Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(13):2915-2926. doi:10.7150/jca.31130 This issue Cite
Research Paper
Glutathione S-transferases genes variants and chemotherapy efficacy in gastrointestinal cancer patients: a meta-analysis based on 50 pharmacogenetic studies
1. Department of General Surgery, The Third Clinical College of Wenzhou Medical University, Wenzhou People 's Hospital, Wenzhou, 325000, People's Republic of China
2. Department of General Surgery, Taishun People's Hospital, Wenzhou, 325000, People's Republic of China
3. Department of Pharmacy, Xiangya Hospital, Central South University, Changsha 410078, People's Republic of China
4. Department of Pharmacy, the Second Xiangya Hospital, Central South University; Institute of Clinical Pharmacy, Central South University, Changsha 410011, People's Republic of China.
Received 2018-10-31; Accepted 2019-4-30; Published 2019-6-2
Abstract
Background: The role of glutathione s-transferase genes (GSTP1, GSTM1 and GSTT1) variants and the GSTP1 expression level on chemotherapy efficacy of gastrointestinal cancer (GIC) patients were inconsistent.
Methods: A meta-analysis about GSTP1, GSTM1 and GSTT1 variants and the GSTP1 expression level on chemotherapy efficacy of GIC patients was performed using data from PubMed, PMC, EMBASE, Web of Science, and Wanfang database.
Results: Our meta-analysis enrolled 50 publications including 6518 patients. We found that patients with GIC harboring GSTP1 (IIe105Val) Val locus had higher objective response rates (ORR) than the IIe/IIe genotypic patients (odds ratio (OR) = 1.580, 95% confidence interval (CI) = 1.159-2.154, P = 0.004). Significant associations were found between the Ile105Val variant and overall survival of Caucasian GIC patients (IIe/Val vs. IIe/IIe: OR = 0.797 (0.674-0.944), P = 0.009). Caucasian GIC patients and gastric cancer patients with GSTT1 null genotype had worse response rates compared to GSTT1 present patients (OR = 0.530 (0.356-0.789), P = 0.002; OR = 0.643 (0.463-0.895), P = 0.009, respectively).
Conclusion: This meta-analysis illustrates that GSTP1 IIe105Val and GSTT1 null/present variants could be useful predictors of chemotherapy efficacy in patients with gastrointestinal cancer.
Keywords: gastrointestinal cancer, glutathione S-transferases genes, variants, chemotherapy, efficacy
Introduction
Esophagus, stomach, small and large intestine, and rectum constitute the digestive tract, which is one of the important parts of the body [1]. Gastrointestinal cancer (GIC), including esophageal cancer (EAC), gastric cancer (GC) and colorectal cancer (CRC) represent a major public health problem worldwide [2]. Chemotherapy is widely used in many patients with postoperative recurrence or distant metastasis. [3, 4]. Platinum (oxaliplatin, cisplatin, carboplatin) combined with fluoropyrimidine (5-fluorouacil, capecitabine, S-1) is most commonly used in chemotherapy of GIC [3-7]. However, the effects of chemotherapy vary widely among GIC patients.
More evidence implied that the inter-individual variability of chemotherapy therapeutic efficacy in GIC patients was influenced by genetic factors including GSTP1, GSTT1, GSTM1, ERCC2, ERCC2, ABCC2, ENOSF1 and CD24 [8-13]. As a basic regiment to chemotherapy in GIC patients, platinum inhibits DNA synthesis and transcription [14].
Although resistance to chemotherapy is multifactorial, the metabolic enzymes of chemotherapeutic drugs play an important role in chemotherapy resistance. As a series of phase II metabolic enzymes, Glutathione S-transferases (GSTs) including GSTM1, GSTP1 and GSTT1 are involved in platinum detoxification [15, 16]. The effects of GSTM1/GSTT1 (null/present) and GSTP1 (rs1695, Ile105Val) genetic polymorphisms on chemotherapy efficacy in GIC patients were not consistent in previous research [9, 13, 17-26].
There were four meta-analyses for the efficacy of glutathione S-transferases (GSTs) variants and chemotherapy in patients with GC or CRC[27-30]. But updated publications were not analyzed in these meta-analyses, which may have biased conclusions. Moreover, there were no meta-analyses of the association between GSTP1 expression and the efficacy of chemotherapy in GC patients. There were no meta-analyses enrolled EAC patients and combined GC, CRC and EAC patients together in meta-analysis. Therefore, we have updated new literatures to investigate the associations between GSTP1 (Ile105Val), GSTM1 (null/present), GSTT1 (null/present) variants and GSTP1 expression and clinical outcomes in GIC patients.
Materials and Methods
Studies selection
All literatures in PubMed, PMC, EMBASE, Web of Science, and Wanfang database until 14th July 2017 were reviewed. Searching key words were “glutathione S-transferase pi 1 or GSTP1”, “glutathione S-transferase mu 1 or GSTM1”, “glutathione S-transferase theta 1 or GSTT1”, “gastrointestinal cancer or carcinoma or tumor”, “colorectal cancer or carcinoma or tumor”, “esophageal cancer or carcinoma or tumor”, “gastric cancer or carcinoma or tumor”, “SNPs or genetic polymorphisms or variations”, “expression” and “chemotherapy”. All literatures were reviewed by Dr. Jian Qu and Yuesheng Sun.
Inclusion and exclusion criteria
The inclusion criteria in our meta-analysis were as follows: (1) GIC patients including CRC, GC and EAC patients; (2) GSTP1 IIe105Val variant, GSTP1 expression condition (high and low), and GSTM1/GSTT1 (null /present) variants information; (3) at least having one clinical indicator (ORR, OS, TTP and PFS, ORs and HRs with corresponding to 95% CIs); (4) treatments with chemotherapy details. We excluded publications according: (1) duplicates and irrelevant studies; (2) no data for meta-analysis; (3) meta-analysis or basic research. All authors discussed literature selections that were enrolled in our meta-analysis.
Data processing and quality assessment
Two investigators extracted data independently. All authors discussed different opinions on data of each literature. Each literature data includes authors' names, the year of publication, the country, ethnicity, the number of patients, chemotherapy, median age (years), evaluation criteria, genotyping methods, quality score (QS), and outcomes (ORs or HRs and 95% CIs of ORR, OS, PFS, and TTP). The QS was evaluated independently by Dr. Qiang Qu and Jianghua Pan using previous methods [31]. Low quality publication was defined as QS ≤ 14 and high quality was QS >14.
Statistical analysis
Meta-analysis was analyzed by STATA version 12 (Stata Corp, College Station, TX, USA). Heterogeneity was analyzed by Cochrane's Q-statistic test and I2 test. If P < 0.05 and I2 > 50%, we defined it as significant heterogeneity, then Mantel-Haenszel random effect model was used in pooling ORs and HRs [32]. Z-test was used to analyze the pooled ORs or HRs and statistical significance was accepted if P < 0.05. Egger's test and Begg's test were used in publication bias and statistical significance was accepted if P < 0.05.
Results
Studies' characteristics and selection
Figure 1 presents the process of research selection. We found 2374 publications after duplicate removal from 5527 publications. We excluded 2324 publications including 1340 irrelevant studies, 39 meta-analyses, 18 case reports, 898 basic studies and 29 studies having no data. Fifty studies including 6518 patients were enrolled for further review. Among them, there were four studies involving in the GSTP1 expression and the chemotherapy efficacy in 264 GIC patients; forty-six studies were enrolled in the meta-analysis of glutathione S-transferase variants (GSTP1 Ile105Val, GSTM1/ GSTT1 null/present) and chemotherapy efficacy in 6254 GIC patients. Forty-six studies including 6254 patients were selected in investigation about GSTP1 Ile105Val; 2408 patients in 17 studies were selected in meta-analysis about GSTM1 null/present variant; and meta-analysis about GSTT1 null/present variant has 17 studies, including 2414 patients.
The characteristics of author name, the year of publication, the country, the ethnicity, the number of patients, chemotherapy, median age (year), evaluation criterion, genotyping methods, and QS were present in Table 1. Seventeen literatures were conducted on GC patients; 28 literatures were related with CRC patients; 4 literatures were involved in EAC patients and one study was conducted on adenocarcinoma of gastroesophageal junction patients. The detail information of objective response rate (ORR), overall survival (OS), median time to progression (TTP) and median progression-free survival (PFS) in each study is shown in Table S1 and Table S2.
