Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(14):3259-3266. doi:10.7150/jca.30079 This issue Cite
Research Paper
Urovysion FISH Could Be Effective and Useful Method to Confirm the Identity of Cultured Circulating Tumor Cells from Bladder Cancer Patients
1. Department of Hospital Pathology, College of Medicine, The Catholic University of Korea, Seoul, Republic of Korea
2. Department of Urology, College of Medicine, The Catholic University of Korea, Seoul, Republic of Korea
3. The Cancer Research Institute, College of Medicine, The Catholic University of Korea, Seoul, Republic of Korea
Received 2018-9-20; Accepted 2019-4-28; Published 2019-6-2
Abstract
Objective: To explore whether cultured CTC from bladder-cancer patients originate from bladder cancer and share chromosomal abnormalities, by means of a fluorescence in situ hybridization (FISH) test.
Methods: A total of 15 ml of blood was collected from the patients with bladder cancer before treatment began. Isolated CTCs were divided into 5 ml for CTC enumeration and 10 ml for CTC culture. CTCs were counted by immunofluorescent staining with vimentin, cytokeratin, CD45, and DAPI antibody. CTCs were cultured using isolated CTCs in 96-well plates of Mesenchymal Stem Cell Growth Medium for 16~18 days. The resulting cultured CTCs from 20 men with bladder cancer were analyzed by Urovysion FISH.
Results: Common gains were on chromosome 3, 7, and 17 in 20 (74.1%), 14 (51.9%), and 20 (74.1%) of 27 patients, respectively. Polysomy was detected on chromosomes 3 and 7 in 9 patients (33.3%). Polysomy involving two chromosomes was observed in 16 (59.3%, chromosome 3 and 17) and 9 patients (33.3%, chromosome 7 and 17) in the same cell. Among the patients with isolated gain, 17 (63.0%) met the positive criteria for Urovysion FISH. Homozygous deletion of 9p21, 5 (18.5%) involved more than 12 cells. Among the different patient cohorts, positive results based on the Urovysion criteria were obtained in cultured CTCs derived from 19 (70.4%) patients.
Conclusion: Application of FISH Urovysion to cultured CTCs from bladder cancer could be an effective first step to confirm their origin and sharing of chromosomal abnormalities
Keywords: Circulating tumor cells, bladder cancer, FISH Technique, chromosomal abnormality
Introduction
Circulating tumor cells (CTCs) have attracted interest as a “liquid biopsy” from the blood, which may ideally replace aggressive tissue biopsies for serial monitoring of tumor characteristics and tailoring personalized therapy [1]. CTCs can be used to guide cancer management and serve as drug targets [2]. Therefore, it may be possible to evaluate drug sensitivity and resistance and predict patient prognosis following therapy using the CTCs obtained [3]. However, there is a hurdle to overcome in order to realize these potential benefits. Circulating tumor cells (CTCs) are present in the blood of cancer patients at extremely low concentrations. The isolation and characterization of CTCs is technically challenging because of their rareness. Despite the highly promising clinical relevance, clinical application of CTCs has not progressed as far as expected, and the rarity of CTC limits their potential role.
To overcome the rarity of CTCs, in vitro expansion of CTCs has been tried and success has been reported by several laboratories, including ours [4-6]. It can be questionable whether expanded cells originate from the primary cancer and retain their original identification. Therefore, accurate characterization of CTCs is essential for clinical application.
UroVysion™ fluorescence in situ hybridization (FISH) is a molecular diagnostic technique approved by the United States Food and Drug administration (FDA), based on DNA probes. It has been used to identify the most common UC-related chromosomal abnormalities including chromosomes 3 (red), 7 (green), and 17 (aqua), and the 9p21 in malignant urothelial cells that are shed in the urine of persons with UCC. Urovysion™ FISH has been shown to have high sensitivity and specificity for the detection of malignant urothelial cells shedding from primary bladder cancer [7, 8] For this reason, we applied the FISH test to cultured CTCs from bladder-cancer patients to explore whether cultured CTCs from the patients originated from the bladder cancer and shared chromosomal abnormalities.
