Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(4):1023-1031. doi:10.7150/jca.26472 This issue Cite
Research Paper
The Role of Polymorphisms in Genes of PI3K/Akt Signaling Pathway on Prostate
1. Department of Urology, The First Affiliated Hospital of Anhui Medical University, Hefei, 230000, China
2. Department of Blood Transfusion, The First Affiliated Hospital of Anhui Medical University, Hefei, 230000, China;
3. Trauma Department of Orthopedics, The Third Affiliated Hospital of Anhui Medical University, Hefei, Anhui, 230000, China;
4. Department of Urology, Xiangya Hospital, Central South University, NO.87 Xiangya Road, Changsha, Hunan, 410008, China
5. Shenzhen Following Precision Medical Institute, Shenzhen Luohu Hospital Group, Shenzhen, 518000, China
# These authors contributed equally to the work.
Received 2018-4-4; Accepted 2019-1-9; Published 2019-1-29
Abstract
Background and Objective: Increasing evidence suggested that polymorphisms in genes of PI3K/Akt pathway were closely related to prostate cancer (PCa) risk. Nevertheless, these results are controversial and inconclusive. Here, we conducted a comprehensive updated meta-analysis and systematic review to precisely illustrate the association between polymorphisms in genes of PI3K/Akt signaling pathway and PCa risk.
Materials and Methods: The gene set of PI3K/Akt pathway was referenced from the Kyoto Encyclopedia of Genes and Genomes (KEGG) website. Relevant studies were identified by the systematically researching on PubMed, Web of Science and Google Scholar databases up to October 1, 2017. The odds ratios (ORs) with a corresponding 95% confidential intervals (95%CIs) were applied to test their associations. All the analyses were conducted by using Stata 12.0 (Stata Corporation, USA).
Results: Finally, 38 articles comprising 62 case-control studies were enrolled for 13 polymorphisms in genes of PI3K/Akt pathway. However, overall results failed to present a positive association between polymorphisms in genes of PI3K/Akt pathway and PCa risk. Nevertheless, in the subgroup analysis by ethnicity, we identified that IL-6-rs1800795 polymorphism was associated with an increased risk of PCa for Caucasian individuals in dominant model (MM + MW vs. WW: OR = 1.245, 95%CI = 1.176-1.318, P < 0.001).
Conclusion: Our work suggests that polymorphisms in genes of PI3K/Akt Signaling Pathway are not risk factor for PCa. Further well-designed studies with larger samples and precise designs are demanded to corroborate our findings.
Keywords: PI3K/Akt, polymorphism, prostate cancer
Introduction
For males worldwide, prostate cancer (PCa) has the second highest incidence of all cancers. Each year, approximately 238,590 new cases and 29,720 deaths are reported according to cancer statistics, 2013. In view of the anfractuous pathogenesis of PCa, interconnected cell signaling pathways and transmissions which manipulates the survival, evolution and apoptosis of cells1-3, would provide us new inspirations of the prevention and treatment of PCa patients.
Among diverse pathways, genes encompassed in phosphoinositide 3-kinase (PI3K)/Akt signaling pathway, such as toll-like receptor-4 (TLR4), vascular endothelial growth factor (VEGF), interleukin 6 (IL-6), insulin receptor substrate 1 (IRS1) and insulin-like growth factor 1 (IGF1), appear with more common mutations or amplifications in PCa (Figure S1 and Figure S2). PI3K is a phosphatidylinositol kinase which is encoded by the PIK3CA gene. It consists of a catalytic subunit p110 and regulatory subunit p85. Akt is a cytoplasmic serine-threonine protein kinase which promotes the progression of cell cycle and inhibits cell apoptosis. The PI3K/Akt signaling pathway is implicated in different cellular functions, including survival, growth, proliferation, metabolism and angiogenesis. Currently, the relationships between polymorphisms in genes of PI3K/Akt signaling pathway and PCa risk have been an area of intense investigations but with mixed results4,5. For instance, Balistreri et al.6 pointed out that there existed a significant association between polymorphisms in TRL4 and an increased risk of PCa, a result consistent with both Wang et al.'s7 and Chen et al.'s8 work. However, Shui et al.9 has conducted a case-control study comprising 1,267 controls and 1,286 PCa cases and found that genetic variation across TLR4 alone is not strongly associated with PCa risk. As for polymorphisms in IGF1, Schildkraut et al.10 revealed the significant association between genetic polymorphisms in IGF1 and PCa risk among Black and White men. On the contrary, Neuhausen et al.11 failed to find any positive connection between IGF1 polymorphisms and PCa risk. In addition, for IL-6-rs1800795, both Kesarwani et al.12 and Mandal et al.'s13 studies supported the role of IL-6-rs1800795 polymorphism in PCa, while the result was inconsistent with Bao et al.'s14 work.
Hence, previous studies had presented inconsistent views between polymorphisms encompassed in genes of PI3K/Akt signaling pathway and PCa risk. Considering that, we conducted the current updated meta-analysis in order to precisely evaluate their associations on the foundation of all available eligible studies, providing with convincible evidence for the prevention and/or targeted therapy for PCa patients.
Material and Methods
Acquisition of the PI3K/Akt Pathway Gene Set
The gene set of PI3K/Akt pathway was referenced to the Kyoto Encyclopedia of Genes and Genomes (KEGG) website (http://www.kegg.jp/kegg-bin/show_pathway?hsa04151). The gene set was originally provided via the KEGG signaling database, and encompassed the following 101 genes: ANGPT2, ANGPT4, IL2RB, CD19, COL1A, IL3, COL2A, IL3RA, COL4A, COL6A, IL6, COL9A, CSF1, CSF1R, CSF3, CSF3R, EFNA, EGF, EGFR, EPHA2, EPO, EPOR, FGF, FGF1, FGF2, FGFR1, FGFR2, FGFR3, FGFR4, FLT1, FLT4, GH, GHR, IL6R, GRB2, HGF, HRAS, IFNA, IFNAR1, IFNAR2, IFNB, IGF1, IGF1R, IGH, IL2, IL2RA, IL2RG, IL4, IL4R, IL7, IL7R, INS, INSR, IRS1, JAK1, JAK2, JAK3, KDR, KIT, KITLG, KRAS, LAMA1_2, LAMA3_5, LAMA4, LAMB1, LAMB2, LAMB3, LAMB4, LAMC1, LAMC2, MAP2K1, MAP2K2, MAPK1, MAPK2, MAPK3, MET, NGFA, NGFB, NGFR, NRAS, OSM, OSMR, PDGFA, PDGFB, PDGFC_D, PDGFRA, PDGFRB, PGF, PIK3AP1, PRL, PRLR, RAC1, RAF1, SOS, SYK, TEK, TLR2, TLR4, VEGFA, VEGFB and VEGFC-D.