ORR of GIC patients harboring GSTP1 Ile105Val variant
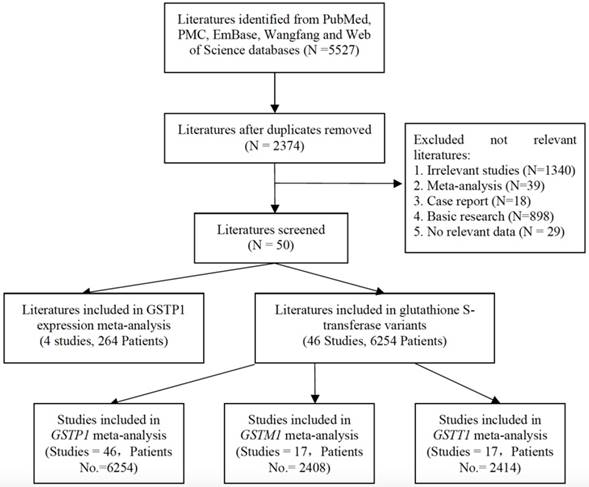
Thirty-one literatures with 3548 patients were selected for meta-analysis about ORR of GIC patients with GSTP1 Ile105Val variant. ORR of GIC patients harboring GSTP1 Ile105Val variants was different (Val carriers vs. IIe/IIe: OR=1.58(1.159-2.154), P=0.004). Tumor type-subgroup analyses found positive result in CRC patients (OR= 1.761(1.075-2.884), P=0.025). Subgroup analyses found association in Asian GIC patients (OR=1.567(1.058-2.319), P=0.025) (Table 2, Figure 2a, 2b). Subgroup analyses based on other index, such as evaluation criterion, chemotherapy, genotyping method, and quality score present in Table S3.
Compared with Asian GIC patients harboring GSTP1 Val105Val genotypic patients, IIe105IIe genotype or IIe carriers have lower ORR (OR=3.400(1.521-7.599), P=0.003; OR=3.466 (1.610-7.463), P=0.001, separately, Table 2). Moreover, different ORRs in GIC patients harboring different Ile105Val genotype were found (Val/Val vs. IIe carriers: OR= 2.256(1.297-3.926), P=0.004). Tumor type-subgroup analyses found the association in GC patients (OR=2.279(1.169-4.443), P=0.016, Figure 2c, 2d).
OS of GIC patients harboring GSTP1 Ile105Val variant
In order to pooling the HRs of the OS in GIC patients harboring different GSTP1 Ile105Val genotypes, we selected 21 literatures including 3509 patients. OS of Caucasian GIC patients were different between GSTP1 IIe/Val genotypic patients and IIe/IIe genotypic patients (HR=0.797(0.674-0.944), P=0.009, Figure 3a, Table 3).
Procedure of literature selection. Figure 1 present the procedure of literature selection from PubMed, PMC, EMBASE, Web of Science, and Wanfang database.
Basic information of publications enrolled in meta-analysis
| Author | Year | Country | Ethnicity | Tumor types | Study type | Patients No. | Chemotherapy | Median age (year) | Evaluation criterion | Outcomes | Genotyping method | Genes | QS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Meulendijks D[41] | 2016 | Mixed | Caucasian | GC | P | 185 | FU/platinum-based | 59 (27-77) | RECIST | ORR, PFS, OS | TaqMan assay, PCR-RFLP | GSTP1 | 17 |
| Liu R[17] | 2016 | China | Asian | GC | R | 108 | epirubicin/oxaliplatin/ FU | - | RECIST | ORR,PFS,OS | TaqMan assay | GSTP1, GSTT1 | 19 |
| Liang J[19] | 2010 | China | Asian | GC | R | 85 | FU/oxaliplatin | 55(32-77) | NA | TTP,OS | TaqMan assay | GSTP1 | 10 |
| Li QF[20] | 2010 | China | Asian | GC | R | 89 | FU/oxaliplatin | 55(32-77) | NA | TTP,OS | TaqMan assay | GSTP1 | 10 |
| Shim HJ[21] | 2010 | Korea | Asian | GC | R | 200 | taxane and cisplatin | 58 (19-76) | RECIST | OS,PFS,ORR | TaqMan assay, HRM | GSTP1,GSTM1.GSTT1 | 21 |
| Huang ZH[22] | 2009 | China | Asian | GC | R | 102 | FU/oxaliplatin | 58 (34-76) | WHO | OS | PCR-LDR | GSTP1, GSTM1 | 16 |
| Ott K[23] | 2008 | Germany | Caucasian | GC | R | 139 | cisplatin-based | 57 (47-67) | NA | OS,TR | TaqMan assay | GSTP1,GSTM1.GSTT1 | 11 |
| Goekkurt E[24] | 2006 | Germany | Caucasian | GC | R | 52 | FU/cisplatin/FA | 56(27-82) | RECIST | OS,TR | PCR-RFLP | GSTP1,GSTM1.GSTT1 | 9 |
| Seo BG[25] | 2009 | Korea | Asian | GC | R | 75 | FOLFOX | 56 (29-84) | RECIST | ORR | PCR-RFLP | GSTP1,GSTM1.GSTT1 | 12 |
| Goekkurt E[26] | 2009 | Germany | Caucasian | GC | P | 134 | platinum-based | 64 (27-86) | NA | OS,ORR | PCR-RFLP | GSTP1,GSTM1.GSTT1 | 17 |
| Ruzzo A[42] | 2006 | Italy | Caucasian | GC | R | 175 | fluorouracil/cisplatin | 61(38-79) | RECIST | OS,ORR,PFS | PCR-RFLP | GSTP1,GSTM1.GSTT1 | 16 |
| Keam B[43] | 2008 | Korea | Asian | GC | P | 73 | modified FOLFOX-6 | 59 (24-77) | WHO | OS,ORR | PCR-RFLP | GSTP1 | 15 |
| Ji M[13] | 2013 | China | Asian | GC | R | 59 | docetaxel, cisplatin, and 5-FU | 58.6(30-75) | NA | OS,ORR | PCR-LDR | GSTP1 | 10 |
| JI YU-ZHI[44] | 2011 | China | Asian | GC | R | 80 | oxaliplatin‑based | 52 (25-69) | RECIST | ORR | TaqMan assay | GSTP1 | 13 |
| Kap EJ[45] | 2014 | Germany | Caucasian | CRC | R | 176 | oxaliplatin | - | NA | OS | Fluorescence-basedmelting curve analysis | GSTP1,GSTM1.GSTT1 | 15 |
| Kumamoto K[46] | 2013 | Japan | Asian | CRC | R | 63 | modified FOLFOX-6 | 65(32‑84) | RECIST | ORR | PCR‑RFLP | GSTP1,GSTM1.GSTT1 | 21 |
| Lai CY[47] | 2013 | China | Asian | CRC | R | 491 | 5-FU-Based | 58.5±12.5 | NA | OS | PCR‑RFLP | GSTP1,GSTM1.GSTT1 | 16 |
| Bohanes P[48] | 2015 | USA | Caucasian | CRC | R | 746 | 5-FU | 61(19-86) | NA | OS | DNA sequencing / PCR-RFLP | GSTP1 | 15 |
| Nishina T[49] | 2013 | Japan | Asian | CRC | P | 68 | modified FOLFOX-6 + bevacizumab | 63(28-81) | RECIST | ORR | TaqMan | GSTP1 | 16 |
| Li HY[50] | 2012 | China | Asian | CRC | 335 | FOLFOX6 | 61.5±6.9 | NA | OS | TaqMan | GSTP1 | 18 | |
| Fariña Sarasqueta A(1)[51] | 2011 | the Netherlands | Caucasian | CRC | R | 50 | fluorouracil/oxaliplatin | 64(30-85) | NA | ORR | DNA sequencing/ PCR-RFLP | GSTP1,GSTM1.GSTT1 | 13 |
| Fariña Sarasqueta A(2)[51] | 2011 | the Netherlands | Caucasian | CRC | R | 42 | fluorouracil/leucovorin | 64(30-85) | NA | ORR | DNA sequencing/PCR-RFLP | GSTP1,GSTM1.GSTT1 | 13 |
| Páez D[52] | 2011 | Spain | Caucasian | CRC | R | 128 | 5-FU/capecitabine/oxaliplatin | 65(32-83) | NA | ORR | DNA sequencing | GSTP1 | 16 |
| Zarate R[53] | 2010 | Spain | Caucasian | CRC | R | 87 | oxaliplatin | 58(37-75) | RECIST | ORR/PFS | PCR-RFLP | GSTP1,GSTM1.GSTT1 | 18 |
| Jones BA[54] | 2009 | USA | Caucasian | CRC | R | 47 | 5-FU, levamisole, leucovorin, methotrexate | - | NA | OS | PCR-RFLP | GSTP1 | 12 |
| Le Morvan V(1)[55] | 2007 | France | Caucasian | CRC | R | 48 | TS inhibiter/irinotecan | 65(45-85) | WHO | ORR | PCR-RFLP | GSTP1 | 18 |
| Le Morvan V(2)[55] | 2007 | France | Caucasian | CRC | R | 59 | TS inhibiter/oxaliplatin | 62(41-86) | WHO | ORR | PCR-RFLP | GSTP1 | 18 |
| Stoehlmacher J[39] | 2004 | USA | Caucasian | CRC | R | 106 | 5-FU/oxaliplatin | 60 (24-84) | NA | TTP | PCR-RFLP | GSTP1,GSTM1.GSTT1 | 13 |
| Stoehlmacher J[56] | 2002 | USA | mainly Caucasian | CRC | R | 107 | 5-FU/oxaliplatin | 60 (24-83) | NA | OS | PCR-RFLP | GSTP1,GSTM1.GSTT1 | 13 |
| Chen Jian Guo[57] | 2016 | China | Asian | CRC | R | 60 | FOLFOX | - | WHO | ORR | PCR-RFLP | GSTP1 | 15 |
| Dong Ning-ning[58] | 2014 | China | Asian | CRC | R | 63 | FOLFIRI | 57 (29-75) | RECIST | ORR | Sequenom Mass ARRAY | GSTP1 | 16 |
| Ying bei-bei[59] | 2009 | China | Asian | CRC | R | 102 | FOLFOX-4 | - | RECIST | ORR/TTP | TaqMan-MGB | GSTP1 | 20 |
| Han-lei[60] | 2015 | China | Asian | CRC | R | 71 | modified FOLFOX6 | - | RECIST | ORR | Sequenom Mass Array | GSTP1 | 16 |
| Dongya Shen[61] | 2015 | China | Asian | CRC | R | 150 | FOLFOX | - | RECIST | ORR | HRM-SNP | GSTP1 | 18 |
| Ruzzo[62] | 2007 | Italy | Caucasian | CRC | R | 167 | FOLFOX-4 | 66(38-79) | NA | PFS | PCR-RFLP | GSTP1 | 15 |
