Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(13):2961-2968. doi:10.7150/jca.31004 This issue Cite
Research Paper
Knockdown of linc01023 restrains glioma proliferation, migration and invasion by regulating IGF-1R/AKT pathway
1. Department of Neurosurgery, Shengjing Hospital of China Medical University, Shenyang 110004, People's Republic of China
2. Liaoning Clinical Medical Research Center in Nervous System Disease, Shenyang 110004, People's Republic of China
3. Key Laboratory of Neuro-oncology in Liaoning Province, Shenyang 110004, People's Republic of China
4. Department of Neurology, Shengjing Hospital of China Medical University, Shenyang 110004, People's Republic of China
5. Exprimental Research center, Shengjing Hospital of China Medical University, Shenyang 110004, People's Republic of China
Received 2018-10-27; Accepted 2019-4-28; Published 2019-6-2
Abstract
LncRNAs have been proved to be involved in the promotion of glioma cell malignant development. However, the exact roles and molecular mechanisms of linc01023 in glioma remain blurred. In this study, we confirm linc01023 is up-regulated in glioma tissues and cell lines. In addition, elevated linc01023 expression indicates shorter survival times in patients with glioma. Moreover, loss-of-function studies reveal that restoration of linc01023 restrains glioma cell proliferation, migration and invasion by regulating IGF1R/AKT pathway in vitro and in vivo. Collectively, the study indicates that linc01023 plays an oncogenic role in glioma through activation of IGF1R/AKT signal pathway, and it could be a candidate therapeutic target.
Keywords: Glioma, Prognosis, Linc01023, IGF1R, AKT
Introduction
Glioma, the vast majority of intracranial malignant tumors, is characterized by rapid proliferation and aggressive invasion [1]. According to the 2016 World Health Organization (WHO) classification of Central Nervous System (CNS) tumors, glioma was divided into low grade glioma (LGG, grades Ⅰ and Ⅱ) and high grade glioma (HGG, grades Ⅲ and Ⅳ)[2]. Despite the advances in therapeutic strategies, combination of surgical resection, radiotherapy and temozolomide chemotherapy, patients suffering from glioma still exhibit poor prognosis. Thus, there have been a growing number of publications focusing on appraising significant index which control glioma risk stratification [3]. Particularly, isocitrate dehydrogenase 1 (IDH1) mutation is written in the amended 2016 WHO classification of CNS tumors [4]. However, the difficulty in defining all patient outcomes based on the traditional biomarkers suggests an urgent need to identify new clinicobiological markers to assess patient prognosis.
Recently, there has been a growing recognition of the vital links between long non-coding RNAs (lncRNAs) and cancers [5, 6]. Remarkably, emerging articles have shown that lncRNAs are involved in regulation of glioma malignant behaviors [7, 8]. By retrieving TCGA database, we discovered that expression of linc01023 was up-regulated in glioma, however, the molecular mechanisms of linc01023 in glioma remain unclear.
Insulin like growth factor I receptor (IGF1R) is an inducible transmembrane tyrosine kinase receptor that often activated in several types of cancer including glioma [9]. Previous studies have demonstrated that IGF-1R activates the phosphoinositide 3- kinase (PI3K)/AKT signaling pathways and promotes glioblastoma cell proliferation and survival by ligand binding [10]. In the current study, we profiled linc01023 expressions in glioma tissues and cells. We also confirmed glioma patients with high expression of linc01023 indicated poor prognosis. Moreover, linc01023 depletion inhibited glioma cell proliferation, migration and invasion by regulating IGF1R/AKT pathway in vitro and in vivo. Above all, restoration of IGF1R/AKT pathway reversed the inhibitory effects of linc01023. The results above offered encouraging targets for glioma therapy.