Study Description
To evaluate the connections between polymorphisms in genes of PI3K/Akt pathway and PCa risk, we conducted the present study by combining all accessible studies together from diverse databases, including Web of Science, PubMed, and China National Knowledge Infrastructure (CNKI) databases. The integrated keywords were: ('genes' OR 'abbreviations of genes') AND ('cancer' OR 'tumor' OR 'carcinoma' OR 'neoplasms') AND ('polymorphism' OR 'mutation' OR 'variant' OR 'SNP' OR 'genotype'). At the same time, we used the integrated keywords (Gene_ID & prostate cancer) to search on Google, and performed the hand screening from all highly connected results. Besides, extra studies were collected via the reference lists of the identified studies. The final date of retrieval was in October 1, 2017. The whole studies in the analysis were firstly published in the primary literature with no reproduction in other studies.
Inclusion and Exclusion Criteria
The inclusion criteria in this analysis were: (1) the cases were PCa patients and the controls were no history of cancers; 2) cohort studies or case-control studies concerning the relationships between polymorphisms in genes of PI3K/Akt signaling pathway and PCa risk; (3) the raw data of genotype frequency can be extracted. The exclusion criteria were as follows: (1) the raw data were not accessible; (2) case-only studies that didn't have control groups; (3) family-based association studies; and (4) Review papers.
Data Extraction
All of the data extraction work should be completed independently by 2 of the authors according to the prelisted inclusion criteria. And the arguments should be solved by another expert(s). You didn't mention the procedure in your article. In addition, we extracted data from each case-control study, including genotype frequencies, name of first author; year of publication; ethnicity and number of cases and controls. In addition, we used The Newcastle-Ottawa Scale (NOS) to evaluate the quality of enrolled studies.
Statistical analysis
The meta-analysis was conducted to assess the associations between polymorphisms in genes of PI3K/Akt pathway and PCa risk. Hardy-Weinberg equilibrium (HWE) in the control group was tested15. To make a more comprehensive meta-analysis, five genetic models were adopted, including allele contrast (M vs. W), codominant (MM vs. WW and MW vs. WW), dominant (MM + MW vs. WW) and recessive models (MM vs. MW + WW). The impact of relationship was evaluated by odds ratio (OR) with a corresponding 95% confidential intervals (95%CI). What's more, when the heterogeneity (P > 0.1 as the standard) 16 was assessed, the I2-based Q statistic was used (I2 = 0-25%: no heterogeneity; I2 = 25%-50%: moderate heterogeneity; I2 = 50%-75%: large heterogeneity; I2 = 75%-100%: extreme heterogeneity) 17, which represented the weighted sum of the squared difference between the overall effect size and the effect size from every study. When I2 > 50% or PQ ≤ 0.1, substantial heterogeneity was existed, then, a random-effects model was used; otherwise, the fixed-effects model was be applied. It has been recognized that when results of the component studies differ among themselves, random effects incorporate an estimate of the inter-study variance and provide wider 95%CIs18. The analyses were conducted using Stata 12.0 (Stata Corporation, USA), and all P values were two-tailed.
Results
Main Characteristics of the Enrolled Studies
After initial screening, there were 1,166 results related to the search words enrolled. After reading the important information such as the titles and abstracts, 51 potential eligible studies were selected for next step full-text view. When a further screening was conducted, 13 of these studies were excluded for not associated with PCa risk. Finally, 38 articles with 62 case-control studies were left for data extraction (Table 1) 12,19-54.
Of them, there were 2,170 cases and 1,587 controls for TLR4-rs1927914 polymorphism (from three studies), 3,842 cases and 3,143 controls for TLR4-rs10759932 polymorphism (from 4 studies), 3,508 cases and 2,960 controls for TLR4-rs2149356 polymorphism (from 4 studies), 1,467 cases and 1,551 controls for TLR4-rs4986790 polymorphism (from 4 studies), 3,985 cases and 3,438 controls for TLR4-rs11536889 polymorphism (from 5 studies), 2,380 cases and 2,357 controls for TLR4-rs7873784 polymorphism (from 3 studies), 632 cases and 685 controls for VEGF-rs833061 polymorphism (from three studies), 1,511 cases and 821 controls for VEGF-rs1570360 polymorphism (from three studies), 1,243 cases and 1,620 controls for IRS1-rs1801278 polymorphism (from four studies), 2,289 cases and 2,114 controls for FGFR4-rs351855 polymorphism (from three studies), 1,805 cases and 3,235 controls for IL-6-rs1800796 polymorphism (from three studies), 10,625 cases and 12,353 controls for IL-6-rs1800795 polymorphism (from eight studies), 2,217 cases and 2,471 controls for IGF1-(CA) 19 polymorphism (from seven studies), respectively. In addition, the study selection processes for these polymorphisms were showed in Figure S3-8.
Furthermore, of the 62 case-control studies, 41 sets were performed on Caucasian populations, seven sets on Asian populations, six sets on African populations, and the other eight were based on mixed ethnic groups (including at least one race). Controls of 42 studies were population-based (P-B), while the other 20 studies were hospital-based (H-B). The quality of the enrolled studies was assessed by NOS and presented in Table S1.
Quantitative synthesis
Results of the association between polymorphisms in genes of PI3K/Akt pathway and PCa risk were showed in Table 2 and Table S2. However, the pooled results suggested negative associations between all the 13 polymorphisms in six genes of PI3K/Akt signaling pathway and PCa risk.