Methods
Patient recruitment and collection of blood
A total of 15 ml of blood was collected from the patients with bladder cancer at Seoul St. Mary's hospital (Seoul, Korea), under an Institutional Review Board-approved protocol with informed consent (clearance no. 2013 1048). Blood was collected before starting transurethral resection of the bladder (TURBT). A total of 15 cc of blood were divided into 5 ml for CTC enumeration and 10 ml for CTC culture. The stages of the cancer were evaluated using the Tumor, Node, and Metastasis (TNM) system based on the recommendations of the 8th American Joint Committee on Cancer [9](M.B. Amin, S.B. Edge, F.L. Greene, et al. AJCC Cancer Staging Manual, (ed. 8), Springer, Cham, Switzerland (2017)). All blood samples and medical data used were anonymous, to ensure patient confidentiality. A total of 20 patients who succeeded in CTCs cultures were recruited in this study.
Isolation of CTCs and enrichment process
For enrichment and enumeration the CTCs, blood from each patient was collected in Acid Citrate Dextrose tubes (BD Vacutainer®; BD Biosciences, San Jose, CA, USA), and then the Smart Biopsy™ System process was performed within 4 h after blood collection. Enriched CTCs were fixed on slides in 4% paraformaldehyde for 5 min at room temperature and kept at 4°C until further processing. CTCs on slides were permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature, blocked with 1% bovine serum albumin in PBS for 60 min, and incubated with primary antibodies for 60 min, followed by secondary antibody incubation in the same way. The primary antibodies used were rabbit anti-CD45 (Cell Signaling Technology), mouse anti-pan cytokeratin (Sigma), and mouse anti-vimentin Alexa Fluor® 488 conjugated (Cell Signaling Technology). The secondary antibody used goat anti-rabbit Alexa Fluor® 647 and goat anti-mouse Alexa Fluor® 546 (Thermo Fisher Scientific, Inc.). The slides were mounted using Fluoroshield with DAPI (ImmunoBioScience), and then detected using the Nikon Eclipse Ti fluorescent microscope.
Primary short-term culture of CTCs
To analyze the FISH, a short-term culture of 10 ml blood was performed using isolated CTCs for 16~18 days. CTCs were isolated in the way mentioned above. Enriched CTCs were collected, washed with PBS, and then cultured in 96-well ULA plates (Corning) using MSCGM™ Mesenchymal Stem Cell Growth Medium (LONZA). The culture medium was replaced every 3-4 days with minimal disturbance to avoid cell loss. The cultured CTCs were collected in 1.5 mL microtubes. Harvested CTCs were washed using PBS and then fixed by 10% formalin for FISH analysis.
Preparation of cell pellets for UroVysion FISH analysis
The resulting cultured CTCs were centrifugated initially for 10 minutes at 1,840 rpm. The supernatant was decanted, and a full pipette of CytoRich Red was added to the resulting cell pellet, which was allowed to sit for at least 30 minutes. The resulting suspension was centrifuged for 10 minutes at 1,840 rpm. The remaining pellet was vortexed, and Tris-buffered water was added to the 10-ml line. The suspension was centrifuged for 5 minutes at 1,866 rpm to concentrate the specimen. The remaining pellet was vortexed and placed in a corresponding slot on the PrepstainTM slide processor system. Coated glass slides were placed onto staining racks. The non-gyn program was run, and instructions prompted by the computer were followed. After the final wash, the resulting cell pellet on the slide was stored in a 4°C refrigerator for FISH processing.
Interpretation of UroVysion FISH analysis
Specimens were processed, hybridized, and analyzed by FISH using a modified version of the UroVysion packet insert protocol using the UroVysion probe set, which contains centromere enumeration probes for chromosomes 3, 7 and 17, and a locus-specific indicator probe for the 9p21 band, which is the site of the P16 tumor suppressor gene. The UroVysion FISH assay was independently captured and scored by two pathologists aided by the FDA-approved automated BioView Duet scanning system, Allegro-Plus (BioView, Rehovot, Israel). The system included a fluorescent microscope (Olympus), a high-resolution progressive-scan charge-coupled digital camera, and a computer equipped with imaging and analysis software. UroVysion FISH interpretations were divided into positive, negative, unsatisfactory, or equivocal cases. A positive result was defined by four or more cells with aneuploidy for two or more probes, and/or 12 or more cells with homozygous deletion of 9p21, as described in the package insert for the UroVysion Bladder Cancer Kit. For clarifying chromosomal traits of enriched CTCs, all scanned cells in each case were manually categorized into multiple gain (aneuploidy of chromosome 3 with 7 and 17, chromosome 3 with 7, chromosome 3 with 17, chromosome 7 with 17), single gain (aneuploidy of each chromosome 3, 7, and 17), loss of 9p21, wild type (an adequate cell not meeting the above criteria), and nondiagnostic (degenerated or necrotic). Positive criteria used in a previous study[10] were applied in this study. A positive test result was defined as one of the following: the presence of 4 or more morphologically abnormal cells out of 25 analyzed cells that showed polysomy of 2 or more of the chromosomes 3, 7, and 17 in the same cell; isolated gain of a single chromosome in 10% or greater of cells; or homozygous deletion of 9p21 in 12 or more cells. The tests with unsatisfactory or equivocal results were excluded from the study.