Materials and Methods
Patients and specimens
We studied 169 glioma tissues and 30 normal brain tissues (NBTs) from patients suffered traffic injury in the Department of Neurosurgery and Pathology, Shengjing Hospital of China Medical University (Liaoning, China). The inclusion criteria used to screen patients were as follows: (1) patients underwent surgery in Shengjing Hospital from October 2008 to October 2017; (2) tumors were confirmed by histopathology as glioma. Patients were excluded if they met any of the following criteria: (1) patients received radiotherapy or chemotherapy before operation; (2) patients combine other tumors; (3) recurrence case; (4) simple pathological biopsy case. After excision, the specimens were put in liquid nitrogen immediately. The clinical data, including pathological grade, ECOG score, sex, age and IDH1 mutation were collected according to the 2016 classification of glioma. Moreover, we registered the date of deaths resulted from glioma. The interval time from the diagnosis date to death was recorded. We gained permissions from all the patients. The study was approved by the Ethics Committee of Shengjing Hospital (No. 2018PS300K). The median of linc01023 expression in glioma tissues was used to distinguish high and low expression of linc01023.
RNA Isolation and Quantitative RT-PCR
Total RNA was isolated from NBTs, gliomas, normal human astrocytes (NHA), U87, and U251 cells using TRIzol (Life Technologies Corporation, CA, USA). We performed a One-Step SYBR PrimeScript RT-PCR Kit (Takara Bio, Inc, Kusatsu, Japan) to detect the expression of linc01023. GAPDH was employed as an endogenous control. We used the comparative Ct method to calculate the relative expression of linc01023. Primer sequences for linc01023: forward 5′-GAAAA CCAGGAGAGCATGGAGCT-3′, reverse 5′-TTTTTTCGCGTTCATTATCTCAT-3′; GAPDH forward 5′-TGCACCACCAACTGCTTAGC-3′, reverse 5′-GGCATGCACTGTGGTCATGAG -3′.
Western Blot Analysis
Western blot was performed as described earlier [11]. In short, total proteins were extracted from U87 and U251 cells using RIPA buffer with protease inhibitors. Protein levels of AKT, p-AKT, IGF-1R and p-IGF-1R then analyzed by SDS-PAGE and transferred for immunoblotting with primary antibodies overnight at 4 °C. Then the membranes were incubated with secondary antibodies for 2 hours. Immunoblots were visualized using ECL chemiluminescent detection system and relative integrated density values (IDVs) were calculated by FLuor Chem2.0 software using GAPDH as a control for protein normalization. The antibodies were listed in the following: IGF-1R, p-IGF-1R, AKT, p-AKT (1:1000; Cell Signaling, Beverly, MA, USA), GAPDH (1:1000, Santa Cruz Biotechnology), Goat anti-rabbit antibody and Goat anti-mouse antibody (1:5000, Proteintech, Hubei, China).
Cell Culture and reagents
U87 and U251 cells lines (Chinese Academy of Medical Sciences, Beijing, China) were cultured in DMEM/high glucose supplemented with 10% fetal bovine serum (FBS, Life Technologies, Carlsbad, CA, USA). NHA were purchased from ScienCell Research Laboratories (Carlsbad, CA, USA) and cultured in RPMI-1640 medium with 10% FBS. All the cells were cultured in a humidified incubator (37 °C, 5% CO2). Human IGF1 was purchased from PeproTech (Rocky Hill, NJ).
Cell Transfections
Cell Transfection was performed as described earlier [12]. Short-hairpin linc01023 (sh-linc01023) and its negative control sequence (sh-NC) were ordered from GenePharma (GenePharma, Suzhou, China). Glioma cells were transfected at roughly 70% confluence using Opti-MEM and Lipofectamine 3000 reagents (Invitrogen, CA, USA) followed the manufacturer's instructions.
Cell Proliferation, Migration and Invasion Detection
As described previously, we analysed cell proliferation, cell migration and invasion using CCK-8 and transwell assay [13].
Tumor xenografts in nude mice
In vivo study, sh-linc01023 and NC were ligated into the LV3-CMVGFP-Puro vector (GenePharma). Then, LV3-CMV-GFPPuro-sh-linc01023 and its NC vectors were generated. We used ViraPower Packaging Mix to generate Lentivirus in 293FT cells. After infection, the cells expressing sh-linc01023 were obtained. Five-week-old BALB/C athymic nude mice were purchased from the National Laboratory Animal Center (Beijing, China). After stably transfected with sh-linc01023 or sh-NC, U87 and U251 cells were respectively injected into the right flanks subcutaneous in a dose of 5×105 per group. Tumor volume was measured every 4 days using the formula: volume (mm3)= length × width2/2. The subcutaneous tumor-bearing mice were executed 40 days after injection. As for intracerebral orthotopic inoculation, 3×105 cells were stereotactically implanted to the right striatum of nude mice (n=8, each group). The number of survivals was counted daily. Kaplan-Meier survival curve was made to conduct the survival analysis.