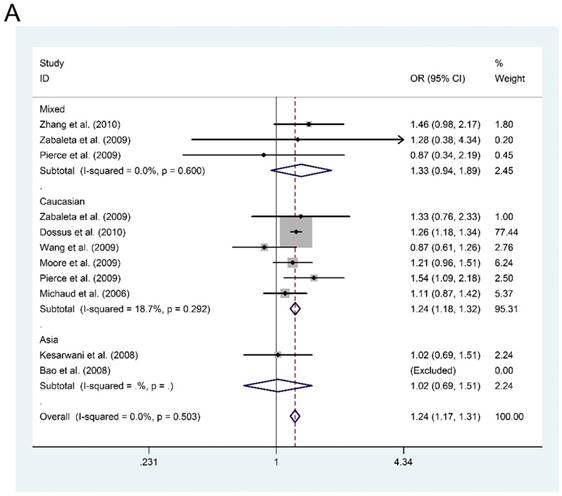
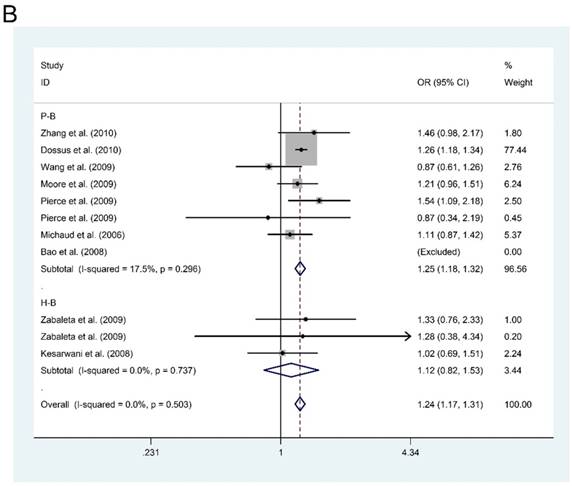
However, in the subgroup analysis by ethnicity, we found that IL-6-rs1800795 polymorphism was associated with an increased risk of PCa in dominant model for Caucasian population (MM + MW vs. WW: OR=1.245, 95%CI = 1.176-1.318, P < 0.001, Figure 1A). Furthermore, in the subgroup analysis by source of control, we also found an increased risk of PCa for P-B groups in dominant model (MM + MW vs. WW; OR = 1.246, 95%CI = 1.177-1.319, P < 0.001, Figure 1B). Although subgroup analyses were also conducted for other polymorphisms in genes of PI3K/Akt signaling pathway, negative results were found.
Sensitivity analysis and Publication bias
Sensitivity analysis was conducted by excluding one single study each time, and no evidence was observed suggesting pooled ORs shift (Table S3). In addition, we used Begg's funnel plot and Egger's regression test to assess potential publication bias. As for TLR4-rs1927914, TLR4-rs10759932, TLR4-rs2149356, TLR4-rs4986790, TLR4-rs11536889, TLR4-rs7873784, VEGF-rs833061, VEGF-rs1570360, IRS1-rs1801278, FGFR4-rs351855, IL-6-rs1800796, IGF1-(CA)19 polymorphisms, no evidence of publication bias was identified by viewing the shape of Begg's funnel plot, which was further validated by Egger's regression test. However, for IL-6-rs1800795 polymorphism, potential publication bias was existed (P = 0.016) (Table S4). In that case, we further conducted sensitivity analysis by using the trim and fill method55, and imputed studies provide a symmetrical funnel plot (data not shown), indicating publication bias was not existed.
Characteristics of the enrolled studies.
| Gene | SNP | First Author | Year | Genotyping Method | Ethnicity | Source of Control | Case | Control | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WW | WM | MM | WW | WM | MM | Y(HWE) | |||||||
| TLR4 | rs1927914 | Chen et al. | 2005 | MassARRAY | Caucasian | P-B | 297 | 301 | 60 | 290 | 288 | 91 | Y |
| rs1927914 | Zheng et al. | 2004 | MassARRAY | Caucasian | P-B | 625 | 596 | 154 | 341 | 354 | 81 | Y | |
| rs1927914 | Song et al. | 2009 | PCR-RFLP | Asian | H-B | 69 | 54 | 14 | 48 | 87 | 7 | N | |
| rs10759932 | Chen et al. | 2005 | MassARRAY | Caucasian | P-B | 511 | 155 | 11 | 472 | 197 | 12 | Y | |
| rs10759932 | Zheng et al. | 2004 | MassARRAY | Caucasian | P-B | 991 | 350 | 34 | 571 | 194 | 13 | Y | |
| rs10759932 | Shui et al. | 2012 | MALDI-TOF | Caucasian | P-B | 897 | 260 | 27 | 908 | 244 | 27 | N | |
| rs10759932 | Cheng et al. | 2007 | Sequencing | Caucasian | H-B | 370 | 117 | 119 | 358 | 143 | 4 | N | |
| rs2149356 | Chen et al. | 2005 | MassARRAY | Caucasian | P-B | 320 | 286 | 61 | 305 | 275 | 91 | N | |
| rs2149356 | Zheng et al. | 2004 | MassARRAY | Caucasian | P-B | 603 | 423 | 136 | 331 | 224 | 74 | N | |
| rs2149356 | Shui et al. | 2012 | MALDI-TOF | Caucasian | P-B | 579 | 489 | 106 | 576 | 460 | 119 | Y | |
| rs2149356 | Cheng et al. | 2007 | Sequencing | Caucasian | H-B | 197 | 223 | 85 | 210 | 213 | 82 | N | |
| rs4986790 | Chen et al. | 2005 | MassARRAY | Caucasian | P-B | 588 | 66 | 3 | 605 | 59 | 5 | N | |
| rs4986790 | Cheng et al. | 2007 | TaqMan | Caucasian | H-B | 439 | 66 | 1 | 456 | 48 | 2 | Y | |
| rs4986790 | Wang et al. | 2009 | TaqMan | Caucasian | P-B | 230 | 24 | 0 | 216 | 35 | 0 | Y | |
| rs4986790 | Balistreri et al. | 2010 | PCR-RFLP | Caucasian | H-B | 49 | 1 | 0 | 111 | 13 | 1 | Y | |
| rs11536889 | Chen et al. | 2005 | MassARRAY | Caucasian | P-B | 515 | 167 | 10 | 513 | 159 | 15 | Y | |
| rs11536889 | Zheng et al. | 2004 | MassARRAY | Caucasian | P-B | 1047 | 318 | 15 | 625 | 141 | 12 | Y | |
| rs11536889 | Shui et al. | 2012 | MALDI-TOF | Caucasian | P-B | 909 | 202 | 32 | 897 | 291 | 27 | Y | |
| rs11536889 | Cheng et al. | 2007 | Sequencing | Caucasian | H-B | 385 | 105 | 16 | 401 | 93 | 12 | N | |
| rs11536889 | Wang et al. | 2009 | TaqMan | Caucasian | P-B | 178 | 79 | 7 | 175 | 71 | 6 | Y | |
| rs7873784 | Chen et al. | 2005 | MassARRAY | Caucasian | P-B | 475 | 178 | 16 | 459 | 180 | 30 | N | |
| rs7873784 | Shui et al. | 2012 | MALDI-TOF | Caucasian | P-B | 887 | 295 | 24 | 861 | 302 | 19 | Y | |
| rs7873784 | Cheng et al. | 2007 | Sequencing | Caucasian | H-B | 362 | 130 | 13 | 346 | 146 | 14 | Y | |