Results
Enumeration and immunophenotype of initial captured CTCs
A total of 27 patients with bladder cancer were examined in this study. Table 1 shows the clinical characteristics, counts, and immunophenotype for CTCs. Pathological staging showed 6 (22.2%) in T2N0M0, 4 (14.8%) in T3N0M0, 2 (7.4%) in T4N0M0, and 6 (22.2%) in metastatic disease. CTC counts are reported at baseline for all cohorts. There was a general tendency to increase the total number of CTC (from pT1 to Pt4), but the immunophenotype of captured CTCs showed very wide distribution even in the same stage.
Characterization of cultured CTC by FISH analysis
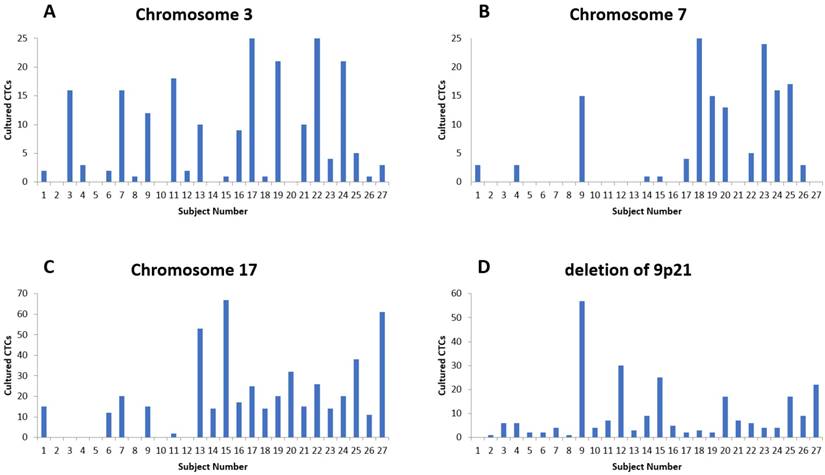
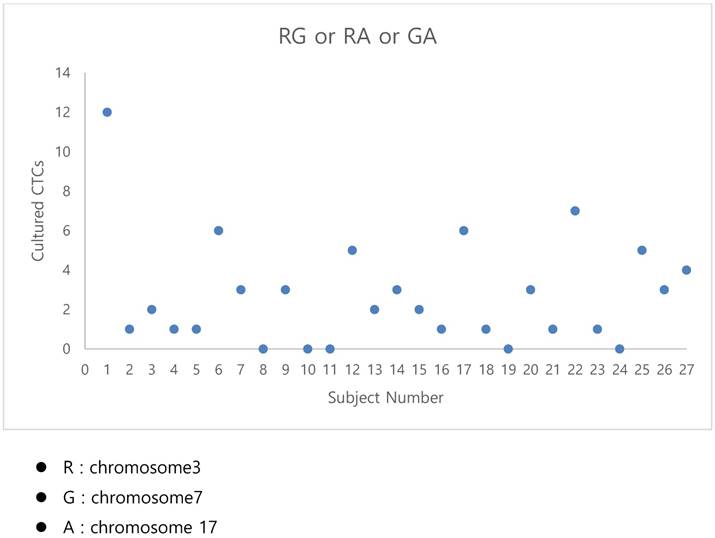
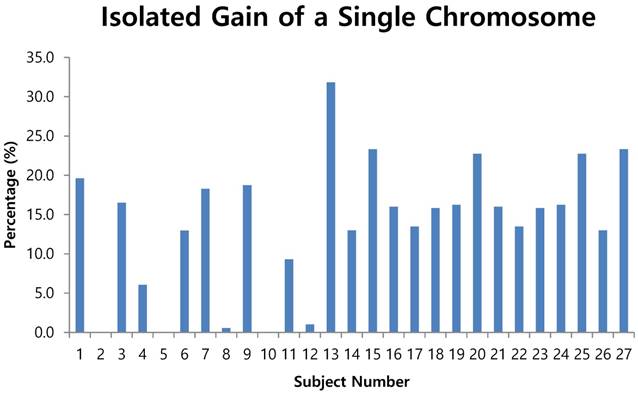
The fluorescent images of the FISH for cultured CTCs displayed the morphological diversity of the cells, with deletion of 9p21, single gain, and multiple chromosome gain (Fig. 1). The numerical results of FISH analysis are summarized in Table 2. Figure 2 is a graphical representation of the individual numerical FISH results for each chromosome. The deletion of 9p21 was observed in all but one enrolled patient. Single gain of chromosome 3, 7, and 17 were detected in 20 (74.1%), 14 (51.9%), and 20 (74.1%) patients among the 27 patients, respectively. Polysomy of chromosome 3 and 7 were detected in 9 patients (33.3%); 16 patients (59.3%, chromosome 3 and 17) and 9 patients (33.3%, chromosome 7 and 17) showed polysomy of two chromosomes in the same cell. Figure 3 is a graphical representation of the polysomy of two or more chromosomes in each patient. Isolated gain of a single chromosome among chromosomes 3, 7, and 17 was detected in 24 patients (88.9%). Among the patients with isolated gain, 17 (63.0%) patients met the positive criteria for Urovysion (Fig. 4). For deletion of 9p21, 5 (25.0%) patients showed more than 12 cells with homozygous deletion of 9p21.
Clinical characteristics and immunophenotype of initial captured CTCs from bladder-cancer patients
| Immunophenotyping & Enumeration (5ml) | |||||||
|---|---|---|---|---|---|---|---|
| Subject number | Sex | Age (years) | pTNM stage | Vimentin (+), CK (-) | Vimentin (-), CK (+) | Vimentin (+), CK (+) | Total CTC |