| Liang Jun[63] | 2009 | China | Asian | CRC | R | 112 | 5-FU/oxaliplatin-based | 58 (34-80) | RECIST | ORR | TaqMan | GSTP1 | 20 |
| Joerger M[64] | 2015 | The Netherlands | Caucasian | CRC | P | 64 | capecitabine+oxaliplatin | 58.4(31.7-72.8) | RECIST | ORR,PFS,OS | DNA sequencing | GSTP1 | 22 |
| Boige V[65] | 2010 | France | Caucasian | CRC | P | 346 | LV5FU2/FOLFOX/FOLFIRI | 68(34-83) | WHO | PFS | PCR/qPCR | GSTP1,GSTM1.GSTT1 | 20 |
| Yen-Chung Chen[66] | 2009 | China | Asian | CRC | R | 166 | FOLFOX-4 | - | RECIST | ORR | PCR-RFLP | GSTP1 | 17 |
| Hong J[67] | 2011 | Korea | Asian | CRC | P | 52 | oxaliplatin + S-1 | 63 (37-76) | RECIST | ORR/PFS | PCR-RFLP | GSTP1 | 15 |
| Huang MY[68] | 2011 | China | Asian | CRC | R | 157 | FOLFOX-4 | 62.5 ± 10.9 | NA | PFS | PCR-RFLP/DNA sequencing | GSTP1 | 14 |
| Lamas MJ[69] | 2011 | Spain | Caucasian | CRC | R | 72 | 5-FU/oxaliplatin | 66.5 (32-80) | RECIST | ORR | Snapshot | GSTP1 | 12 |
| Rumiato E[70] | 2013 | Italy | Caucasian | EAC | R | 63 | cisplatin/5-FU-based | 62(25-80) | RECIST | ORR | (ARMS)-PCR,PCR-RFLP | GSTP1,GSTM1.GSTT1 | 15 |
| Wang Y[71] | 2011 | China | Asian | EAC | R | 256 | cisplatin-based | - | RECIST | ORR | DNA sequencing | GSTP1 | 16 |
| Gui Yan[72] | 2016 | China | Asian | EAC | R | 168 | 5-FU and cisplatin | - | RECIST | ORR,OS | DNA sequencing | GSTP1 | 17 |
| Joerger M[64] | 2015 | The Netherlands | Caucasian | EAC | R | 76 | capecitabine+cisplatin+epirubicin | 57.2(35-75.3) | RECIST | ORR,OS | DNA sequencing | GSTP1 | 22 |
| Kwon HC[73] | 2007 | Korea | Asian | GC | R | 64 | 5-FU/oxaliplatin | 51(31-74) | RECIST | expression | immunohistochemistry | GSTP1 | 18 |
| Boku N[74] | 2007 | Japan | Asian | GC | R | 66 | 5-FU/cisplatin | 63 (19-75) | WHO | expression | immunohistochemistry | GSTP1 | 20 |
| In Sil Choi[75] | 2011 | Korea | Asian | GC | R | 41 | S-1 plus cisplatin | 62 (33-73) | RECIST | expression | immunohistochemistry | GSTP1 | 16 |
| Li S[76] | 2017 | China | Asian | AGEJ | R | 93 | oxaliplatin based | 66.6(40-76) | RECIST | expression | immunohistochemistry | GSTP1 | 18 |
NR: not reported; QS, quality score; HR: hazard ratio; ORR: objective response rate; OS, overall survival; PFS, progression-free survival; MST, median survival time; TTP, time to progression; PCR, polymerase chain reaction; PCR-RFLP, PCR-restriction fragment length polymorphism; RECIST, Response Evaluation Criteria in Solid Tumors; WHO, World Health Organization; PCR-LDR, PCR-ligase detection reaction; PCR-CTPP, duplex PCR with the confronting-two-pair primer; HRM, high resolution melt; 5-FU, 5-fluorouracil; FOLFOX, leucovorin+5-fluorouracil+oxaliplatin; TS, thymidylate synthase; FOLFIRI, leucovorin+5-fluorouracil+irinotecan; LV5FU2, leucovorin+5-fluorouracil; EAC , esophageal cancer; GC, gastric cancer; CRC , colorectal cancer; AGEJ, Adenocarcinoma of Gastroesophageal Junction. P, prospective study; R, retrospective study.
The pooling ORs of ORR in GIC patients with different GSTP1 IIe105Val, GSTM1/GSTT1 variants and GSTP1 expression levels.
| Genetic comparisons | No. of studies | Study groups | Test of association | Test of heterogeneity | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR/HR (95% CI) | Z | P-value | Model | χ2 | P-value | I2 (%) | Tau-squared | |||
| GSTP1 | ||||||||||
| Val carriers vs. IIe/IIe | 31 | Overall | 1.580(1.159-2.154) | 2.9 | 0.004 | R | 102.39 | <0.001 | 70.70% | 0.5184 |
| 11 | GC | 1.598(0.998-2.560) | 1.95 | 0.051 | R | 31.75 | <0.001 | 68.50% | 0.4165 | |
| 16 | CRC | 1.761(1.075-2.884) | 2.25 | 0.025 | R | 53.12 | <0.001 | 71.80% | 0.702 | |
| 4 | EAC | 1.080(0.482-2.418) | 0.19 | 0.852 | R | 13.16 | 0.004 | 77.20% | 0.5119 | |
| 20 | Asian | 1.567(1.058-2.319) | 2.24 | 0.025 | R | 69.3 | <0.001 | 72.60% | 0.554 | |
| 11 | Caucasian | 1.607(0.944-2.736) | 1.75 | 0.08 | R | 33.1 | <0.001 | 69.80% | 0.5374 | |
| Val/Val vs. IIe/IIe | 14 | Overall | 2.265(0.937-5.475) | 1.82 | 0.069 | R | 52.01 | <0.001 | 75.00% | 1.9912 |
| 8 | GC | 1.982(0.599-6.563) | 1.12 | 0.263 | R | 36.26 | <0.001 | 80.70% | 2.2709 | |
| 4 | CRC | 3.151(0.315-31.545) | 0.98 | 0.329 | R | 13.75 | 0.003 | 78.20% | 4.248 | |
| 2 | EAC | 2.480(0.784-7.842) | 1.55 | 0.122 | F | 1.5 | 0.22 | 33.50% | - | |
| 5 | Asian | 3.400(1.521-7.599) | 2.98 | 0.003 | F | 8.11 | 0.088 | 50.70% | - | |
| 9 | Caucasian | 2.059(0.661-6.412) | 1.25 | 0.213 | R | 41.28 | <0.001 | 80.60% | 2.3382 | |
| IIe/Val vs. IIe/IIe | 14 | Overall | 1.130(0.607-2.103) | 0.38 | 0.701 | R | 69.89 | <0.001 | 81.40% | 1.0835 |
| 8 | GC | 0.825(0.347-1.961) | 0.44 | 0.663 | R | 48.96 | <0.001 | 85.70% | 1.2721 | |
| 4 | CRC | 2.780(0.710-10.892) | 1.47 | 0.142 | R | 14.58 | 0.002 | 79.40% | 1.51 | |
| 2 | EAC | 0.845(0.415-1.721) | 0.46 | 0.642 | F | 2.44 | 0.118 | 59.10% | - | |
| 5 | Asian | 1.270(0.836-1.929) | 1.12 | 0.263 | F | 8.23 | 0.083 | 51.40% | - | |
| 9 | Caucasian | 1.061(0.420-2.677) | 0.12 | 0.901 | R | 61.01 | <0.001 | 86.90% | 1.6738 | |
| Val/Val vs. IIe carriers | 15 | Overall | 2.256(1.297-3.926) | 2.88 | 0.004 | R | 29.71 | 0.008 | 52.90% | 0.5742 |
| 8 | GC | 2.279(1.169-4.443) | 2.42 | 0.016 | R | 18.72 | 0.016 | 57.30% | 0.5481 | |
| 4 | CRC | 1.991(0.325-12.197) | 0.74 | 0.457 | R | 10.1 | 0.018 | 70.30% | 2.3401 | |
| 2 | EAC | 2.715(0.890-8.285) | 1.76 | 0.079 | F | 0.68 | 0.409 | 0.00% | - | |
| 5 | Asian | 3.466(1.610-7.463) | 3.18 | 0.001 | F | 7.9 | 0.095 | 49.30% | - | |
| 9 | Caucasian | 2.011(1.324-3.052) | 3.28 | 0.026 | R | 19.65 | 0.02 | 54.20% | 0.5014 | |
| GSTT1 Null vs. Present | 10 | Overall | 0.657(0.489-0.883) | 2.78 | 0.005 | F | 5.86 | 0.753 | 0.00% | - |
| 7 | GC | 0.643(0.463-0.895) | 2.62 | 0.009 | F | 3.57 | 0.735 | 0.00% | - | |
| 2 | CRC | 1.014(0.447-2.302) | 0.03 | 0.973 | F | 0.17 | 0.68 | 0.00% | - | |
| 3 | Asian | 0.873(0.557-1.368) | 0.59 | 0.553 | F | 0.29 | 0.865 | 0.00% | - | |
| 7 | Caucasian | 0.530(0.356-0.789) | 3.13 | 0.002 | F | 2.96 | 0.814 | 0.00% | - | |
| GSTM1 Null vs. Present | 10 | Overall | 1.120(0.872-1.440) | 0.89 | 0.375 | F | 6.29 | 0.71 | 0.00% | - |
| 7 | GC | 1.209(0.918-1.593) | 1.35 | 0.177 | F | 3.07 | 0.8 | 0.00% | - | |
| 2 | CRC | 0.772(0.351-1.701) | 0.64 | 0.521 | F | 1.58 | 0.209 | 36.50% | - | |
| 3 | Asian | 1.067(0.669-1.700) | 0.27 | 0.786 | F | 0.07 | 0.966 | 0.00% | - | |
| 7 | Caucasian | 1.143(0.849-1.540) | 0.88 | 0.379 | F | 6.15 | 0.406 | 2.50% | - | |
| GSTP1 expression | 4 | Overall | 0.854(0.527-1.384) | 0.64 | 0.521 | F | 7.5 | 5.70% | 60.00% | - |
| 3 | GC | 0.671(0.369-1.221) | 1.31 | 0.191 | F | 5.74 | 5.70% | 65.20% | - | |
OR, odds ratio; HR: hazard ratio; CI, confidence interval; vs., versus; F, fixed effect model; R, random effect model.