| IL-6 | rs1800796 | Wang et al. | 2009 | TaqMan | Caucasian | P-B | 233 | 19 | 1 | 225 | 25 | 0 | Y |
| rs1800796 | Pierce et al. | 2009 | TaqMan | Caucasian | P-B | 156 | 19 | 0 | 1740 | 192 | 2 | Y | |
| rs1800796 | Pierce et al. | 2009 | TaqMan | Mixed | P-B | 37 | 2 | 1 | 251 | 41 | 6 | N | |
| rs1800796 | Sun et al. | 2004 | Microarray | Caucasian | P-B | 1226 | 109 | 2 | 675 | 74 | 4 | Y | |
| rs1800795 | Mandal et al. | 2014 | PCR | Mixed | H-B | 108 | 44 | 12 | 74 | 44 | 22 | N | |
| rs1800795 | Zhang et al. | 2010 | Sequenom | Mixed | P-B | 80 | 86 | 27 | 100 | 75 | 22 | Y | |
| rs1800795 | Zabaleta et al. | 2009 | TaqMan | Caucasian | H-B | 19 | 34 | 21 | 126 | 163 | 112 | N | |
| rs1800795 | Zabaleta et al. | 2009 | TaqMan | Mixed | H-B | 10 | 2 | 3 | 41 | 10 | 6 | N | |
| rs1800795 | Dossus et al. | 2010 | GoldenGate | Caucasian | P-B | 3594 | 3218 | 1125 | 3832 | 3402 | 274 | N | |
| rs1800795 | Wang et al. | 2009 | TaqMan | Caucasian | P-B | 91 | 116 | 43 | 84 | 128 | 40 | Y | |
| rs1800795 | Moore et al. | 2009 | TaqMan | Caucasian | P-B | 191 | 485 | 281 | 196 | 401 | 250 | Y | |
| rs1800795 | Pierce et al. | 2009 | TaqMan | Caucasian | P-B | 48 | 96 | 31 | 648 | 805 | 305 | N | |
| rs1800795 | Pierce et al. | 2009 | TaqMan | Mixed | P-B | 34 | 5 | 1 | 216 | 43 | 1 | Y | |
| rs1800795 | Kesarwani et al. | 2008 | PCR | Asia | H-B | 102 | 84 | 14 | 103 | 87 | 10 | Y | |
| rs1800795 | Bao et al. | 2008 | TaqMan | Asia | P-B | 136 | 0 | 0 | 120 | 0 | 0 | N | |
| rs1800795 | Michaud et al. | 2006 | TaqMan | Caucasian | P-B | 170 | 223 | 91 | 230 | 293 | 90 | Y | |
| IGF1 | (CA)19 | Chu et al. | 2006 | Sequenom | Caucasian | P-B | 75 | 28 | 21 | 73 | 76 | 25 | Y |
| (CA)19 | Chu et al. | 2006 | Sequenom | Mixed | P-B | 4 | 17 | 17 | 2 | 20 | 16 | Y | |
| (CA)19 | Neuhausen et al. | 2005 | PCR | Caucasian | H-B | 78 | 86 | 29 | 107 | 124 | 32 | Y | |
| (CA)19 | Schildkraut et al. | 2005 | PCR | Mixed | P-B | 20 | 39 | 35 | 28 | 33 | 20 | Y | |
| (CA)19 | Norihiko et al. | 2005 | PCR | Asian | H-B | 155 | 130 | 18 | 289 | 172 | 20 | Y | |
| (CA)19 | Friedrichsen et al. | 2005 | PCR-RFLP | Mixed | P-B | 73 | 289 | 219 | 64 | 237 | 219 | Y | |
| (CA)19 | Nam et al. | 2003 | PCR-RFLP | Mixed | P-B | 64 | 230 | 189 | 103 | 253 | 192 | Y | |
| (CA)19 | Wenndy et al. | 2007 | PCR | Mixed | H-B | 324 | 28 | 49 | 289 | 26 | 51 | N | |
| VEGF | rs833061 | Fukuda et al. | 2007 | PCR-RFLP | Asian | H-B | 143 | 103 | 24 | 132 | 97 | 23 | Y |
| rs833061 | Onen et al. | 2008 | PCR-RFLP | Mixed | P-B | 33 | 89 | 11 | 50 | 94 | 13 | N | |
| rs833061 | Lin et al. | 2003 | PCR-RFLP | Asian | H-B | 60 | 32 | 4 | 43 | 72 | 4 | N | |
| rs833061 | Onen et al. | 2008 | PCR-RFLP | Caucasian | P-B | 33 | 89 | 11 | 50 | 94 | 13 | N | |
| rs1570360 | Sfar et al. | 2006 | RFLP-PCR | Caucasian | H-B | 58 | 37 | 6 | 36 | 50 | 14 | Y | |
| rs1570360 | Jacobs et al. | 2008 | TaqMan | Caucasian | P-B | 557 | 489 | 126 | 210 | 194 | 54 | Y | |
| rs1570360 | McCarron et al. | 2013 | TaqMan | Caucasian | P-B | 114 | 109 | 15 | 120 | 109 | 34 | Y | |
| IRS1 | rs1801278 | Andrea et al. | 2011 | PCR | Caucasian | H-B | 56 | 5 | 0 | 106 | 12 | 1 | Y |
| rs1801278 | Fall et al. | 2008 | PCR | Mixed | H-B | 489 | 73 | 2 | 662 | 90 | 6 | Y | |
| rs1801278 | Li et al. | 2013 | PCR | Mixed | P-B | 386 | 50 | 2 | 422 | 65 | 1 | Y | |
| rs1801278 | Neuhausen et al. | 2005 | PCR | Caucasian | P-B | 118 | 50 | 12 | 160 | 81 | 14 | Y | |
| FGFR4 | rs351855 | FitzGerald et al. | 2009 | SNPlex™ | Caucasian | P-B | 587 | 544 | 123 | 631 | 496 | 124 | Y |
| rs351855 | FitzGerald et al. | 2009 | SNPlex™ | Mixed | P-B | 104 | 39 | 3 | 60 | 18 | 2 | Y | |
| rs351855 | Lee et al. | 2010 | TaqMan | Caucasian | P-B | 183 | 182 | 32 | 235 | 167 | 37 | Y | |
| rs351855 | Zhiyong et al. | 2010 | PCR | Asian | H-B | 133 | 196 | 163 | 67 | 152 | 125 | Y | |
Note: Hardy-Weinberg equilibrium (HWE); population-based (P-B); hospital-based (H-B); Mixed: more than two descendants; W: wild allele; M: mutated allele; PCR: Polymerase chain reaction; RFLP-PCR: restriction fragment length polymorphism-Polymerase chain reaction
Details of the association between IL-6-rs1800795 polymorphism and prostate cancer risk.