| 1 | F | 37 | pT4aN1M0 | 2 | 0 | 0 | 2 |
| 2 | F | 80 | T1N0M0 | 3 | 0 | 0 | 3 |
| 3 | M | 60 | T2bN0M0 | 2 | 11 | 3 | 16 |
| 4 | M | 65 | T4aN0M0 | 1 | 17 | 0 | 18 |
| 5 | M | 59 | T1N0M0 | 2 | 2 | 1 | 6 |
| 6 | M | 62 | T1N0M0 | 0 | 5 | 0 | 5 |
| 7 | M | 77 | T3aN0M0 | 0 | 17 | 1 | 18 |
| 8 | M | 74 | T2bN0M0 | 6 | 1 | 21 | 28 |
| 9 | F | 87 | T3aN0M0 | 0 | 0 | 15 | 15 |
| 10 | M | 81 | T1N0M0 | 0 | 2 | 41 | 43 |
| 11 | M | 73 | T2bN0M1 | 1 | 17 | 11 | 29 |
| 12 | M | 80 | T1N0M0 | 0 | 1 | 31 | 32 |
| 13 | M | 65 | T2bN0M0 | 5 | 4 | 6 | 15 |
| 14 | M | 76 | T3aN0M1 | 0 | 11 | 0 | 11 |
| 15 | M | 70 | T4aN0M0 | 0 | 12 | 5 | 17 |
| 16 | M | 75 | T1N0M0 | 0 | 3 | 26 | 29 |
| 17 | M | 62 | T2aN0M0 | 9 | 1 | 4 | 14 |
| 18 | F | 61 | T3aN0M0 | 3 | 11 | 18 | 32 |
| 19 | M | 50 | T1N0M0 | 0 | 3 | 4 | 7 |
| 20 | F | 73 | T3aN2M0 | 0 | 6 | 2 | 8 |
| 21 | M | 76 | T3aN0M1 | 0 | 9 | 0 | 9 |
| 22 | M | 70 | T2aN0M0 | 0 | 3 | 4 | 7 |
| 23 | M | 75 | T1N0M0 | 0 | 3 | 0 | 3 |
| 24 | M | 62 | T2aN0M0 | 3 | 5 | 0 | 8 |
| 25 | M | 61 | T3aN0M0 | 0 | 6 | 0 | 6 |
| 26 | M | 50 | T1N0M0 | 1 | 3 | 1 | 5 |
| 27 | M | 73 | T3aN2M0 | 1 | 3 | 2 | 6 |
The analysis according to the criteria for the Urovysion test
Among the cohort patients, positive results, judging from the Urovysion criteria to be applied to urine specimens, were obtained in cultured CTCs from 15 patients (75%). The distribution and summary for each criterion is as follows:
- By criterion 1 (polysomy of two or more of chromosomes 3, 7, and 17), positive results were obtained from the CTCs of patients 1, 6, 12, 17, 22, and 25 (Fig. 3);
- by criterion 2 (isolated gain of a single chromosome in 10% or more of the cells, positive results were obtained from the CTCs of patients 1, 3, 6, 7, 9, 11, 13, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 and 27 (Fig. 4);
- by criterion 3 (homozygous deletion of 9p21 in 12 or more cells), positive results were obtained from the CTCs of patients 9, 12, 15, 20, 25, and 27 (Fig. 2D)
Discussion
The main findings of this experimental pilot study are as follows:
(1) The FISH method could be applied to cultured CTCs from bladder-cancer patients.