Forest plots of ORR in GIC patients with GSTP1 IIe105Val variants (Val carriers vs. IIe/IIe, Val/Val vs. IIe carriers models). ORs and 95% CI of ORR stratified by (a) tumor types in Val carriers vs. IIe/IIe model; (b) ethnicity in Val carriers vs. IIe/IIe; (c) tumor types in Val/Val vs. IIe carriers model; (d) ethnicity in Val/Val vs. IIe carriers model.
The pooling HRs of OS, PFS and TTP in GIC patients with different GSTP1 IIe105Val and GSTM1/GSTT1 variants.
| Genetic comparisons | No. of studies | Study groups | Test of association | Test of heterogeneity | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR/HR(95% CI) | Z | P-value | Models | χ2 | P-value | I2 (%) | Tau-squared | |||
| GSTP1-OS | ||||||||||
| Val carriers vs. IIe/IIe | 10 | Overall | 0.903(0.613-1.332) | 0.51 | 0.608 | R | 48.2 | <0.001 | 81.30% | 0.3021 |
| 5 | GC | 0.797(0.437-1.455) | 0.74 | 0.461 | R | 24.64 | <0.001 | 83.80% | 0.3891 | |
| 4 | CRC | 0.967(0.515-1.819) | 0.1 | 0.918 | R | 14.89 | 0.002 | 79.90% | 0.3181 | |
| 7 | Asian | 0.975(0.606-1.569) | 0.11 | 0.916 | R | 41.13 | <0.001 | 85.40% | 0.3385 | |
| 3 | Caucasian | 0.738(0.347-1.570) | 0.79 | 0.43 | R | 6.56 | 0.038 | 69.50% | 0.3093 | |
| IIe/Val vs. IIe/IIe | 13 | Overall | 0.900(0.794-1.020) | 1.65 | 0.098 | F | 18.31 | 0.107 | 34.50% | - |
| 11 | CRC | 0.878(0.763-1.010) | 1.83 | 0.068 | F | 16.37 | 0.089 | 38.90% | - | |
| 3 | Asian | 1.044(0.865-1.259) | 0.45 | 0.655 | F | 1.34 | 0.512 | 0.00% | - | |
| 10 | Caucasian | 0.797(0.674-0.944) | 2.63 | 0.009 | F | 12.59 | 0.182 | 28.50% | - | |
| Val/Val vs. IIe/IIe | 13 | Overall | 0.646(0.398-1.046) | 1.78 | 0.076 | R | 43.88 | <0.001 | 72.70% | 0.5021 |
| 11 | CRC | 0.619(0.342-1.120) | 1.59 | 0.113 | R | 43.59 | <0.001 | 77.10% | 0.6777 | |
| 3 | Asian | 0.774(0.495-1.210) | 1.13 | 0.216 | F | 2.86 | 0.239 | 30% | - | |
| 10 | Caucasian | 0.605(0.315-1.164) | 1.51 | 0.132 | R | 40.86 | <0.001 | 78.00% | 0.7601 | |
| GSTP1-PFS | ||||||||||
| Val carriers vs. IIe/IIe | 4 | Overall | 0.855(0.410-1.781) | 0.42 | 0.675 | R | 17.22 | 0.001 | 82.60% | 0.4565 |
| 2 | GC | 1.509(1.059-2.150) | 2.28 | 0.023 | F | 1.7 | 0.192 | 41.30% | - | |
| 2 | CRC | 0.420(0.247-0.715) | 3.19 | 0.001 | F | 0.13 | 0.714 | 0.00% | - | |
| 3 | Asian | 1.115(0.552-2.253) | 0.3 | 0.762 | R | 8.54 | 0.014 | 76.60% | 0.2913 | |
| IIe/Val vs. IIe/IIe | 5 | Overall | 0.990(0.827-1.185) | 0.11 | 0.911 | F | 3.97 | 0.41 | 0.00% | - |
| 4 | CRC | 0.978(0.790-1.210) | 0.21 | 0.835 | F | 3.92 | 0.27 | 23.50% | - | |
| 2 | Asian | 0.996(0.719-1.379) | 0.03 | 0.979 | F | 0.47 | 0.493 | 0.00% | - | |
| 3 | Caucasian | 0.987(0.796-1.225) | 0.12 | 0.907 | F | 3.49 | 0.174 | 42.80% | - | |
| Val/Val vs. IIe/IIe | 5 | Overall | 0.709(0.384-1.308) | 1.1 | 0.27 | R | 16.5 | 0.002 | 75.80% | 0.3262 |
| 4 | CRC | 0.664(0.306-1.441) | 1.04 | 0.3 | R | 16.49 | 0.001 | 81.80% | 0.4454 | |
| 2 | Asian | 0.759(0.367-1.570) | 0.74 | 0.457 | F | 0.55 | 0.459 | 0.00% | - | |
| 3 | Caucasian | 0.707(0.306-1.638) | 0.81 | 0.419 | R | 15.79 | <0.001 | 87.30% | 0.4745 | |
| GSTP1-TTP | ||||||||||
| Val carriers vs. IIe/IIe | 4 | Overall | 0.961(0.356-2.591) | 0.08 | 0.937 | R | 43.02 | <0.001 | 93.00% | 0.9525 |
| 3 | GC | 0.628(0.310-1.274) | 1.29 | 0.198 | R | 10.03 | 0.007 | 80.10% | 0.3121 | |
| 4 | Asian | 0.961(0.356-2.591) | 0.08 | 0.937 | R | 43.02 | <0.001 | 93.00% | 0.9525 | |
| GSTT1-PFS | 5 | Overall | 1.102(0.918-1.322) | 1.04 | 0.299 | F | 1.42 | 0.841 | 0.00% | - |
| 3 | GC | 1.017(0.807-1.282) | 0.14 | 0.885 | F | 0.2 | 0.904 | 0.00% | - | |
| 2 | CRC | 1.257(0.934-1.692) | 1.51 | 0.132 | F | 0 | 0.963 | 0.00% | - | |
| 4 | Caucasian | 1.178(0.937-1.480) | 1.4 | 0.161 | F | 0.52 | 0.915 | 0 | - | |
| GSTT1-OS | 8 | Overall | 1.104(0.889-1.370) | 0.89 | 0.371 | R | 15.36 | 0.032 | 54.40% | 0.0482 |
| 4 | GC | 1.136(0.689-1.872) | 0.5 | 0.618 | R | 8.83 | 0.012 | 77.40% | 0.1492 | |
| 4 | CRC | 0.998(0.844-1.180) | 0.03 | 0.979 | F | 4.32 | 0.229 | 30.50% | - | |
| GSTM1-PFS | 5 | Overall | 0.957(0.823-1.114) | 0.57 | 0.572 | F | 1.96 | 0.743 | 0.00% | - |
| 3 | GC | 1.034(0.838-1.275) | 0.31 | 0.755 | F | 0.88 | 0.645 | 0.00% | - | |
| 2 | CRC | 0.880(0.707-1.095) | 1.14 | 0.253 | F | 0 | 1 | 0.00% | - | |
| 2 | Asian | 1.054(0.810-1.371) | 0.39 | 0.695 | F | 0.82 | 0.365 | 0.00% | - | |
| 3 | Caucasian | 0.913(0.758-1.098) | 0.97 | 0.333 | F | 0.37 | 0.83 | 0.00% | - | |
| GSTM1-OS | 7 | Overall | 1.001(0.862-1.163) | 0.01 | 0.992 | F | 5.74 | 0.453 | 0.00% | - |
| 3 | GC | 1.103(0.889-1.368) | 0.89 | 0.372 | F | 1.14 | 0.565 | 0.00% | - | |
| 3 | CRC | 0.900(0.722-1.121) | 0.94 | 0.346 | F | 2.89 | 0.236 | 30.80% | - | |
| 2 | Asian | 1.174(0.891-1.545) | 1.14 | 0.254 | F | 0.64 | 0.424 | 0.00% | - | |
| 5 | Caucasian | 0.936(0.783-1.119) | 0.73 | 0.466 | F | 3.27 | 0.514 | 0.00% | - | |
OR, odds ratio; HR: hazard ratio; CI, confidence interval; vs., versus; F, fixed effect model; R, random effect model.