Statistical analysis
The proliferation, migration, invasion and western blot data were expressed as means ± standard deviation (SD) from at least three independent experiments. Significant differences were determined using the GraphPad 7.0 software (USA). Survival curves were calculated using the Kaplan-Meier method. A chi-square test was applied to calculate the correlation of linc01023 with clinical data. Univariable and multivariate Cox analysis were applied to analyze the prognostic value of clinical data, respectively. We applied SPSS 17.0 software for chi-square test, univariable and multivariate Cox analysis. P < 0.05 was regarded as statistically significant.
Results
Upregulation of linc01023 in glioma tissues and glioma cell lines
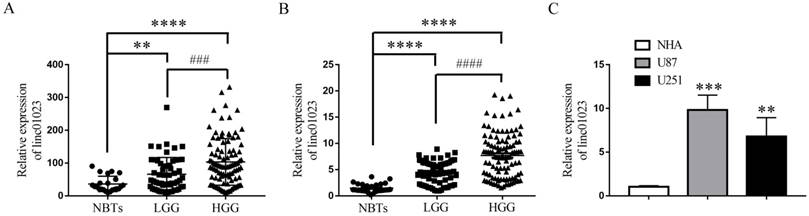
To detect the clinical significance of linc01023 in glioma, we analyzed publicly available lncRNA datasets of glioma from The Cancer Genome Atlas (TCGA). As shown in Figure 1A, linc01023 level elevated in high grade glioma tissues (HGG) and low grade glioma tissues (LGG) compared with normal brain tissues (NBTs). We then detected linc01023 expression levels in our clinical glioma tissues and glioma cells by qPCR. The results indicated that linc01023 expression was positively correlated with the progression of glioma pathological grades (Figure 1B). And linc01023 was up-regulated in U87 and U251 glioma cell lines compared with normal human astrocytes (NHA) (Figure 1C). Thus, we hypothesized that high expression of linc01023 may be associated with the progression of glioma.
Expression of linc01023 correlated with the clinical characteristics of patients with glioma
We further detected the correlation of linc01023 expression levels with clinical characteristics in glioma patients in our glioma tissues. We considered the median of linc01023 expression as the dividing line between the high and low expression of linc01023. As shown in Table 1 and 2, linc01023 expression levels correlated with WHO grade of glioma (P < 0.001) and IDH1 mutation (P = 0.007). However, linc01023 expression had no correlation with gender (P = 0.585), age (P = 0.317) and ECOG (P = 0.824).
Association of linc01023 with WHO grade
| Number of patients | linc01023 expression | r | P value | ||
|---|---|---|---|---|---|
| low | high | ||||
| Total | 169 | 84 | 85 | 0.362 | <0.001 |
| WHO grade | |||||
| Ⅰ | 24 | 19 | 5 | ||
| Ⅱ | 41 | 26 | 15 | ||
| Ⅲ | 42 | 19 | 23 | ||
| Ⅳ | 62 | 20 | 42 | ||
P value was evaluated by spearman's correlation test
Up-regulation of linc01023 in glioma tissues and glioma cell lines. A Expression of linc01023 in normal brain tissues vs. gliomas. Normal brain tissues (NBTs), n=23; low grade gliomas (LGG), n=57; high grade gliomas (HGG), n=100. Data source: GSE4290. B qRT-PCR analysis of linc01023 expression in gliomas. NBTs, n=30; LGG, n=65; HGG, n=104. For A and B, **P < 0.01 versus NBTs; ****P < 0.0001 versus NBTs; ###P < 0.001 versus LGG; ####P < 0.0001 versus LGG. C qRT-PCR analysis of linc01023 expression in NHA, U87 and U251 glioma cells. n= 5 in each group, ***P < 0.001 versus NHA, **P < 0.01 versus NHA. Data are presented as the mean ± SD.