| Comparison | Subgroup | N | PH | PZ | Random | Fixed |
|---|---|---|---|---|---|---|
| M vs. W | Overall | 11 | 0.000 | 0.207 | 1.108 (0.945-1.300) | 1.347 (1.292-1.403) |
| M vs. W | Asia | 1 | 1.000 | 0.692 | 1.065 (0.780-1.453) | 1.065 (0.780-1.453) |
| M vs. W | Caucasian | 6 | 0.000 | 0.071 | 1.171 (0.986-1.391) | 1.370 (1.313-1.429) |
| M vs. W | H-B | 4 | 0.020 | 0.772 | 0.946 (0.648-1.380) | 0.926 (0.765-1.122) |
| M vs. W | P-B | 7 | 0.000 | 0.038 | 1.188 (1.010-1.397) | 1.371 (1.315-1.431) |
| M vs. W | N | 5 | 0.000 | 0.528 | 1.106 (0.809-1.514) | 1.430 (1.365-1.499) |
| M vs. W | Y | 6 | 0.710 | 0.053 | 1.090 (0.999-1.190) | 1.090 (0.999-1.190) |
| WM vs. WW | Overall | 11 | 0.103 | 0.269 | 1.080 (0.951-1.227) | 1.033 (0.975-1.096) |
| WM vs. WW | Asia | 1 | 1.000 | 0.903 | 0.975 (0.650-1.463) | 0.975 (0.650-1.463) |
| WM vs. WW | Caucasian | 6 | 0.061 | 0.177 | 1.112 (0.953-1.296) | 1.036 (0.975-1.100) |
| WM vs. WW | H-B | 4 | 0.380 | 0.678 | 0.941 (0.709-1.250) | 0.943 (0.716-1.243) |
| WM vs. WW | P-B | 7 | 0.054 | 0.156 | 1.116 (0.959-1.299) | 1.038 (0.978-1.102) |
| WM vs. WW | N | 5 | 0.048 | 0.512 | 1.101 (0.826-1.466) | 1.022 (0.958-1.089) |
| WM vs. WW | Y | 6 | 0.353 | 0.217 | 1.088 (0.935-1.266) | 1.092 (0.950-1.256) |
| MM vs. WW | Overall | 11 | 0.000 | 0.211 | 1.411 (0.823-2.421) | 2.609 (2.359-2.885) |
| MM vs. WW | Asia | 1 | 1.000 | 0.428 | 1.414 (0.600-3.329) | 1.414 (0.600-3.329) |
| MM vs. WW | Caucasian | 6 | 0.000 | 0.214 | 1.529 (0.783-2.986) | 2.778 (2.502-3.085) |
| MM vs. WW | H-B | 4 | 0.043 | 0.960 | 0.982 (0.473-2.036) | 0.912 (0.606-1.372) |
| MM vs. WW | P-B | 7 | 0.000 | 0.119 | 1.671 (0.877-3.184) | 2.790 (2.513-3.097) |
| MM vs. WW | N | 5 | 0.000 | 0.464 | 1.431 (0.548-3.734) | 3.601 (3.177-4.082) |
| MM vs. WW | Y | 6 | 0.682 | 0.025 | 1.233 (1.028-1.479) | 1.231 (1.027-1.477) |
| WM + MM vs. WW | Overall | 11 | 0.047 | 0.040 | 1.147 (1.006-1.308) | 1.228 (1.162-1.298) |
| WM + MM vs. WW | Asia | 1 | 1.000 | 0.920 | 1.020 (0.689-1.510) | 1.020 (0.689-1.510) |
| WM + MM vs. WW | Caucasian | 6 | 0.292 | <0.001 | 1.224 (1.113-1.346) | 1.245 (1.176-1.318) |
| WM + MM vs. WW | H-B | 4 | 0.114 | 0.505 | 0.936 (0.631-1.390) | 0.917 (0.710-1.184) |
| WM + MM vs. WW | P-B | 7 | 0.296 | <0.001 | 1.227 (1.117-1.349) | 1.246 (1.177-1.319) |
| WM + MM vs. WW | N | 5 | 0.017 | 0.359 | 1.150 (0.853-1.552) | 1.252 (1.178-1.331) |
| WM + MM vs. WW | Y | 6 | 0.500 | 0.082 | 1.124 (0.985-1.283) | 1.124 (0.985-1.282) |
| MM vs. WM + WW | Overall | 11 | 0.000 | 0.315 | 1.331 (0.762-2.323) | 2.292 (2.093-2.509) |
| MM vs. WM + WW | Asia | 1 | 1.000 | 0.402 | 1.430 (0.620-3.300) | 1.430 (0.620-3.300) |
| MM vs. WM + WW | Caucasian | 6 | 0.000 | 0.366 | 1.388 (0.682-2.826) | 2.404 (2.189-2.640) |
| MM vs. WM + WW | H-B | 4 | 0.087 | 0.866 | 0.949 (0.516-1.746) | 0.899 (0.618-1.308) |
| MM vs. WM + WW | P-B | 7 | 0.000 | 0.218 | 1.543 (0.774-3.074) | 2.429 (2.211-2.669) |
| MM vs. WM + WW | N | 5 | 0.000 | 0.585 | 1.323 (0.485-3.606) | 3.390 (3.015-3.812) |
| MM vs. WM + WW | Y | 6 | 0.449 | 0.171 | 1.114 (0.956-1.298) | 1.112 (0.955-1.296) |
Note: Hardy-Weinberg equilibrium (HWE); P-B: population-based; H-B: hospital-based; Y: Studies conformed to HWE; N: studies did not conform to HWE; Mixed: more than two descendant; *P value less than [0.05/ (5*13)] means statistically significant.
Forest plots of the association between IL-6-rs1800795 polymorphism and prostate cancer risk. Subgroup analysis by ethnicity (A) and source of control (B).