(2) We could confirm that cultured CTCs maintain the chromosomal characteristics of bladder-cancer cells.
(3) Application of FISH to CTCs could be an effective first step to confirm that cultured CTCs originated from primary bladder cancer before clinical application.
Numerical FISH analysis for cultured CTCs from bladder-cancer patients
| Single gain | Multiple gain | |||||||
|---|---|---|---|---|---|---|---|---|
| Subject number | Wild type cell | 9p21 | Chr 3 | Chr 7 | Chr 17 | Chr. 3+7 | Chr. 3+17 | Chr. 7+17 |
| 1 | 70 | 0 | 2 | 3 | 15 | 1 | 11 | |
| 2 | 9 | 1 | 1 | |||||
| 3 | 75 | 6 | 16 | 1 | 1 | |||
| 4 | 86 | 6 | 3 | 3 | 1 | |||
| 5 | 3 | 2 | 1 | |||||
| 6 | 86 | 2 | 2 | 12 | 6 | |||
| 7 | 154 | 4 | 16 | 20 | 2 | 1 | ||
| 8 | 178 | 1 | 1 | |||||
| 9 | 122 | 57 | 12 | 15 | 15 | 3 | ||
| 10 | 198 | 4 | ||||||
| 11 | 172 | 7 | 18 | 2 | 2 | |||
| 12 | 167 | 30 | 2 | |||||
| 13 | 131 | 3 | 10 | 53 | 1 | 1 | ||
| 14 | 162 | 9 | 1 | 14 | 3 | |||
| 15 | 196 | 25 | 1 | 1 | 67 | 2 | ||
| 16 | 160 | 5 | 9 | 17 | 1 | |||
| 17 | 341 | 2 | 25 | 4 | 25 | 1 | 4 | 1 |
| 18 | 209 | 3 | 1 | 25 | 14 | 1 | ||
| 19 | 287 | 2 | 21 | 15 | 20 | |||
| 20 | 133 | 17 | 13 | 32 | 1 | 2 | ||
| 21 | 160 | 7 | 10 | 15 | 1 | |||
| 22 | 302 | 6 | 25 | 5 | 26 | 2 | 4 | 1 |
| 23 | 201 | 4 | 4 | 24 | 14 | 1 | ||
| 24 | 284 | 4 | 21 | 16 | 20 | |||
| 25 | 134 | 17 | 5 | 17 | 38 | 1 | 2 | 2 |
| 26 | 166 | 9 | 1 | 3 | 11 | 3 | ||
| 27 | 181 | 22 | 3 | 61 | 1 | 1 | 2 | |
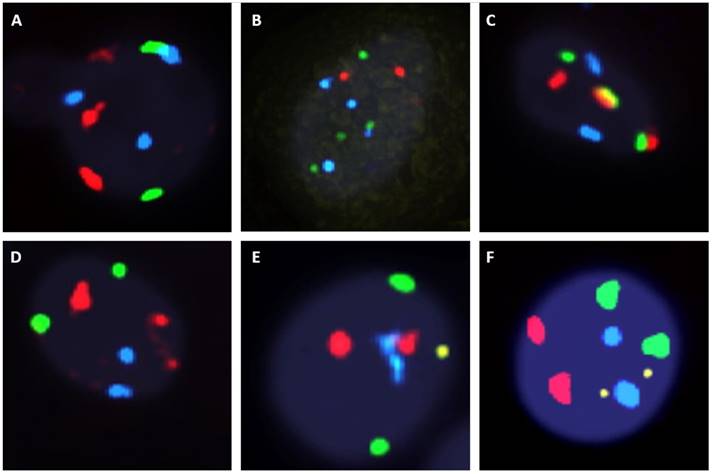
Fluorescent in situ hybridization (UroVysion) staining reveals aneuploidy by gain of chromosome 3 (red), 7 (green) and 17 (aqua) or loss of 9p21 (gold) A. Multiple gain (chromosome 3 with 17). B. multiple gain (chromosome 7 with 17). C. multiple gain (chromosome 3 with 7). D. single gain (chromosome 3). E. single gain (chromosome 17). F. deletion of 9p21 G. Wild type.