Association of linc01023 expression with clinopathological characteristics
| Clinicopathological parameters | number | linc01023 expression | P value | |
|---|---|---|---|---|
| Low | High | |||
| Total | 169 | 84 | 85 | |
| Gender | 0.585 | |||
| Male | 91 | 47 | 44 | |
| Female | 78 | 37 | 41 | |
| Age | 0.317 | |||
| ≤50 | 86 | 46 | 40 | |
| >50 | 83 | 38 | 45 | |
| ECOG | 0.824 | |||
| <2 | 96 | 47 | 49 | |
| ≥2 | 73 | 37 | 36 | |
| IDH1 mutation | 0.007 | |||
| + | 71 | 44 | 27 | |
| - | 98 | 40 | 58 | |
P value was evaluated by chi-square test
Elevated linc01023 expression indicates shorter survival times in patients with glioma
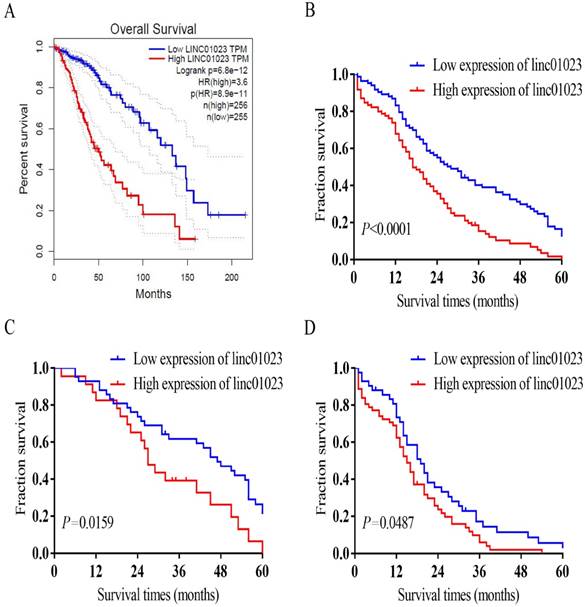
The correlation of linc01023 with WHO grade and IDH1 mutation revealed the potential role of linc01023 in the prognosis of glioma. As shown in Fig. 2A, survival analysis of Gene Expression Profiling Interactive Analysis (GEPIA) glioma cohorts indicated low linc01023 expression patients had obviously longer survivals than those with high linc01023 expression (P < 0.001). Kaplan-Meier survival analysis of our data demonstrated that patients with glioma in the high linc01023 expression group manifested a worse prognosis compared with those in the low linc01023 expression group (P < 0.001, Fig. 2B). In addition, low linc01023 expression group manifested longer survivals in both LGG and HGG group (P < 0.05, Fig. 2C-D).
Besides the high expression of linc01023, we analysed other factors might correlate with the survival time. The results revealed that age > 50 (P = 0.007), HGG (P < 0.001), as well as IDH mutation (P < 0.001) were the significant factors correlated with the survival time (Table 3). Multivariable Cox analysis indicated that the high expression of linc01023 was an independent prognostic factor for shorter survival in patients with glioma (P = 0.011). Furthermore, WHO grade (P < 0.001) and IDH mutation (P = 0.012), were obviously associated with the survival of patients with glioma (Table 4).
Association of linc01023 expression with glioma patients' survival. A Patients with low expression of linc01023 manifested a significantly higher probability of survival. (log rank test, n=511, P < 0.0001) Data source: GEPIA. B-D Kaplan-Meier survival analysis and log rank test for all glioma patients (B, n=169), LGG (C, n=65) and HGG (D, n=104) with different linc01023 expression. linc01023-high had worse prognosis (log rank test, P < 0.05).