Discussion
Recently, enormous studies suggested that polymorphisms in genes of PI3K/Akt pathway may play an important role in the prevention, diagnosis and treatment of PCa. For example, TLR4 is the main component of TLRs and has been positively investigated in inflammation and cancer. Previous studies had confirmed that two polymorphisms in TLR4 (rs4986790 and rs4986791) owned susceptibility to various type of cancers, including PCa56. VEGF is the most significant regulator of angiogenesis in human, and it plays a significant role in the occurrence and development of PCa49,50. It had been identified that there were many genetic variants in the VEFG gene57, but the conclusions were remained inconsistent23,52-57. The IRS1 gene Gly972Arg (rs1801278) polymorphisms had been found a significant association with increased cancer risk58. In vitro studies have proved that the IRS1 gene rs1801278 polymorphism impaired insulin-stimulated signaling pathway, especially through the PI3-kinase pathway59. What's more, as a docking protein for the insulin-like growth factor receptor 1 (IGF1R) 60,61, IRS1 controls IGF-1 mediated cell growth and survival60. Thus the polymorphisms of IGF-1 gene also related to the cancer risks including PCa61. Fibroblast growth factor receptor 4 (FGFR4) is one member of the family of fibroblast growth factor receptors (FGFR1-4), which displays complicated biological activities such as angiogenic and mitogenic activity. Previous study had presented that its gene polymorphism was related to PCa risks62. The human IL-6 gene encodes IL-6, a cytokine which adjusts the level of inflammation. Two polymorphisms on the promoter region of IL-6, rs1800795 (-174G/C) and rs1800796 (-572C/G) have been identified to be associated with IL-6 production63. And these association with risks of cancer have been published in a previous meta-analysis64,65. Furthermore, although these studies and meta-analysis provided some clues for separate polymorphisms in one or more genes of PI3K/Akt pathway and PCa risk, these results were not fully consistent, or even contradictory at sometimes. Therefore, we performed current meta-analysis in order to provide a comprehensive accurate assessment of the associations of these polymorphisms in genes of PI3K/Akt pathway with PCa risk. To the best of our knowledge, this is the first pooled study that analyzed the associations between 13 polymorphisms in six pivotal genes of PI3K/Akt pathway and PCa risk. Meanwhile, further analyses were conducted in different subgroups to explore the potential associations or heterogeneity sources.
Nevertheless, overall results revealed that none of these polymorphisms was associated with PCa risk. Then, we performed subgroup analysis based on ethnicity, source of control (population-based or hospital-based) and HWE status (conform or not conform). For IL-6-rs1800795 polymorphism, when the stratification analysis was conducted by ethnicity, we found that a statistically significant increased risk of PCa was identified in the dominant model for Caucasians. However, in the meta-analysis conducted by Liu et al.66, they did not reveal a significant connection between IL-6-rs1800795 polymorphism and PCa risk in Caucasian. For other polymorphisms, null association was uncovered when the stratified analyses were conducted based on ethnicity, source of control or HWE status.
Although we were surprised by these negative results, the high quality of these included studies and the substantial amount of data strengthened the possibility that the lack of association was not caused by chance. For those comparisons that did not exhibit a statistically significant association, may be as a result of the characteristics of low-penetrance genes. Moreover, although these polymorphisms assessed were appropriate candidates, they only account for some of the factors, and ignored other factors such as obesity, diet and environment. We summarized the advantages of current work. Firstly, although many meta-analyses provided some clues for separate polymorphisms in one or more genes of PI3K/Akt pathway and PCa risk, the current one provide a more comprehensive accurate assessment of the associations of all available polymorphisms in genes of PI3K/Akt pathway with PCa risk. To the best of our knowledge, this is the first pooled study that analyzed the associations between 13 polymorphisms in six pivotal genes of PI3K/Akt pathway and PCa risk. Secondly, we applied classic formula to adjust the P-values, which removed most of the marginal or false-positive P-values, making the final pool results more convincing. Thirdly, we found IL-6-rs1800795 polymorphism could be served as a risk prediction marker for Caucasian PCa patients. Our results provided some clues for the future clinical research that polymorphisms in genes of this pathway may not suitable for high-risk prostate cancer patients' screening. There are also several deficiencies that should be addressed. Firstly, other factors such as the density of prostate-specific antigen (PSA), living conditions and histological types, the stage and grades of PCa should be included to get more precise results. Secondly, for many polymorphisms of these inclusive genes, relatively small samples were included for the assessment, such as rs1927914 polymorphism. Finally, we ignored that there were many individual characters such as age, obesity, alcohol, consumption and other lifestyle risk factors which could influence our conclusions.
Overall, our meta-analysis provided no statistically significant association between the 13 polymorphisms in six genes of PI3K/Akt signaling pathway and PCa risk. However, a significantly increased risk of PCa in Caucasian individuals was identified for IL-6-rs1800795 polymorphism in the dominant model. Due to the limitations of these included studies, as well as the risk factors we ignored, further well-designed studies with larger samples are warranted to verify our findings.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This study was supported by the Natural Science Foundation of Guangdong Province, China (2017A030313800).
Authors' contributions
W.X., M.Z., J.M. and Z.N. designed the studies and drafted the manuscript. M.Z. and Z.N. performed the statistical analysis. L.Z., S.W. and C.L. managed the experimental design, reviewed the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zhao GY, Cai CK, Yang TT, Qiu XC, Liao B, Li W. et al. MicroRNA-221 Induces Cell Survival and Cisplatin Resistance through PI3K/Akt Pathway in Human Osteosarcoma. PloS one. 2013;8:e53906