PFS and TTP of GIC patients harboring GSTP1 Ile105Val genotypes
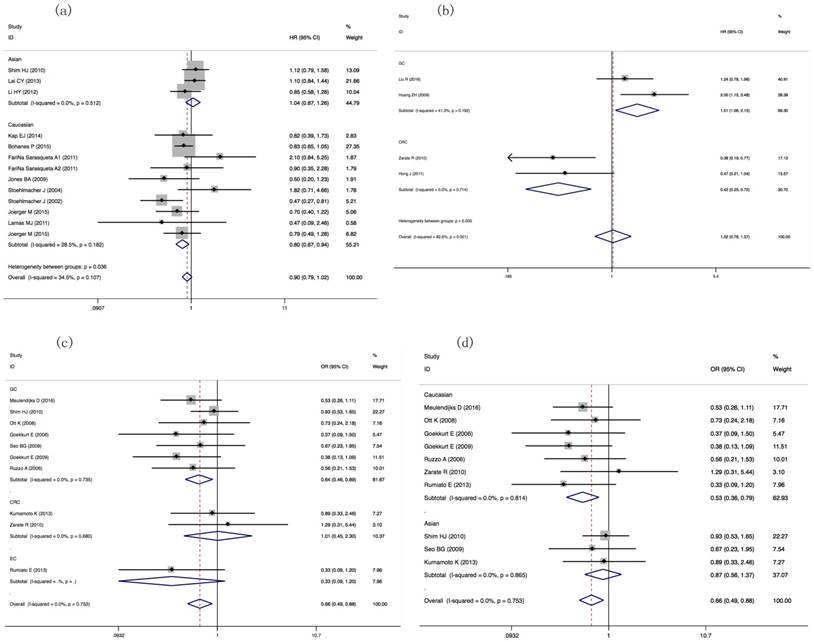
For comparing the PFS, we selected 9 studies having 1378 patients in our meta-analysis. The HRs of PFS in GC or CRC patients harboring different Ile105Val genotypes was different (Val carriers vs. IIe/IIe: HR= 1.509(1.059-2.150), P=0.023; HR= 0.420(0.247-0.715), P=0.001, respectively, Table 3, Figure 3b).
In order to compare TTP in GIC patients with different GSTP1 Ile105Val variants, four publications including 349 patients were enrolled and found no association between Val carriers and IIe/IIe patients (HR= 0.961(0.356-2.591), P=0.937). Tumor types or ethnicity subgroup analyses were also negative results (for GC patients: HR= 0.628(0.310-1.274), P=0.198; for Asian patients: HR= 0.961(0.356-2.591), P=0.937, Table 3).
ORR of GIC patients harboring GSTM1/GSTT1 (null/present) variants
For comparing ORR in GSTT1 null/present patients, 10 publications including 1104 patients' data showed positive result between GSTT1 null and present patients (OR= 0.657(0.489-0.883), P=0.005). Ethnicity- and tumor type-subgroup analyses suggested that, for the Caucasian group, GSTT1 null/present was associated with ORR (OR= 0.530(0.356-0.789), P=0.002); for GC patients, GSTT1 null/present was associated with ORR (OR=0.643(0.463-0.895), P=0.009) (Table 2, Figure 3c, 3d).
Our meta-analysis about comparing the ORRs in GSTM1 null/present patients included 10 literatures with 1102 patients. No association was found between patients harboring GSTM1 null/present variant and ORRs in GIC patients (OR=1.120 (0.872-1.440), P= 0.375). Ethnicity-subgroup and tumor type-subgroup analyses also suggested the negative results (Table 2).
OS and PFS of GIC patients harboring GSTM1/GSTT1 (null/present) variants
We compared the pooling OS in GSTM1 null/present genotypic patients using 7 publications including 892 patients' data, no statistically significant associations were found in GSTM1 null/present genotypic patients (HR= 1.001(0.862-1.163), P=0.992). No significant difference of OS was found between different GSTM1 null/present variant patients (Table 3).
Forest plots of ORR, OS and PFS in GIC patients with GSTP1 IIe105Val and GSTT1 null/present variants. (a) HRs and 95%CI of OS stratified by ethnicity in GSTP1 IIe105Val IIe/Val vs. IIe/IIe model; (b) HRs and 95%CI of PFS stratified by tumor types in GSTP1 IIe105Val Val carriers vs. IIe/IIe model; (c) ORs and 95%CI of ORR stratified by tumor types in GSTT1 null/present variant; (d) ORs and 95%CI of ORR stratified by ethnicity in GSTT1 null/present variant.
We compared the pooling PFS in GSTM1 null/present genotypic patients using five publications including 996 patients' data and found no significant association (HR= 0.957(0.823-1.114), P=0.572). Ethnicity- and tumor type-subgroup tests also showed negative results (Table 3).
We compared the pooled OS and PFS in GSTT1 null/present genotypic patients using 8 publications including 1366 patients' data and found no significant associations (null vs. present: HR= 1.104(0.889-1.370), P=0.371; HR= 1.102(0.918-1.322), P=0.299, respectively). Ethnicity- and tumor type-subgroup tests also showed negative results (Table 3).
ORR and expression level of GSTP1 in GIC patients
No association was found between GSTP1 expression level and ORR after we pooling data from four publications including 264 patients (low expression vs. high expression: OR= 0.854 (0.527-1.384), P=0.64). After tumor type-subgroup analysis, GSTP1 expression level and ORR in GC patients were negative results (low expression vs. high expression: OR= 0.671 (0.369-1.221), P=0.191, Table 2).
Sensitivity analysis
Sensitivity analysis found that the OR and HR of every enrolled study didn't influence the final significant associations between GSTP1, GSTM1 and GSTT1 variants and chemotherapy efficacy in GIC patients (Figure S1), except the relationship between GSTP1 (IIe105Val) Val carriers vs. IIe/IIe model and the pooled HRs of PFS, which was just two literatures enrolled in tumor type-subgroup analysis.
Moreover, changing the effect models could change the significant association to negative results about GSTP1 Val/Val vs. IIe/IIe and Val/Val vs. IIe carriers models and the ORs of ORR in GIC Asian patients (Figure S2). It implies that the associations between GSTP1 Val/Val vs. IIe/IIe, Val/Val vs. IIe carriers and the ORR of GIC Asian patients were not robust.
Publication bias
Egger's test and Begg's test was used in Publication bias. As shown in Figure S3, Begg's and Egger's funnel plots found no publications bias under GSTP1 (IIe105Val) any genetic models (all P>0.05, Figure S4), and under GSTT1/GSTM1 (null/present) variants (GSTT1: P=0.788, P=0.247, GSTM1: P=0.421, P=0.272, respectively, Figure S5).
Begg's and Egger's tests showed no publication bias in pooling OS analysis among Caucasian genotypic GIC patients under GSTP1 (IIe105Val) IIe/Val vs. IIe/IIe model (P=0.421, P=0.724, respectively, Figure S6) and in pooling PFS analysis among GC patients or CRC patients under Val carriers vs. IIe/IIe model (P=0.317, P=0.317, respectively, Figure S6).
Discussion
We performed a meta-analysis to investigate the association between glutathione S-transferase gene (GSTP1 (Ile105Val), GSTM1/GSTT1 (null/present) variants and GSTP1 expression and clinical outcomes in patients with GIC. The results showed that Asian GIC patients with GSTP1 (IIe105Val) Val carriers had better anticancer efficacy than IIe/IIe patients. Caucasian GIC patients carrying the GSTP1 Val/Val genotype, especially those with stomach disease, have better chemotherapy efficacy than patients with IIe carriers. Caucasian GIC patients bearing IIe/Val genotype have longer survival time than patients with IIe/IIe genotype. Caucasian GIC patients or gastric patients having GSTT1 present genotype have higher ORR compared to GSTT1 null genotypic patients. While GSTM1 present/null variant and the expression level of GSTP1 were not associated with the chemotherapy efficacy to GIC patients. We found that GSTP1 IIe105Val and GSTT1 null/present polymorphisms could predict chemotherapy efficacy in GIC patients. Based on the individual genetic profile, the oncologists will have new possibilities to make treatment decisions for their patients, to predictive efficacy of chemotherapy and to redefine scheduling and dosage.