Univariate analysis of prognostic factors in glioma for overall survival
| P value | Hazard Ratio | 95% Confidence Interval | |
|---|---|---|---|
| linc01023 expression High versus low | <0.001 | 2.008 | 1.431-2.818 |
| Gender male versus female | 0.243 | 1.213 | 0.877-1.679 |
| Age >50 versus ≤ 50 | 0.007 | 1.582 | 1.135-2.204 |
| ECOG ≥2 versus <2 | 0.136 | 0.778 | 0.559-1.082 |
| WHO grade Ⅲ+ⅣversusⅠ+Ⅱ | <0.001 | 3.044 | 2.119-4.372 |
| IDH mutation + versus - | <0.001 | 0.456 | 0.324-0.642 |
Multivariate analysis of prognostic factors in glioma for overall survival
| P value | Hazard Ratio | 95% Confidence Interval | |
|---|---|---|---|
| Linc01023 expression High versus low | 0.011 | 1.584 | 1.111-2.258 |
| Age >50 versus ≤ 50 | 0.835 | 0.962 | 0.669-1.383 |
| WHO grade Ⅲ+ⅣversusⅠ+Ⅱ | <0.001 | 2.541 | 1.701-3.796 |
| IDH mutation + versus - | 0.012 | 0.623 | 0.432-0.900 |
Linc01023 depletion inhibits glioma cell proliferation, migration and invasion by regulating IGF1R/AKT pathway
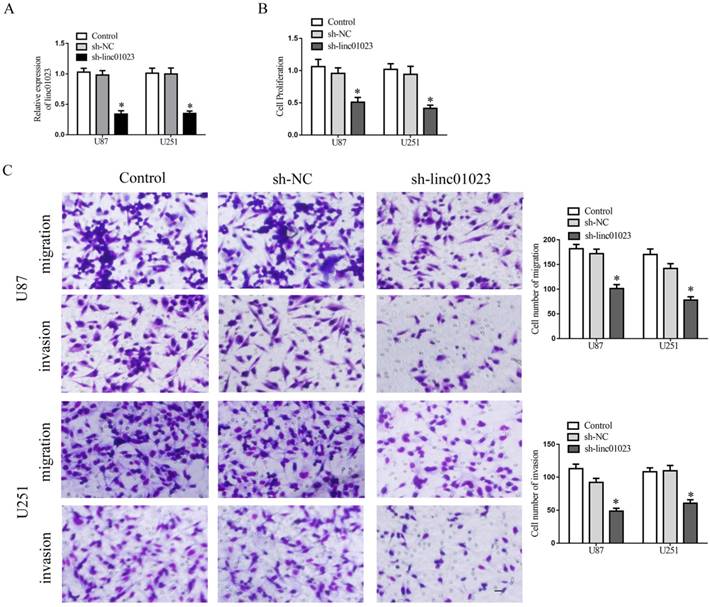
We next sought to investigate the mechanism of linc01023 affecting the prognosis of glioma and performed loss-of-function studies in U87 and U251 glioma cells. After the detection of transfection efficiency (Fig. 3A), we investigated the roles linc01023 exerted in glioma cells. As Figure 3B-C showed, depletion of linc01023 manifested a depression in proliferation, migration and invasion of glioma cells.
Inhibition of linc01023 restrained glioma cell proliferation, migration and invasion. A Relative expression of linc01023 after U87 and U251 glioma cells transfected with sh-linc01023 plasmids and scrambled vectors (NC). B CCK-8 assay to investigate the effect of linc01023 knockdown on proliferation of U87 and U251 cells. C Quantification number of migration and invasion cells treated with inhibition of linc01023. Representative images and accompanying statistical plots were presented. Data are presented as the mean ± SD (n = 3 in each group). *P < 0.05 versus sh-NC group (empty vector). Scale bars represent 20 mm.
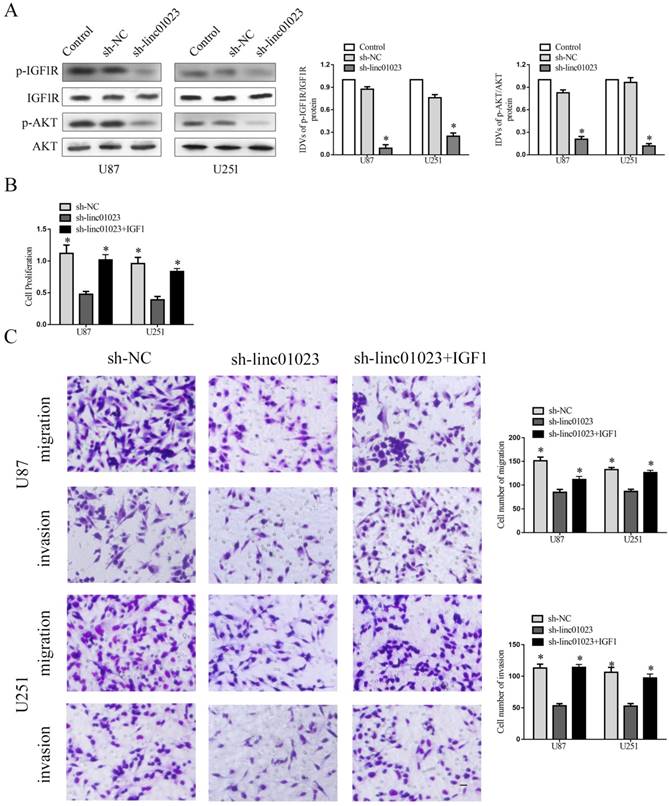
To determine the molecular mechnism of linc01023 inhibiting proliferation, migration and invasion in glioma, we detected the pathway changes in sh-linc01023 glioma cells. As shown in Fig. 4A, knock-down of linc01023 significantly restrained the activity of IGF1R/AKT pathway.