The numerical distribution of FISH analysis for cultured CTCs (A) Results for chromosome 3. (B) Results for chromosome 7. (C) Results for chromosome 17. (D). Results for chromosome 9p21.
The graphical representation of the polysomy of two or more of the chromosomes 3, 7, and17.
Isolated gain of a single chromosome in 10% or more of cells
A lot of research on liquid biopsy for clinical applications has been going on. CTCs may be used as a substitute for tissue biopsy to evaluate drug responsiveness and predict an optimal therapy [11, 12]. Sequencing and analyzing CTCs are also actively under way. Although single-cell analysis technically might be possible, DNA extraction from CTCs is still challenging because of the rarity of CTCs. Isolated CTCs are also difficult to apply directly to clinical applications, such as drug sensitivity and gene expressional profiling. The technological challenge of assessing CTCs is that in most cases only a few cells are available and repeated sampling or sampling of a larger blood volume are not realistic possibilities. So, the expansion of CTCs is required, and expansion of CTC could also make clinical applications of CTCs much easier.
It would be ideal if a few CTCs could be amplified into a larger population by in vitro expansion. Several laboratories, including our own, have reported successful in vitro expansion for CTCs. But few reports have verified the cultured CTCs.
To expand CTCs, intact CTCs were isolated by size and were subsequently cultured to obtain enough cells, as mentioned above. Several studies, done at about the same time as ours, using the CTC expansion method have been reported. Several other laboratories have also reported successful in vitro culture for CTCs. But few reports have verified the cultured CTCs. It can be questionable whether cultured CTC originate from primary cancer and retain their original identification. Moreover, the molecular characterization of cells expanded from CTCs is possibly affected by their prior phenotypic heterogeneity [13, 14]. Further, the altered markers of the epithelial-mesenchymal transition (EMT) including EpCAM, cytokeratin and vimentin interfere with the detection of CTCs [15]. Therefore, we enriched CTCs to generate potential subpopulations and genetically unique CTC subsets using multitargeted biomarkers or combination of biomarkers and established putative CTCs that are distinct from malignant urothelial cells in urine. Since morphological identification of urothelial cancer cells is subjective and has low sensitivity [16, 17], multiple molecular markers of urothelial cancer (UC) have been developed for diagnostic purposes [18].
Karyotyping, comparative genomic hybridization, and DNA ploidy studies have shown that most UCC possess chromosomal abnormalities, with some aneuploidy and structural chromosomal abnormalities [19-21]. UroVysion (Abbott Molecular Inc., Ill., USA), was developed to detect the common chromosome abnormalities seen in UCC. It consists of fluorescently labeled DNA probes to the pericentromeric regions of chromosomes 3 (red), 7 (green), and 17 (aqua) and to the 9p21 band (gold) location of the P16 tumor suppressor gene in urine samples [7]. UroVysion was approved by the US Food and Drug Administration initially for surveillance of recurrent urothelial cancer cells and later was extended to detect bladder cancer in patients with hematuria [22-24]. Most of the studies for UroVysion concluded that UroVysion FISH appeared to have good sensitivity and specificity for detecting urothelial cancer cells in urinary specimens [8, 25, 26]. This suggested that the detection of chromosomally abnormal cells might be a good way to identify UCC.
Our study explored the feasibility of performing FISH (Urovysion) to confirm if the cultured CTCs retain the original identification of primary bladder cancer. In this study, we demonstrated that the morphological appearance of the cultured CTCs provides FISH signals, which successfully enable detection of the chromosomal abnormalities associated with bladder cancer. Until now, there have not been identification standards and criteria to apply to cultured CTCs from bladder cancer. In applying the criteria in urine specimens, about 80% of the cultured CTCs from bladder cancer met the positive criteria. These results might prove that the cultured CTCs originated from primary bladder cancer.