Platinum (oxaliplatin, cisplatin, carboplatin) combined with fluoropyrimidines (5-fluorouacil, capecitabine, S-1) was most commonly used in chemotherapy of gastrointestinal cancers[6, 33, 34]. However, the chemotherapy efficacy of GIC patients is different. Previous studies suggested that variants in GSTP1, GSTM1, GSTT1, XPCC1, MTHFR, TYMS and ABCC2 influence the chemotherapy efficacy in GC or/and CRC patients [5, 6, 12, 34-36]. GSTP1, GSTM1 and GSTT1 belong to human glutathione S-transferases super family members and are involved in the inactivation of chemotherapeutic drugs such as platinum through the glutathione metabolic pathway [9, 28, 37]. GSTP1 IIe105Val and GSTM1/GSTT1 (null/present) polymorphisms decrease enzyme activity, resulting in the lower intracellular concentration of drugs such as cisplatin [29, 38-40]. Therefore, patients harboring GSTP1 IIe105Val mutant variants may reduce the ability to detoxify drug metabolites, and then have better chemotherapy efficacy.
Our meta-analysis showed that the GSTP1 IIe105Val variant was associated with ORR of GIC patients (Table 2, Figure 2a, 2b). Heterogeneity is an important problem in meta-analysis. We also carried out heterogeneity analysis and we found significant heterogeneity when pooling the ORs of ORR in different GSTP1 IIe105Val variant patients, so we used the Mantel-Haenszel random model to analyze the associations. Changing effect models (Mantel-Haenszel random model and fixed model) didn't change the final results. Moreover, sensitivity analysis results found that excluded any studies has no impact on the overall effective size in GIC patients (Figure S1). GIC is a series of complex cancer diseases. Tumor subtypes, patients' ethnicity, different chemotherapy regimens and clinical stage may affect the anti-cancer efficacy in GIC patients. Moreover, the evaluation criterion, genotyping method or quality assessment of literatures may also affect the heterogeneity of meta-analysis. Herein, we performed subgroup-analysis according to different evaluation criterion, genotyping methods and quality assessment of literatures. The subgroup-analysis also supported the significant association between GSTP1 IIe105Val and chemotherapy efficacy in GC and EC patients, or Caucasian GIC patients under dominant genetic model (Table 2, Table S3). It implies that tumor type and ethnicity may contribute to the associations. The study-type subgroup analysis showed prospective study groups have no difference between GSTP1 Val carriers vs. IIe/IIe and ORR of chemotherapy in GIC patients. But there were significant difference on GSTP1 Val carriers vs. IIe/IIe and ORR of chemotherapy in GIC patients based on retrospective study.
Compared with Asian GIC patients harboring GSTP1 IIe105IIe genotypes, Val105Val genotypic patients have better response rates to chemotherapy under the fixed model (OR=3.400(1.521-7.599), P=0.003, Table 2). However, the random model analysis showed no significant (Figure S2a), which implied that the positive result was unstable and we could not draw a robust conclusion.
Compared with GIC patients harboring GSTP1 IIe carriers, Val105Val genotypic patients have better chemotherapy efficacy (Figure 2). While changing the analytical models could change the significant association of GSTP1 variants (Val/Val vs. IIe carriers) and ORR in Asian GIC patients (Figure S2). Sensitivity analysis confirmed the positive results (Figure S1). Tumor type-subgroup analysis showed that, compared with IIe carriers variants, GC patients harboring Val/Val variant, not CRC patients, have better chemotherapy efficacy (Figure 2c, 2d). Therefore, tumor types and ethnicity both influence the meta-analysis results about GSTP1 variants (Val/Val vs. IIe carriers) and ORR of GIC patients.
We also found that GSTP1 (Ile105Val) IIe/Val patients had longer survival time than wild-type patients. Exclusion studies did not influence the pooling HR of OS in Caucasian GIC patients. Although significant associations were found between GSTP1 (Val carriers vs. IIe/IIe) and the HRs of PFS in GC or CRC patients, the enrolled studies were two, which could not draw the robust conclusion.
It is the first meta-analysis to investigate the relationship between GSTP1 expression level and anti-cancer efficacy in GIC patients. There were four literatures enrolled for this meta-analysis and we did not find any significant associations. Further updated meta-analysis should be done to confirm our negative results.
Our meta-analysis suggests that GIC patients harboring GSTT1 present genotype have better chemotherapy efficacy compared to Caucasian patients harboring GSTT1 null genotype, but not in Asian patients (Table 2, Figure 3c, 3d). Subgroup analysis with tumor types showed GSTT1 null/present variant associated with ORR in GC patients (OR= 0.643(0.463-0.895), P=0.009, Table 2, Figure 3c, 3d). There was no significant association between GSTM1 and chemotherapy efficacy. These varies results may be attributed to differences in the distribution of GST families and enzymatic activity of drug detoxification. Our results were consistent with previous meta-analyses, which was just enrolled seven literatures about the GSTT1 null/present variant and the ORs of ORR in GC patients [28].
Heterogeneity and publication bias are important parts of meta-analysis. In order to draw a robust and confidential conclusion, heterogeneity analyzed by Q test and I2 statistics; publication bias analyzed by Egger's test and Begg's test; sensitivity analysis and subgroup analysis were also performed. There were heterogeneities when we pooled ORs or HRs of ORR, OS, PFS and TTP in patients harboring GSTP1 IIe105Val different variants (Table 2, Table 3). Therefore, we used fixed and random models to confirm the positive results. We also used subgroup analysis by other index to find the source of heterogeneity. However, there were still heterogeneities after subgroup-analysis (Table S3). Publication bias test also showed no publication bias (Figure S3- S5).
Previously, there were four meta-analyses involved in Glutathione S-transferases genes' variants and chemotherapy efficacy in CRC or GC patients [27, 28, 30, 36]. No meta-analysis is involved in the association between GSTP1 expression and chemotherapy efficacy in GC patients. No meta-analysis is enrolled with esophageal cancer patients and is combined GC, CRC and EC patients together in analysis. Moreover, previous meta-analyses' results are conflicting. Therefore, we systematically analyzed all available literatures related to GSTP1 expression levels and GSTP1 IIe105Val, GSTM1/GSTT1 deletion variants and chemotherapy efficacy in GIC patients.
There were several limitations about our meta-analysis. First, sample sizes and enrolled studies are still limited. Second, therapeutic indexes such as TTP or PFS were abandoned for analysis. Third, although subgroup analysis, sensitivity analysis, and publication bias were carried out to find the source of heterogeneity, there was still heterogeneity in pooled analysis for GSTP1. Fourth, among 50 publications enrolled in the meta-analysis, there were only seven prospective studies. After subgroup-analysis according to tumor type and genetic model, there were less prospective studies to carry out pooling ORs/HRs, or there were publication bias and heterogeneity.
Conclusion
In conclusion, we carried out the meta-analysis including 50 publications with 6518 gastrointestinal cancer patients. We found that GSTP1 IIe105Val and GSTT1 deletion variants were associated with chemotherapy efficacy in gastrointestinal cancer patients. A larger sample of further research is needed in different ethnic populations to confirm our conclusions.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We thank the supported grants of the National Natural Scientific Foundation of China (No. 81503166, 81603208), and the Science and Technology Foundation of Wenzhou, China (Grant No. Y20170448).