Although IGF1 can activate IGF1R/AKT pathway in glioma cells are well recognized, it has not been well documented whether linc01023 could affect IGF1R/AKT pathway in glioma [10]. Furthermore, earlier study has shown that IGF-1 could stimulate the phosphorylation and activation of IGF-1R [14]. To further verify the functional connection between linc01023 and IGF1R/AKT pathway, we rescued inhibition of IGF1R/AKT pathway by linc01023 depletion using addition 10ng/ml IGF1 in the medium of glioma cells. As Fig. 4B-C showed, the inhibition of cell proliferation, migration, and invasion induced by sh-linc01023 was rescued by IGF1R/AKT pathway activation (Fig. 4B-C). Collectively, the data revealed that linc01023 depletion inhibited malignant behavior of glioma cells by regulating IGF1R/AKT pathway.
Knock-down of linc01023 restrained glioma cell growth in vivo
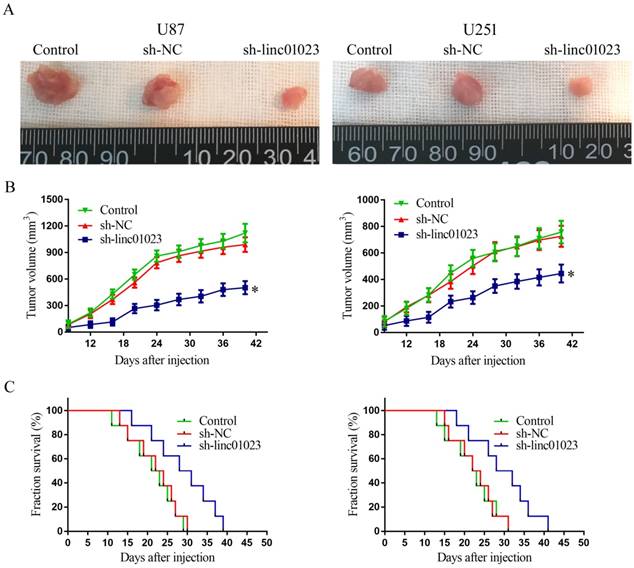
To further validate the glioma growth inhibition of linc01023 in vivo, we injected U87 and U251 cells transfected with sh-linc01023/sh-NC into BALB/c nude mice. As shown in Fig. 5A-B, in contrast to sh-NC group, sh-linc01023 group produced smaller tumor. Moreover, the survival analysis showed that sh-linc01023 group exhibited longer survival time (Figure 5C).
Inhibition of linc01023 restrained glioma cell proliferation, migration and invasion by regulating IGF1R/AKT pathway. A Knock-down of linc01023 inhibited activity of IGF1R/AKT pathway. B-C Inhibition of proliferation, migration and invasion by deletion of linc01023 could be rescued by addition of IGF1 in glioma cells. Data are presented as the mean ± SD (n = 3 in each group). *P < 0.05 versus sh-linc01023 group. Scale bars represent 20 mm.
Tumor xenografts study. A Tumors carried by nude mice of respective groups were shown. B Tumor growth curves in respective groups (n = 8), *P < 0.05 vs. sh-NC group. C Survival curves of nude mice injected tumors (n = 8), Mice were monitored for up to 50 days (n = 8).