The distinctive feature of our study is that, to the best of our knowledge, it is the first study to use the FISH method to show that cultured CTCs originated from primary bladder cancer. Although there were a few previous tests that applied FISH to CTCs, those were only karyotypic characterization of CTCs. The application of FISH to CTCs from bladder cancer could be a novel and easy method to confirm the identification of CTCs, if standards and criteria are established.
The other distinctive feature of our study is that application of FISH to CTCs could provide a useful clinical tool to investigate the biologic targets for therapy by way of peripheral blood sampling. Recently, a few overexpressed genes, such as epidermal growth factor receptor, human epidermal growth factor receptor 2, and vascular endothelial growth factor receptor in bladder cancer, could be a targetable marker for treating and improving the survival of bladder-cancer patients [27]. FISH assessment for the genomic status of target genes in CTCs could be a valuable and attractive tool for future selection of target therapy. Our study has demonstrated the feasibility of FISH in CTCs as a non-invasive and continuously monitoring method.
One limitation of this study was that that UroVysion is not a validated method for detection of CTCs. However, several FISH approaches for cancer diagnosis such as those based on ALK or HER2 have been used to detect CTCs and most of them have been validated for formalin-fixed paraffin-embedded tissue [28, 29]. By contrast, UroVysion is validated for application with urine specimens, which are compatible with the liquid environment of CTCs. Another limitation relates to application of the criteria used for urine specimens to CTCs. CTCs isolated from the blood can be expected to show a different distribution from cells from the urine, which makes it difficult to apply the criteria for urine directly. Considering CTCs' distribution characteristics, the interpretation of the results may change and the positive rate may be higher. Therefore, standards need to be established for the application of FISH Urovysion to CTCs based on further prospective studies. Finally, the relatively small sample size of the study yields weak statistical power to draw any definitive conclusions.
Conclusion
This current study explored the feasibility and usefulness of the Urovysion FISH method for confirming the origin and chromosomal abnormalities of cultured CTCs from bladder cancer. Positive results were obtained for most of the cultured CTCs from bladder-cancer patients, although they were judged by the criteria for urine specimens. Our study suggests that Urovysion FISH could be an effective and useful method to confirm the identity of cultured CTCs before clinical application. To realize and make the possibilities come to life, further studies have to be supplemented.
Acknowledgements
This Research was supported by National Research Foundation of Korea (Grant number: NRF-2015R1C1A1A01051802).
Funding Information
This Research was supported by National Research Foundation of Korea (Grant number: NRF-2015R1C1A1A01051802).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Maheswaran S, Sequist LV, Nagrath S. et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366-77
2. Haber DA, Gray NS, Baselga J. The evolving war on cancer. Cell. 2011;145:19-24
3. Zer A, Leighl N. Promising Targets and Current Clinical Trials in Metastatic Non-Squamous NSCLC. Front Oncol. 2014;4:329
4. Khoo BL, Grenci G, Lim YB. et al. Expansion of patient-derived circulating tumor cells from liquid biopsies using a CTC microfluidic culture device. Nat Protoc. 2018;13:34-58
5. Hwang E, Uh JH, Lee HS. et al. Cancer gene panel analysis of cultured circulating tumor cells and primary tumor tissue from patients with breast cancer. Oncol Lett. 2017;13:4627-32
6. Sheng W, Ogunwobi OO, Chen T. et al. Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip. Lab Chip. 2014;14:89-98
7. Halling KC, Kipp BR. Bladder cancer detection using FISH (UroVysion assay). Adv Anat Pathol. 2008;15:279-86
8. Skacel M, Fahmy M, Brainard JA. et al. Multitarget fluorescence in situ hybridization assay detects transitional cell carcinoma in the majority of patients with bladder cancer and atypical or negative urine cytology. J Urol. 2003;169:2101-5
9. Paner GP, Stadler WM, Hansel DE. et al. Updates in the Eighth Edition of the Tumor-Node-Metastasis Staging Classification for Urologic Cancers. Eur Urol. 2018;73:560-9