2. Jung HY, Joo HJ, Park JK, Kim YH. The Blocking of c-Met Signaling Induces Apoptosis through the Increase of p53 Protein in Lung Cancer. Cancer Res Treat. 2012;44:251-61
3. Shih MC, Chen JY, Wu YC, Jan YH, Yang BM, Lu PJ. et al. TOPK/PBK promotes cell migration via modulation of the PI3K/PTEN/AKT pathway and is associated with poor prognosis in lung cancer. Oncogene. 2012;31:2389-400
4. Al-Azayzih A, Gao F, Goc A, Somanath PR. TGFbeta1 induces apoptosis in invasive prostate cancer and bladder cancer cells via Akt-independent, p38 MAPK and JNK/SAPK-mediated activation of caspases. Biochemical and biophysical research communications. 2012;427:165-70
5. Park YH, Seo SY, Lee E, Ku JH, Kim HH, Kwak C. Simvastatin Induces Apoptosis in Castrate Resistant Prostate Cancer Cells by Deregulating Nuclear Factor-kappa B Pathway. J Urology. 2013;189:1547-52
6. Balistreri CR, Caruso C, Carruba G, Miceli V, Campisi I, Listi F. et al. A pilot study on prostate cancer risk and pro-inflammatory genotypes: pathophysiology and therapeutic implications. Current pharmaceutical design. 2010;16:718-24
7. Wang MH, Helzlsouer KJ, Smith MW, Hoffman-Bolton JA, Clipp SL, Grinberg V. et al. Association of IL10 and other immune response- and obesity-related genes with prostate cancer in CLUE II. The Prostate. 2009;69:874-85
8. Chen YC, Giovannucci E, Lazarus R, Kraft P, Ketkar S, Hunter DJ. Sequence variants of Toll-like receptor 4 and susceptibility to prostate cancer. Cancer Res. 2005;65:11771-8
9. Shui IM, Stark JR, Penney KL, Schumacher FR, Epstein MM, Pitt MJ. et al. Genetic variation in the toll-like receptor 4 and prostate cancer incidence and mortality. The Prostate. 2012;72:209-16
10. Schildkraut JM, Demark-Wahnefried W, Wenham RM, Grubber J, Jeffreys AS, Grambow SC. et al. IGF1 (CA)19 repeat and IGFBP3 -202 A/C genotypes and the risk of prostate cancer in Black and White men. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2005;14:403-8
11. Neuhausen SL, Slattery ML, Garner CP, Ding YC, Hoffman M, Brothman AR. Prostate cancer risk and IRS1, IRS2, IGF1, and INS polymorphisms: strong association of IRS1 G972R variant and cancer risk. The Prostate. 2005;64:168-74
12. Kesarwani P, Ahirwar DK, Mandhani A, Mittal RD. Association between -174 G/C promoter polymorphism of the interleukin-6 gene and progression of prostate cancer in North Indian population. Dna & Cell Biology. 2008;27:505-10
13. Mandal S, Abebe F, Chaudhary J. -174G/C polymorphism in the interleukin-6 promoter is differently associated with prostate cancer incidence depending on race. Genetics & Molecular Research. 2014;13:139-51
14. Bao S, Yang W, Zhou S, Ye Z. Relationship between Single Nucleotide Polymorphisms in -174G/C and -634C/G Promoter Region of Interleukin-6 and Prostate Cancer. J Huazhong Univ Sci Technolog Med Sci. 2008;28:693-6
15. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315:629-34
16. Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Annals of internal medicine. 1997;127:820-6
17. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. British medical journal. 2003;327:557-60
18. Yong G, Pan X, Su T, Mo Z, Cao Y, Feng G. Glutathione S-transferase P1 Ile105Val polymorphism and colorectal cancer risk: a meta-analysis and HuGE review. European Journal of Cancer. 2009;45:3303-14
19. Balistreri CR, Ccarruba C. A pilot study on prostate cancer risk and pro-inflammatory genotypes: pathophysiology and therapeutic implications. Current Pharmaceutical Design. 2010;16:718-24
20. Chen C, Freeman R, Voigt LF, Fitzpatrick A, Plymate S, Weiss N. Prostate cancer risk in relation to selected genetic polymorphisms in insulin-like growth factor-I, insulin-like growth factor binding protein-3, and insulin-like growth factor-I receptor. Cancer Epidemiology Biomarkers & Prevention. 2006;15:2461-6
21. Cheng I, Plummer SJ, Casey G, Witte JS. Toll-like receptor 4 genetic variation and advanced prostate cancer risk. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007;16:352-5
22. Dossus L, Kaaks R, Canzian F, Albanes D, Berndt SI, Boeing H. et al. PTGS2 and IL6 genetic variation and risk of breast and prostate cancer: results from the Breast and Prostate Cancer Cohort Consortium (BPC3). Carcinogenesis. 2010;31:455-61
23. Fall K, Stark JR, Mucci LA, Chan J, Stampfer MJ, Kurth T. et al. No association between a polymorphic variant of the IRS-1 gene and prostate cancer risk. Prostate. 2008;68:1416-20
24. Fitzgerald LM, Karlins E, Karyadi DM, Kwon EM, Koopmeiners JS, Stanford JL. et al. Association of FGFR4 genetic polymorphisms with prostate cancer risk and prognosis. Prostate Cancer & Prostatic Diseases. 2008;12:192-7
25. Friedrichsen DM, Hawley S, Shu J, Humphrey M, Sabacan L, Iwasaki L. et al. IGF-I and IGFBP-3 polymorphisms and risk of prostate cancer. Prostate. 2005;65:44-51
26. Fukuda H, Tsuchiya N, Narita S, Kumazawa T, Horikawa Y, Inoue T. et al. Clinical implication of vascular endothelial growth factor T-460C polymorphism in the risk and progression of prostate cancer. Oncology Reports. 2007;18:1155-63
27. Hernandez W, Grenade C, Santos ER, Bonilla C, Ahaghotu C, Kittles RA. IGF-1 and IGFBP-3 gene variants influence on serum levels and prostate cancer risk in African-Americans. Carcinogenesis. 2007;28:2154-9
28. Giachini FR, Sullivan JC, Lima VV, Carneiro FS, Fortes ZB, Pollock DM. et al. Extracellular signal-regulated kinase 1/2 activation, via downregulation of mitogen-activated protein kinase phosphatase 1, mediates sex differences in desoxycorticosterone acetate-salt hypertension vascular reactivity. Hypertension. 2010;55:172-9
29. Jacobs EJ, Hsing AW, Bain EB, Stevens VL, Wang Y, Chen J. et al. Polymorphisms in angogenesis-related genes and prostate cancer. Cancer Epidemiology Biomarkers & Prevention. 2008;17:972-7
30. Li L, Cicek MS, Casey G, Witte JS. No association between genetic polymorphisms in insulin and insulin receptor substrate-1 and prostate cancer. Cancer Epidemiology Biomarkers & Prevention. 2005;14:2462-3
31. Lin CC, Wu HC, Tsai FJ, Chen HY, Chen WC. Vascular endothelial growth factor gene-460 C/T polymorphism is a biomarker for prostate cancer. Urology. 2003;62:374-7
32. Ma Z, Tsuchiya N, Yuasa T, Narita S, Horikawa Y, Kumazawa T. et al. Polymorphisms of fibroblast growth factor receptor 4 have association with the development of prostate cancer and benign prostatic hyperplasia and the progression of prostate cancer in a Japanese population. International Journal of Cancer. 2008;123:2574-9
33. Mandal S, Abebe F, Chaudhary J. -174G/C polymorphism in the interleukin-6 promoter is differently associated with prostate cancer incidence depending on race. Genetics & Molecular Research Gmr. 2014;13:139-51