Discussion
Glioma is the most common neoplasm that threatens people's health for the malignant progression and poor prognosis in the brain [15]. According to the WHO CNS tumors classification of 2016, the prognosis of glioma patients is significantly determined by both histological parameters and specific molecular markers. In the present study, we discovered that linc01023 expression elevated in in glioma tissues and cells. In addition, elevated linc01023 expression indicates short survival times in patients with glioma. Furthermore, knock-down of linc01023 restrained the proliferation, migration, and invasion of glioma cells. The in vivo study indicated the sh-linc01023 group manifested smaller tumor and longer survival time.
Currently, mounting evidence has indicated that lncRNAs are involved in regulation of glioma malignant behaviors by modulating signaling pathways [16]. However, the roles of linc01023 in glioma remains blurred. In this study, we discovered that linc01023 was highly expressed in glioma tissues and cells for the first time. Then, we clarified that linc01023 expression was definitely correlated with the WHO grades and IDH1 mutation. In addition, Kaplan-Meier survival analysis illustrated patients with high expression of linc01023 manifested short overall survival. Consistently, many earlier study also have illuminated that lncRNAs correlates with overall survival in glioma. For example, long non-coding RNA AFAP1-AS1 is up-regulated in glioma, and its expression is correlated with glioma grading and KPS scores [17]. Moreover, long non-coding RNA PVT1 is up-regulated in glioma and serves as an indicator of glioma prognosis [18].
To further confirm the roles of linc01023 in glioma malignant behavior, we conducted the CCK-8 and transwell analysis in sh-linc01023 glioma cells. The results indicated that restoration of linc01023 restrained the proliferation, migration and invasion of glioma cells. Besides, knock-down of linc01023 significantly reduced xenografts tumor growth. In addition, previous studies have shown that lncRNAs participated in the promotion of glioma cell proliferation and malignant development. For instance, knockdown of OIP5-AS1 resulted in inhibition of glioma cells malignant progression by regulating miR-367-3p/CEBPA feedback loop [19].
It has been extensively documented in earlier reports that inhibition of IGF-1R pathway inhibits glioma tumor growth and migration [20]. Furthermore, inhibition of IGF1R restrained glioma cell viability and represses subcutaneous xenograft growth [10]. Our results showed IGF1R/AKT pathway was inhibited in sh-linc01023 group. More importantly, the inhibition of cell proliferation, migration, and invasion induced by sh-linc01023 was rescued by IGF1R/AKT pathway activation.
Conclusions
In summary, our study revealed that linc01023 acted as an oncogene in glioma. Elevated linc01023 expression was a biomarker of poor prognosis in glioma patients. Knock-down of linc01023 restrained the proliferation, migration, and invasion of glioma cells by regulating IGF1R/AKT pathway. The study demonstrated the crucial roles of linc01023 in the pathogenesis of glioma, accordingly, provide a new potential therapeutic target for glioma therapy and a prognostic marker.
Acknowledgements
This work is supported by grants from the Natural Science Foundation of China (81672511 and 81372484), Natural Science Foundation of Liaoning Province (201800885) and Scientific Fund of Shengjing hospital (MF43).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wen PY, Kesari S. Malignant gliomas in adults. The New England journal of medicine. 2008;359(5):492-507
2. Xi Z, Xue Y, Zheng J, Liu X, Ma J, Liu Y. WTAP Expression Predicts Poor Prognosis in Malignant Glioma Patients. Journal of molecular neuroscience: MN. 2016;60(2):131-136
3. Zhao Z, Xiao S, Yuan X, Yuan J, Zhang C, Li H. et al. AHNAK as a Prognosis Factor Suppresses the Tumor Progression in Glioma. Journal of Cancer. 2017;8(15):2924-2932