Author Contributions
J.Q. and Q.Q designed the study. Y.-S.S, J.Q. and Q.Q wrote the main manuscript text, J.-H.P., X.-C.T. and E.-D.C. performed figures and tables, W.-X.Y. and M.-P.W. analyzed data, all authors reviewed the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Prasad S, Tyagi AK. Ginger and its constituents: role in prevention and treatment of gastrointestinal cancer. Gastroenterol Res Pract. 2015;2015:142979
2. Procaccio L, Schirripa M, Fassan M. et al. Immunotherapy in Gastrointestinal Cancers. Biomed Res Int. 2017;2017:4346576
3. Sasaki Y, Iwasa S, Okazaki S. et al. A phase II study of combination therapy with oral S-1 and cisplatin in elderly patients with advanced gastric cancer. Gastric Cancer. 2017;21:439-45
4. Satake H, Miki A, Kondo M. et al. Phase I study of neoadjuvant chemotherapy with S-1 and oxaliplatin for locally advanced gastric cancer (Neo G-SOX PI). ESMO Open. 2017;2:e000130
5. Kap EJ, Popanda O, Chang-Claude J. Nucleotide excision repair and response and survival to chemotherapy in colorectal cancer patients. Pharmacogenomics. 2016;17:755-94
6. Mizrak Kaya D, Harada K, Shimodaira Y. et al. Advanced gastric adenocarcinoma: optimizing therapy options. Expert Rev Clin Pharmacol. 2017;10:263-71
7. Hagiwara Y, Ohashi Y, Okusaka T. et al. Health-related quality of life in a randomised phase III study of gemcitabine plus S-1, S-1 alone and gemcitabine alone for locally advanced or metastatic pancreatic cancer: GEST study. ESMO Open. 2017;2:e000151
8. Kjersem JB, Thomsen M, Guren T. et al. AGXT and ERCC2 polymorphisms are associated with clinical outcome in metastatic colorectal cancer patients treated with 5-FU/oxaliplatin. Pharmacogenomics J. 2016;16:272-9
9. Meulendijks D, Rozeman EA, Cats A. et al. Pharmacogenetic variants associated with outcome in patients with advanced gastric cancer treated with fluoropyrimidine and platinum-based triplet combinations: a pooled analysis of three prospective studies. Pharmacogenomics J. 2017;17:441-51
10. Jia ZF, Wang LZ, Cao XY. et al. CD24 genetic variants contribute to overall survival in patients with gastric cancer. World J Gastroenterol. 2016;22:2373-82
11. Li Y, Liu Z, Liu H. et al. ERCC1 and ERCC2 variants predict survival in gastric cancer patients. PLoS One. 2013;8:e71994
12. Li Z, Xing X, Shan F. et al. ABCC2-24C > T polymorphism is associated with the response to platinum/5-Fu-based neoadjuvant chemotherapy and better clinical outcomes in advanced gastric cancer patients. Oncotarget. 2016;7:55449-57
13. Ji M, Xu B, Jiang JT. et al. Relationship between glutathione S-transferase P1 (GSTP1), X-ray repair cross complementing group 1 (XRCC1) and 5,10-methylenetetrahydrofolate reductase (5,10-MTHFR) gene polymorphisms and response to chemotherapy in advanced gastric cancer. Onkologie. 2013;36:335-40
14. Zhou F, Yu Z, Jiang T. et al. Genetic polymorphisms of GSTP1 and XRCC1: prediction of clinical outcome of platinum-based chemotherapy in advanced non-small cell lung cancer (NSCLC) patients. Swiss Med Wkly. 2011;141:w13275
15. Yang Y, Xian L. The association between the GSTP1 A313G and GSTM1 null/present polymorphisms and the treatment response of the platinum-based chemotherapy in non-small cell lung cancer (NSCLC) patients: a meta-analysis. Tumour Biol. 2014;35:6791-9
16. Sawers L, Ferguson MJ, Ihrig BR. et al. Glutathione S-transferase P1 (GSTP1) directly influences platinum drug chemosensitivity in ovarian tumour cell lines. Br J Cancer. 2014;111:1150-8
17. Liu R, Zhao X, Liu X. et al. Influences of ERCC1, ERCC2, XRCC1, GSTP1, GSTT1, and MTHFR polymorphisms on clinical outcomes in gastric cancer patients treated with EOF chemotherapy. Tumour Biol. 2016;37:1753-62
18. Liu Y, Li Y, Tan BB. et al. Technique appraisement of comparative proteomics and screening of differentiation-related protein in gastric carcinoma. Hepatogastroenterology. 2013;60:633-7
19. Liang J, Li QF, Yao RY. et al. [Association between genetic polymorphisms of ERCC1, XRCC1, GSTP1 and survival of advanced gastric cancer patients treated with oxaliplatin/5-Fu-based chemotherapy]. Zhonghua Zhong Liu Za Zhi. 2010;32:515-9
20. Li QF, Yao RY, Liu KW. et al. Genetic polymorphism of GSTP1: prediction of clinical outcome to oxaliplatin/5-FU-based chemotherapy in advanced gastric cancer. J Korean Med Sci. 2010;25:846-52
21. Shim HJ, Yun JY, Hwang JE. et al. BRCA1 and XRCC1 polymorphisms associated with survival in advanced gastric cancer treated with taxane and cisplatin. Cancer Sci. 2010;101:1247-54
22. Huang ZH, Hua D, Du X. Polymorphisms in p53, GSTP1 and XRCC1 predict relapse and survival of gastric cancer patients treated with oxaliplatin-based adjuvant chemotherapy. Cancer Chemother Pharmacol. 2009;64:1001-7
23. Ott K, Lordick F, Becker K. et al. Glutathione-S-transferase P1, T1 and M1 genetic polymorphisms in neoadjuvant-treated locally advanced gastric cancer: GSTM1-present genotype is associated with better prognosis in completely resected patients. Int J Colorectal Dis. 2008;23:773-82
24. Goekkurt E, Hoehn S, Wolschke C. et al. Polymorphisms of glutathione S-transferases (GST) and thymidylate synthase (TS)-novel predictors for response and survival in gastric cancer patients. Br J Cancer. 2006;94:281-6
25. Seo BG, Kwon HC, Oh SY. et al. Comprehensive analysis of excision repair complementation group 1, glutathione S-transferase, thymidylate synthase and uridine diphosphate glucuronosyl transferase 1A1 polymorphisms predictive for treatment outcome in patients with advanced gastric cancer treated with FOLFOX or FOLFIRI. Oncol Rep. 2009;22:127-36
26. Goekkurt E, Al-Batran SE, Hartmann JT. et al. Pharmacogenetic analyses of a phase III trial in metastatic gastroesophageal adenocarcinoma with fluorouracil and leucovorin plus either oxaliplatin or cisplatin: a study of the arbeitsgemeinschaft internistische onkologie. J Clin Oncol. 2009;27:2863-73
27. Shen X, Wang J, Yan X. et al. Predictive value of GSTP1 Ile105Val polymorphism in clinical outcomes of chemotherapy in gastric and colorectal cancers: a systematic review and meta-analysis. Cancer Chemother Pharmacol. 2016;77:1285-302
28. Wang Z, Chen JQ, Liu JL. et al. Polymorphisms in ERCC1, GSTs, TS and MTHFR predict clinical outcomes of gastric cancer patients treated with platinum/5-Fu-based chemotherapy: a systematic review. BMC Gastroenterol. 2012;12:137
29. Geng P, Li J, Wang N. et al. Genetic Contribution of Polymorphisms in Glutathione S-Transferases to Brain Tumor Risk. Molecular neurobiology. 2016;53:1730-40
30. Ye F, Liu Z, Tan A. et al. XRCC1 and GSTP1 polymorphisms and prognosis of oxaliplatin-based chemotherapy in colorectal cancer: a meta-analysis. Cancer Chemother Pharmacol. 2013;71:733-40
31. Yuan Z, Li J, Hu R. et al. Predictive assessment in pharmacogenetics of XRCC1 gene on clinical outcomes of advanced lung cancer patients treated with platinum-based chemotherapy. Sci Rep. 2015;5:16482
32. Ford AC, Forman D, Hunt RH. et al. Helicobacter pylori eradication therapy to prevent gastric cancer in healthy asymptomatic infected individuals: systematic review and meta-analysis of randomised controlled trials. Bmj. 2014;348:g3174
33. Xu W, Beeharry MK, Liu W. et al. Preoperative Chemotherapy for Gastric Cancer: Personal Interventions and Precision Medicine. Biomed Res Int. 2016;2016:3923585
34. Harada K, Mizrak Kaya D, Shimodaira Y. et al. Translating genomic profiling to gastrointestinal cancer treatment. Future Oncol. 2017;13:919-34
35. Strand MS, Lockhart AC, Fields RC. Genetics of Gastric Cancer. Surg Clin North Am. 2017;97:345-70
36. Zhang K, Li Y, Fan F. et al. Court Decisions on Medical Malpractice in China After the New Tort Liability Law. Am J Forensic Med Pathol. 2016;37:149-51
37. Ye H, Shao M, Shi X. et al. Predictive assessment in pharmacogenetics of Glutathione S-transferases genes on efficacy of platinum-based chemotherapy in non-small cell lung cancer patients. Sci Rep. 2017;7:2670
38. Pasello M, Michelacci F, Scionti I. et al. Overcoming glutathione S-transferase P1-related cisplatin resistance in osteosarcoma. Cancer Res. 2008;68:6661-8
39. Stoehlmacher J, Park DJ, Zhang W. et al. A multivariate analysis of genomic polymorphisms: prediction of clinical outcome to 5-FU/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer. 2004;91:344-54
40. Tew KD. Glutathione-Associated Enzymes In Anticancer Drug Resistance. Cancer Res. 2016;76:7-9
41. Meulendijks D, Rozeman EA, Cats A. et al. Pharmacogenetic variants associated with outcome in patients with advanced gastric cancer treated with fluoropyrimidine and platinum-based triplet combinations: a pooled analysis of three prospective studies. Pharmacogenomics J. 2017;17:441-451
