic variations in key microRNA processing genes and risk of head and neck cancer: a case-control study in Chinese population. PLoS One. 2012;7:e47544
49. Sung H, Lee KM, Choi JY. et al. Common genetic polymorphisms of microRNA biogenesis pathway genes and risk of breast cancer: a case-control study in Korea. Breast Cancer Res Treat. 2011;130:939-951
50. Liang D, Meyer L, Chang DW. et al. Genetic variants in MicroRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res. 2010;70:9765-9776
51. Kim JS, Choi YY, Jin G. et al. Association of a common AGO1 variant with lung cancer risk: a two-stage case-control study. Mol Carcinog. 2010;49:913-921
52. Horikawa Y, Wood CG, Yang H. et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956-7962
53. Yang H, Dinney CP, Ye Y. et al. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68:2530-2537
54. Ye Y, Wang KK, Gu J. et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila). 2008;1:460-469
55. Kruschke JK, Liddell TM. The Bayesian New Statistics: Hypothesis Testing, Estimation, Meta-Analysis, and Power Analysis From a Bayesian Perspective. Psychonomic Bulletin & Review. 2018;25:178-206
56. Szucs D, Ioannidis JPA. When Null Hypothesis Significance Testing is Unsuitable for Research: A Reassessment. Frontiers in Human Neuroscience. 2017;11:390
57. Spiegelhalter DJ. Incorporating Bayesian Ideas Into Health-Care Evaluation. Statistical Science. 2004;19:156-174
58. Ambros V. The functions of animal micrornas. Nature. 2004;431:350-355
59. Bartel DP. Micrornas: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297
60. Melo SA, Moutinho C, Ropero S. et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleous of cancer cells. Cancer Cell. 2010;18:303-315
61. Yi R, Doehle BP, Qin Y. et al. Overexpression of Exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11:220-226
62. Zheng Y, Pu W, Li J. et al. Discovery of a Prenylated Flavonol Derivative as a Pin1 Inhibitor to Suppress Hepatocellular Carcinoma by Modulating MicroRNA Biogenesis. Chem Asian J. 2019;14:130-134
63. Wen J, Gao Q, Wang N. et al. Association of microRNA-related gene XPO5 rs11077 polymorphism with susceptibility to thyroid cancer. Medicine. 2017;96:e6351
64. Chiosea S, Jelezcova E, Chandran U. et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345-2350
65. Mansini AP, Lorenzo Pisarello MJ, Thelen KM. et al. MicroRNA (miR)-433 and miR-22 dysregulations induce histone-deacetylase-6 overexpression and ciliary loss in cholangiocarcinoma. Hepatology. 2018;68:561-573
66. Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185-191
67. Xia F, Lee CW, Altieri DC. Tumor cell dependence on Ran-GTP-directed mitosis. Cancer Res. 2008;68:1826-1833
68. Abe H, Kamai T, Shirataki H. et al. High expression of Ran GTPase is associated with local invasion and metastasis of human clear cell renal cell carcinoma. Int J Cancer. 2008;122:2391-2397
69. Barrès V, Ouellet V, Lafontaine J. et al. An essential role for Ran GTPase in epithelial ovarian cancer cell survival. Mol Cancer. 2010;9:272
Author contact
![]() Corresponding author: Fen Liu, Department of Epidemiology and Health Statistics, School of Public Health, Beijing Municipal Key Laboratory of Clinical Epidemiology, Capital Medical University, Beijing, China, 100069. Phone: 86-010-83911497, E-mail: liufen05edu.cn
Corresponding author: Fen Liu, Department of Epidemiology and Health Statistics, School of Public Health, Beijing Municipal Key Laboratory of Clinical Epidemiology, Capital Medical University, Beijing, China, 100069. Phone: 86-010-83911497, E-mail: liufen05edu.cn

 Global reach, higher impact
Global reach, higher impact