4. Diamandis P, Aldape K. World Health Organization 2016 Classification of Central Nervous System Tumors. Neurologic clinics. 2018;36(3):439-447
5. Wu S, Liu J, Wang X, Li M, Chen Z, Tang Y. Aberrant Expression of the Long Non-coding RNA GHRLOS and Its Prognostic Significance in Patients with Colorectal Cancer. Journal of Cancer. 2017;8(19):4040-4047
6. Zhou K, Zhang C, Yao H, Zhang X, Zhou Y, Che Y. et al. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Molecular cancer. 2018;17(1):105
7. Xue W, Chen J, Liu X, Gong W, Zheng J, Guo X. et al. PVT1 regulates the malignant behaviors of human glioma cells by targeting miR-190a-5p and miR-488-3p. Biochimica et biophysica acta Molecular basis of disease. 2018;1864(5 Pt A):1783-1794
8. Liu X, Zheng J, Xue Y, Qu C, Chen J, Wang Z. et al. Inhibition of TDP43-Mediated SNHG12-miR-195-SOX5 Feedback Loop Impeded Malignant Biological Behaviors of Glioma Cells. Molecular therapy Nucleic acids. 2018;10:142-158
9. Osuka S, Sampetrean O, Shimizu T, Saga I, Onishi N, Sugihara E. et al. IGF1 receptor signaling regulates adaptive radioprotection in glioma stem cells. Stem cells. 2013;31(4):627-640
10. Gong Y, Ma Y, Sinyuk M, Loganathan S, Thompson RC, Sarkaria JN. et al. Insulin-mediated signaling promotes proliferation and survival of glioblastoma through Akt activation. Neuro-oncology. 2016;18(1):48-57
11. Yu M, Xue Y, Zheng J, Liu X, Yu H, Liu L. et al. Linc00152 promotes malignant progression of glioma stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway. Molecular cancer. 2017;16(1):110
12. Yu S, Yu M, He X, Wen L, Bu Z, Feng J. KCNQ1OT1 promotes autophagy by regulating miR-200a/FOXO3/ATG7 pathway in cerebral ischemic stroke. Aging Cell. 2019 e12940
13. Yu M, Yu S, Xue Y, Yu H, Chen D, Wei X. et al. Over-Expressed FEZF1 Predicts a Poor Prognosis in Glioma and Promotes Glioma Cell Malignant Biological Properties by Regulating Akt-ERK Pathway. Journal of molecular neuroscience: MN. 2018;65(4):411-419
14. Zhu H, Wang DD, Yuan T, Yan FJ, Zeng CM, Dai XY. et al. Multikinase Inhibitor CT-707 Targets Liver Cancer by Interrupting the Hypoxia-Activated IGF-1R-YAP Axis. Cancer research. 2018;78(14):3995-4006
15. Zhao X, Wang P, Liu J, Zheng J, Liu Y, Chen J. et al. Gas5 Exerts Tumor-suppressive Functions in Human Glioma Cells by Targeting miR-222. Molecular therapy: the journal of the American Society of Gene Therapy. 2015;23(12):1899-1911
16. Yu H, Xue Y, Wang P, Liu X, Ma J, Zheng J. et al. Knockdown of long non-coding RNA XIST increases blood-tumor barrier permeability and inhibits glioma angiogenesis by targeting miR-137. Oncogenesis. 2017;6(3):e303
17. Wang Y, Lan Q. Long non-coding RNA AFAP1-AS1 accelerates invasion and predicts poor prognosis of glioma. European review for medical and pharmacological sciences. 2018;22(16):5223-5229
18. Yang A, Wang H, Yang X. Long non-coding RNA PVT1 indicates a poor prognosis of glioma and promotes cell proliferation and invasion via target EZH2. Bioscience reports. 2017:37 (6)
19. Liu X, Zheng J, Xue Y, Yu H, Gong W, Wang P. et al. PIWIL3/OIP5-AS1/miR-367-3p/CEBPA feedback loop regulates the biological behavior of glioma cells. Theranostics. 2018;8(4):1084-1105
20. Zhou Q, Zhang J, Cui Q, Li X, Gao G, Wang Y. et al. GSK1904529A, an insulin-like growth factor-1 receptor inhibitor, inhibits glioma tumor growth, induces apoptosis and inhibits migration. Molecular medicine reports. 2015;12(3):3381-3385
Author contact
![]() Corresponding author: Yunhui Liu, Department of Neurosurgery, Shengjing Hospital of China Medical University, Shenyang 110004, People's Republic of China. Telephone number: +86 024 96615-36111; Fax number: +86 024 2295 8989; Email: liuyunhui17com
Corresponding author: Yunhui Liu, Department of Neurosurgery, Shengjing Hospital of China Medical University, Shenyang 110004, People's Republic of China. Telephone number: +86 024 96615-36111; Fax number: +86 024 2295 8989; Email: liuyunhui17com

 Global reach, higher impact
Global reach, higher impact