34. Michaud DS, Daugherty SE, Berndt SI, Platz EA, Yeager M, Crawford ED. et al. Genetic polymorphisms of interleukin-1B (IL-1B), IL-6, IL-8, and IL-10 and risk of prostate cancer. Cancer Research. 2006;66:4525-30
35. Moore S, Leitzmann MF, Albanes D, Weinstein SJ, Snyder K, Virtamo J, Ahn J. et al. Adipokine genes and prostate cancer risk. International Journal of Cancer. 2009;124:869-76
36. Nam RK, Zhang WW, Trachtenberg J, Jewett MA, Emami M, Vesprini D. et al. Comprehensive assessment of candidate genes and serological markers for the detection of prostate cancer. Cancer Epidemiology Biomarkers & Prevention. 2003;12:1429-37
37. Neuhausen SL, Slattery MLGarner CP, Ding YC, Hoffman M, Brothman AR. Prostate cancer risk and IRS1, IRS2, IGF1, and INS polymorphisms: strong association of IRS1 G972R variant and cancer risk. Prostate. 2005;64:168-74
38. Onen IH, Konac E, Eroglu M, Guneri C, Biri H, Ekmekci A. No association between polymorphism in the vascular endothelial growth factor gene at position-460 and sporadic prostate cancer in the Turkish population. Molecular Biology Reports. 2008;35:17-22
39. Pierce BL, Biggs ML, Decambre M, Reiner AP, Li C, Fitzpatrick A. et al. C-reactive protein, interleukin-6, and prostate cancer risk in men aged 65 years and older. Cancer Causes & Control. 2009;20:1193-203
40. Ratliff TL. Sequence Variants of Toll-Like Receptor 4 are Associated With Prostate Cancer Risk: Results From the CAncer Prostate in Sweden Study. Cancer Research. 2004;172:2918-22
41. Saracevic A, Nikolac N, Reljic A, Simundic AM. Insulin receptor H1085H C>T and insulin receptor substrate 1 G972R polymorphisms and prostate cancer risk: a pilot study. Genetic Testing & Molecular Biomarkers. 2011;15:127-31
42. Schildkraut JM, Demarkwahnefried W, Wenham RM, Grubber J, Jeffreys AS, Grambow SC. et al. IGF1 (CA)19 Repeat and IGFBP3 -202 A/C Genotypes and the Risk of Prostate Cancer in Black and White Men. Cancer Epidemiology Biomarkers & Prevention. 2005;14:403-8
43. Sfar S, Hassen E, Saad H, Mosbah F, Chouchane L. Association of VEGF genetic polymorphisms with prostate carcinoma risk and clinical outcome. Cytokine. 2006;35:21-8
44. Shui IM. et al. Genetic variation in the toll-like receptor 4 and prostate cancer incidence and mortality. Prostate. 2012;72:209-16
45. Song J. et al. The association between Toll-like receptor 4 (TLR4) polymorphisms and the risk of prostate cancer in Korean men. Cancer Genet Cytogenet. 2009;190:88-92
46. McCarron SL. et al. Influence of cytokine gene polymorphisms on the development of prostate cancer. Cancer Res. 2002;62:3369-72
47. Sun J. et al. Interleukin-6 sequence variants are not associated with prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2004;13:1677-9
48. Tsuchiya N. et al. CA repeat polymorphism in the insulin-like growth factor-I gene is associated with increased risk of prostate cancer and benign prostatic hyperplasia. Int J Oncol. 2005;26:225-31
49. Wang MH. et al. Association of IL10 and other immune response- and obesity-related genes with prostate cancer in CLUE II. Prostate. 2009;69:874-85
50. Wang MH. et al. Association of IL10 and other immune response- and obesity-related genes with prostate cancer in CLUE II. Prostate. 2009;69:874-85
51. Zabaleta J. et al. Cytokine genetic polymorphisms and prostate cancer aggressiveness. Carcinogenesis. 2009;30:1358-62
52. Zhang J. et al. Polymorphisms in inflammatory genes, plasma antioxidants, and prostate cancer risk. Cancer Causes Control. 2010;21:1437-44
53. Bao S. et al. Relationship between single nucleotide polymorphisms in -174G/C and -634C/G promoter region of interleukin-6 and prostate cancer. J Huazhong Univ Sci Technolog Med Sci. 2008;28:693-6
54. Zheng SL. et al. Sequence variants of toll-like receptor 4 are associated with prostate cancer risk: results from the CAncer Prostate in Sweden Study. Cancer Res. 2004;64:2918-22
55. Duval S, Tweedie R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455-63
56. Zhu LB, Yuan H, Jiang T, Wang RX, Ma HX, Zhang SY. Association of TLR2 and TLR4 Polymorphisms with Risk of Cancer: A Meta-Analysis. PloS one. 2013;8:e82858
57. Giacca M, Zacchigna S. VEGF gene therapy: therapeutic angiogenesis in the clinic and beyond. Gene Ther. 2012;19:622-9
58. Zhang HT, Wang AD, Ma H, Xu Y. Association between insulin receptor substrate 1 Gly972Arg polymorphism and cancer risk. Tumor Biol. 2013;34:2929-36
59. Porzio O, Federici M, Hribal ML, Lauro D, Accili D, Lauro R. et al. The Gly972->Arg amino acid polymorphism in IRS-1 impairs insulin secretion in pancreatic beta cells. The Journal of clinical investigation. 1999;104:357-64
60. Baserga R. The contradictions of the insulin-like growth factor 1 receptor. Oncogene. 2000;19:5574-81
61. Quan HY, Tang H, Fang L, Bi JJ, Liu YS, Li HL. IGF1(CA)19 and IGFBP-3-202A/C Gene Polymorphism and Cancer Risk: A Meta-analysis. Cell Biochem Biophys. 2014;69:169-78
62. Xu B, Tong N, Chen SQ, Hua LX, Wang ZJ, Zhang ZD. et al. FGFR4 Gly388Arg polymorphism contributes to prostate cancer development and progression: a meta-analysis of 2618 cases and 2305 controls. BMC cancer. 2011;11:84
63. Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S. et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. The Journal of clinical investigation. 1998;102:1369-76
64. Liu RY, Song XX, Chen P, Lei Z, Miao JC, Yi NJ. et al. Association between IL6-174G/C and cancer: A meta-analysis of 105,482 individuals. Experimental and therapeutic medicine. 2012;3:655-64
65. Du Y, Gao L, Zhang K, Wang J. Association of the IL6 polymorphism rs1800796 with cancer risk: a meta-analysis. Genetics and Molecular Research. 2015;14:13236-46
66. Liu RY, Song X, Chen P, Lei Z, Miao J, Yi N. et al. Association between IL6 -174G/C and cancer: A meta-analysis of 105,482 individuals. Experimental and therapeutic medicine. 2012;3:655-64
Author contact
![]() Corresponding authors: Chaozhao Liang, Department of Urology, the First Affiliated Hospital of Anhui Medical University, Hefei 230022, China. Tel.: +86 55162923440; Email address: liang_chaozhaoedu.cn and Song Wu, Shenzhen Following Precision Medical Institute, Shenzhen Luohu Hospital Group, Shenzhen 518000, China. Tel.: +86 755 8220 3083; Email address: doctor_wusongcom
Corresponding authors: Chaozhao Liang, Department of Urology, the First Affiliated Hospital of Anhui Medical University, Hefei 230022, China. Tel.: +86 55162923440; Email address: liang_chaozhaoedu.cn and Song Wu, Shenzhen Following Precision Medical Institute, Shenzhen Luohu Hospital Group, Shenzhen 518000, China. Tel.: +86 755 8220 3083; Email address: doctor_wusongcom

 Global reach, higher impact
Global reach, higher impact