10. Dimashkieh H, Wolff DJ, Smith TM. et al. Evaluation of urovysion and cytology for bladder cancer detection: a study of 1835 paired urine samples with clinical and histologic correlation. Cancer Cytopathol. 2013;121:591-7
11. van de Stolpe A, Pantel K, Sleijfer S. et al. Circulating tumor cell isolation and diagnostics: toward routine clinical use. Cancer Res. 2011;71:5955-60
12. Giuliano M, Giordano A, Jackson S. et al. Circulating tumor cells as early predictors of metastatic spread in breast cancer patients with limited metastatic dissemination. Breast Cancer Res. 2014;16:440
13. Jakabova A, Bielcikova Z, Pospisilova E. et al. Molecular characterization and heterogeneity of circulating tumor cells in breast cancer. Breast Cancer Res Treat. 2017;166:695-700
14. Agnoletto C, Minotti L, Brulle-Soumare L. et al. Heterogeneous expression of EPCAM in human circulating tumour cells from patient-derived xenografts. Biomark Res. 2018;6:31
15. Polioudaki H, Agelaki S, Chiotaki R. et al. Variable expression levels of keratin and vimentin reveal differential EMT status of circulating tumor cells and correlation with clinical characteristics and outcome of patients with metastatic breast cancer. BMC Cancer. 2015;15:399
16. Glas AS, Roos D, Deutekom M. et al. Tumor markers in the diagnosis of primary bladder cancer. A systematic review. J Urol. 2003;169:1975-82
17. Brown FM. Urine cytology. It is still the gold standard for screening? Urol Clin North Am. 2000;27:25-37
18. Black PC, Brown GA, Dinney CP. Molecular markers of urothelial cancer and their use in the monitoring of superficial urothelial cancer. J Clin Oncol. 2006;24:5528-35
19. Fadl-Elmula I, Gorunova L, Mandahl N. et al. Karyotypic characterization of urinary bladder transitional cell carcinomas. Genes Chromosomes Cancer. 2000;29:256-65
20. Richter J, Jiang F, Gorog JP. et al. Marked genetic differences between stage pTa and stage pT1 papillary bladder cancer detected by comparative genomic hybridization. Cancer Res. 1997;57:2860-4
21. Bittard H, Lamy B, Billery C. Clinical evaluation of cell deoxyribonucleic acid content measured by flow cytometry in bladder cancer. J Urol. 1996;155:1887-91
22. Sarosdy MF, Kahn PR, Ziffer MD. et al. Use of a multitarget fluorescence in situ hybridization assay to diagnose bladder cancer in patients with hematuria. J Urol. 2006;176:44-7
23. Halling KC, King W, Sokolova IA. et al. A comparison of cytology and fluorescence in situ hybridization for the detection of urothelial carcinoma. J Urol. 2000;164:1768-75
24. Sokolova IA, Halling KC, Jenkins RB. et al. The development of a multitarget, multicolor fluorescence in situ hybridization assay for the detection of urothelial carcinoma in urine. J Mol Diagn. 2000;2:116-23
25. Caraway NP, Khanna A, Fernandez RL. et al. Fluorescence in situ hybridization for detecting urothelial carcinoma: a clinicopathologic study. Cancer Cytopathol. 2010;118:259-68
26. Galvan AB, Salido M, Espinet B. et al. A multicolor fluorescence in situ hybridization assay: A monitoring tool in the surveillance of patients with a history of non-muscle-invasive urothelial cell carcinoma: A prospective study. Cancer Cytopathol. 2011;119:395-403
27. Mohammed AA, El-Tanni H, El-Khatib HM. et al. Urinary Bladder Cancer: Biomarkers and Target Therapy, New Era for More Attention. Oncol Rev. 2016;10:320
28. Tan CL, Lim TH, Lim T. et al. Concordance of anaplastic lymphoma kinase (ALK) gene rearrangements between circulating tumor cells and tumor in non-small cell lung cancer. Oncotarget. 2016;7:23251-62
29. Jaeger BAS, Neugebauer J, Andergassen U. et al. The HER2 phenotype of circulating tumor cells in HER2-positive early breast cancer: A translational research project of a prospective randomized phase III trial. PLoS One. 2017;12:e0173593
Author contact
![]() Corresponding author: U-Syn Ha, Department of Urology, Seoul St. Mary's Hospital, College of Medicine, The Catholic University of Korea, 222, Banpo-daero, Seocho-gu, Seoul, 06591, Republic of Korea. Tel: +82-2-2258-6224; Fax: +82-2-599-7839; E-mail: ushamdac.kr
Corresponding author: U-Syn Ha, Department of Urology, Seoul St. Mary's Hospital, College of Medicine, The Catholic University of Korea, 222, Banpo-daero, Seocho-gu, Seoul, 06591, Republic of Korea. Tel: +82-2-2258-6224; Fax: +82-2-599-7839; E-mail: ushamdac.kr

 Global reach, higher impact
Global reach, higher impact