42. Ruzzo A, Graziano F, Kawakami K. et al. Pharmacogenetic profiling and clinical outcome of patients with advanced gastric cancer treated with palliative chemotherapy. J Clin Oncol. 2006;24:1883-91
43. Keam B, Im SA, Han SW. et al. Modified FOLFOX-6 chemotherapy in advanced gastric cancer: Results of phase II study and comprehensive analysis of polymorphisms as a predictive and prognostic marker. BMC Cancer. 2008;8:148
44. Ji YZ, Liang J, Yao RY. et al. Correlation of genetic polymorphism of XPG and GSTP1 with oxaliplatin-based chemotherapy for advanced gastric cancer. Med J Qilu. 2011;26:390-2
45. Kap EJ, Richter S, Rudolph A. et al. Genetic variants in the glutathione S-transferase genes and survival in colorectal cancer patients after chemotherapy and differences according to treatment with oxaliplatin. Pharmacogenet Genomics. 2014;24:340-7
46. Kumamoto K, Ishibashi K, Okada N. et al. Polymorphisms of GSTP1, ERCC2 and TS-3'UTR are associated with the clinical outcome of mFOLFOX6 in colorectal cancer patients. Oncol Lett. 2013;6:648-54
47. Lai CY, Hsieh LL, Sung FC. et al. Tumor site- and stage-specific associations between allelic variants of glutathione S-transferase and DNA-repair genes and overall survival in colorectal cancer patients receiving 5-fluorouracil-based chemotherapy. PLoS One. 2013;8:e69039
48. Bohanes P, Rankin CJ, Blanke CD. et al. Pharmacogenetic Analysis of INT 0144 Trial: Association of Polymorphisms with Survival and Toxicity in Rectal Cancer Patients Treated with 5-FU and Radiation. Clin Cancer Res. 2015;21:1583-90
49. Nishina T, Takano Y, Denda T. et al. A phase II clinical study of mFOLFOX6 plus bevacizumab as first-line therapy for Japanese advanced/recurrent colorectal cancer patients. Jpn J Clin Oncol. 2013;43:1080-6
50. Li HY, Ge X, Huang GM. et al. GSTP1, ERCC1 and ERCC2 polymorphisms, expression and clinical outcome of oxaliplatin-based adjuvant chemotherapy in colorectal cancer in Chinese population. Asian Pac J Cancer Prev. 2012;13:3465-9
51. Farina Sarasqueta A, van Lijnschoten G, Lemmens VE. et al. Pharmacogenetics of oxaliplatin as adjuvant treatment in colon carcinoma: are single nucleotide polymorphisms in GSTP1, ERCC1, and ERCC2 good predictive markers? Mol Diagn Ther. 2011;15:277-83
52. Paez D, Salazar J, Pare L. et al. Pharmacogenetic study in rectal cancer patients treated with preoperative chemoradiotherapy: polymorphisms in thymidylate synthase, epidermal growth factor receptor, GSTP1, and DNA repair genes. Int J Radiat Oncol Biol Phys. 2011;81:1319-27
53. Zarate R, Rodriguez J, Bandres E. et al. Oxaliplatin, irinotecan and capecitabine as first-line therapy in metastatic colorectal cancer (mCRC): a dose-finding study and pharmacogenomic analysis. Br J Cancer. 2010;102:987-94
54. Jones BA, Christensen AR, Wise JP Sr. et al. Glutathione S-transferase polymorphisms and survival in African-American and white colorectal cancer patients. Cancer Epidemiol. 2009;33:249-56
55. Le Morvan V, Smith D, Laurand A. et al. Determination of ERCC2 Lys751Gln and GSTP1 Ile105Val gene polymorphisms in colorectal cancer patients: relationships with treatment outcome. Pharmacogenomics. 2007;8:1693-703
56. Stoehlmacher J, Park DJ, Zhang W. et al. Association between glutathione S-transferase P1, T1, and M1 genetic polymorphism and survival of patients with metastatic colorectal cancer. J Natl Cancer Inst. 2002;94:936-42
57. Chen JG. Relationship between FOLFOX chemotherapy of colorectal cancer patients and GSTP1, XRCC1 gene polymorphism. J Mod Lab Med. 2016;31:66-9
58. Dong NN, Luo XY, He Z. Genetic polymorphisms of CYP3A5 and Glutathione S-Stransferase P1 predict clinical outcomes of chemotherapy in metastatic colorectal cancer. J Clin Exp Med. 2014;13:607-10
59. Yin BB, Liang J, Gu J. et al. The relationship between the genetic polymorphism of GSTP1 and the chemotherapeutic effect of oxaliplatin in patients with advanced colorectal cancer. Med J Qilu. 2009;24:192-5
60. Han L, Dong NN. Genetic polymorphism of glutahione S-transferase P1 predict clinical outcomes of advanced colorectal cancer treated with mFOLFOX6 chemotherapy. J Clin Exper Med. 2015;14:542-5
61. Shen DY, Xie HT, Chen Y. et al. Association of MTHFR, GSTP1, ERCC1 polymorphisms and response to FOLFOX in colorectal cancer patients. Chin J Clin Pharmacol Ther. 2015;20:75-85
62. Ruzzo A, Graziano F, Loupakis F. et al. Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFOX-4 chemotherapy. J Clin Oncol. 2007;25:1247-54
63. Jun L, Haiping Z, Beibei Y. Genetic polymorphisms of GSTP1 related to response to 5-FU-oxaliplatin-based chemotherapy and clinical outcome in advanced colorectal cancer patients. Swiss Med Wkly. 2009;139:724-8
64. Joerger M, Huitema AD, Boot H. et al. Germline TYMS genotype is highly predictive in patients with metastatic gastrointestinal malignancies receiving capecitabine-based chemotherapy. Cancer Chemother Pharmacol. 2015;75:763-72
65. Boige V, Mendiboure J, Pignon JP. et al. Pharmacogenetic assessment of toxicity and outcome in patients with metastatic colorectal cancer treated with LV5FU2, FOLFOX, and FOLFIRI: FFCD 2000-05. J Clin Oncol. 2010;28:2556-64
66. Chen YC, Tzeng CH, Chen PM. et al. Influence of GSTP1 I105V polymorphism on cumulative neuropathy and outcome of FOLFOX-4 treatment in Asian patients with colorectal carcinoma. Cancer Sci. 2010;101:530-5
67. Hong J, Han SW, Ham HS. et al. Phase II study of biweekly S-1 and oxaliplatin combination chemotherapy in metastatic colorectal cancer and pharmacogenetic analysis. Cancer Chemother Pharmacol. 2011;67:1323-31
68. Huang MY, Huang ML, Chen MJ. et al. Multiple genetic polymorphisms in the prediction of clinical outcome of metastatic colorectal cancer patients treated with first-line FOLFOX-4 chemotherapy. Pharmacogenet Genomics. 2011;21:18-25
69. Lamas MJ, Duran G, Balboa E. et al. Use of a comprehensive panel of biomarkers to predict response to a fluorouracil-oxaliplatin regimen in patients with metastatic colorectal cancer. Pharmacogenomics. 2011;12:433-42
70. Rumiato E, Boldrin E, Amadori A. et al. DMET (Drug-Metabolizing Enzymes and Transporters) microarray analysis of colorectal cancer patients with severe 5-fluorouracil-induced toxicity. Cancer Chemother Pharmacol. 2013;72:483-8
71. Wang Y, Chen J, Li X. et al. Genetic polymorphisms of ERCC1 and their effects on the efficacy of cisplatin-based chemotherapy in advanced esophageal carcinoma. Oncol Rep. 2011;25:1047-52
72. Gui Y, Xie L, Shu XH. et al. Genetic polymorphism of GSTP1, MTHFR correlated with response to cisplatin and fluorouracil chemotherapy in esophagus cancer. Mod oncol. 2015;24:906-9
73. Kwon HC, Roh MS, Oh SY. et al. Prognostic value of expression of ERCC1, thymidylate synthase, and glutathione S-transferase P1 for 5-fluorouracil/oxaliplatin chemotherapy in advanced gastric cancer. Ann Oncol. 2007;18:504-9
74. Boku N, Ohtsu A, Yoshida S. et al. Significance of biological markers for predicting prognosis and selecting chemotherapy regimens of advanced gastric cancer patients between continuous infusion of 5-FU and a combination of 5-FU and cisplatin. Jpn J Clin Oncol. 2007;37:275-81
75. Choi IS, Lee HS, Lee KW. et al. Biomarker analysis in patients with advanced gastric cancer treated with S-1 plus cisplatin chemotherapy: orotate phosphoribosyltransferase expression is associated with treatment outcomes. Med Oncol. 2011;28:991-8
76. Li S, Li B, Wang J. et al. Identification of Sensitivity Predictors of Neoadjuvant Chemotherapy for the Treatment of Adenocarcinoma of Gastroesophageal Junction. Oncol Res. 2017;25:93-7
Author contact
![]() Corresponding authors: Dr. Jian Qu, Department of Pharmacy, the Second Xiangya Hospital, Central South University; Institute of Clinical Pharmacy, Central South University. No. 139 Middle Renmin Road, Changsha, China, 410011; Tel.: +86-15973190614, Fax number: +86-0731-85292128, E-mail: qujianstanleyedu.cn; Dr. Qiang Qu, Department of Pharmacy, Xiangya Hospital, Central South University. No.87 Xiangya Road, Changsha, China, 410008; Tel.: +8673184805380; Email: quqiangedu.cn
Corresponding authors: Dr. Jian Qu, Department of Pharmacy, the Second Xiangya Hospital, Central South University; Institute of Clinical Pharmacy, Central South University. No. 139 Middle Renmin Road, Changsha, China, 410011; Tel.: +86-15973190614, Fax number: +86-0731-85292128, E-mail: qujianstanleyedu.cn; Dr. Qiang Qu, Department of Pharmacy, Xiangya Hospital, Central South University. No.87 Xiangya Road, Changsha, China, 410008; Tel.: +8673184805380; Email: quqiangedu.cn

 Global reach, higher impact
Global reach, higher impact