Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(3):577-582. doi:10.7150/jca.28905 This issue Cite
Research Paper
LRP1B Polymorphisms Are Associated with Multiple Myeloma Risk in a Chinese Han Population
1. Department of Clinical Laboratory, The First Affiliated Hospital of Zhengzhou University, Key Laboratory of Laboratory Medicine of Henan Province, Zhengzhou 450052, Henan , P.R. China
2. Department of Clinical Laboratory, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100730, P.R.China.
3. Department of Laboratory Medicine, Shengjing Hospital of China Medical University, Department of Laboratory Medicine, Shengjing Hospital of China Medical University, Shenyang 110004, Liaoning, P.R.China
* These authors contributed equally to this work.
Received 2018-7-31; Accepted 2018-12-8; Published 2019-1-1
Abstract
Multiple myeloma (MM) is an extremely complex plasma cell malignancy that is genetically heterogeneous. A recent Genome-wide association study (GWAS) indicated that variation at 2q22 (rs61070260) influences MM risk. This association has not been validated to date in a Chinese Han population. In this study, we evaluated the association between rs61070260 in LRP1B and MM risk in a Chinese Han population involving 739 MM patients and 592 healthy controls. Our results indicated that rs61070260 in LRP1B was significantly associated with MM susceptibility (P=3.937×10-37). Furthermore, the linkage disequilibrium (LD) analysis of rs61070260 revealed an LD block encompassing exons 26, 27 and 28 of the LRP1B gene, and a subsequent sequencing analysis identified three SNPs (rs762074421, rs756168629, rs113600691) in exons 26 and 28 of LRP1B. For the SNP rs756168629 in exon 26, a missense mutation which results in a transition from arginine to histidine at position 1661 of the LRP1B protein, has not been found in Chinese populations according to the Chinese Millionome Database and Genome Aggregation Database (EAS), and this mutation was predicted to be deleterious or damaging by SIFT and PolyPhen. These findings firmly establish the role of LRP1B in contributing to MM susceptibility. In addition, the identification of a rare coding mutation (p.R1661H) in LRP1B detected in MM individuals was suggested to be harmful to the encoded protein, which was characterized as a candidate tumour suppressor; thus, LRP1B is likely to be a disease-associated gene that is implicated in the development and progression of MM.
Keywords: multiple myeloma, LRP1B, susceptibility, linkage disequilibrium, mutation
Introduction
Multiple myeloma (MM) is a complex B-lineage neoplasm that accounts for 1% of all malignancies [1] and 1.9% of all cancer deaths [2]. It is characterized by malignant plasma cell proliferation that is accompanied by supererogatory abnormal monoclonal proteins. MM exhibits considerable genetic heterogeneity, which can be divided into distinct groups such as hyperdiploid, hypodiploid, pseudodiploid and near-tetraploid MM, with a low or high prevalence of IgH translocations [3]. Considerable progress has been made in the treatment of MM, but the aetiology underlying this malignant disease has not been clearly delineated to date [4, 5].
Genome-wide association studies (GWASs) have often focused on identifying significant genetic components for various complex human diseases, encompassing MM. A number of reported risk loci have the consistent effects of predisposing individuals to the development of MM [6]. A recent GWAS confirmed that a polymorphism in 6q25.1 (rs12748648) was associated with MM and suggested an association at 2q22 (rs61070260) in LRP1B [7]. LRP1B is composed of 91 exons, and the encoded protein is a multifunctional cell surface receptor that belongs to the low-density lipoprotein (LDL) receptor family [8-10]. LRP1B was characterized as a candidate tumour suppressor, which significantly restrains cancer growth and invasion and, in particular, might modulate the amount of several cytokines in the tumour microenvironment through its endocytic activity [11]. Deletions of LRP1B contributed to the low expression in several types of carcinomas, which was suggested to be associated with aggressive growth [9, 12-14].
The association between the identified risk loci of rs61070260 in LRP1B with MM has not been validated to date in a Chinese Han population. The aim of this study was to determine the allele and genotype frequencies of rs61070260 in the LRP1B gene in a Chinese Han population and to evaluate the significance of their impact on MM. Furthermore, most of the trait/disease-associated SNPs (TASs) typically identified by GWASs do not clearly identify causal variant that may be the TAS itself or a known SNP in strong linkage disequilibrium (LD) with the TAS [15, 16]. It remains a major challenge to identify such causal variants. In this study, we simultaneously screened the exons in high LD with rs61070260 of the LRP1B gene to search for the potential causal variation for MM.
Materials and Methods
In this study, 739 MM patients and 592 healthy controls were used to replicate the association of rs61070260 in LRP1B with MM. Another group of 178 MM patients was used for identifying the potential causal variant(s) in LRP1B that in high LD with rs61070260. The ethics committees of the three hospitals approved the study, and informed consent was acquired from each participant.
Patient recruitment
The current study included 1331 participants (739 MM patients and 592 healthy controls) consecutively recruited from the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China), Beijing Union Medical College Hospital (Beijing, China) and Shengjing Hospital of China Medical University (Shenyang, China) between June 2015 and May 2017. The MM cases were diagnosed based on the bone marrow histopathological examination, clinical manifestations of serious end organ damage, including hypercalcemia, renal failure, anaemia and bone lesions (CRAB), and laboratory features [17]. Basic demographic and clinical information, including age at diagnosis, sex, ISS stage, haemoglobin (Hb), serum albumin (Alb), serum calcium (Ca), serum creatinine (Crea), and β2-microglobulin (β2-MG), was collected. A total of 592 healthy controls that were free of monoclonal gammopathy of undetermined significance or MM were recruited from the groups of patients who had physical examinations in these hospitals. Blood samples (2 ml) were collected from each subject as a source of DNA.
Genotyping
The genomic DNA of 739 MM patients and 592 healthy controls was extracted by using a TIANamp Blood DNA Kit (Tiangen Biotech Co., Ltd., Beijing, China) according to the manufacturer's instructions. Primer was designed using the Sequenom MassARRAY Assay Design 4.0 Software for the multiplex polymerase chain reaction (PCR) and for locus-specific single-base extension. PCR was conducted in a 384-well plate, and the products were subjected to locus-specific single-base extension reactions. After desalting, the products were dispensed onto a 384-format SpectroCHIP array (Sequenom), and genotyping was performed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS). The mass spectrograms and genotype data were analysed using MassArray Typer software version 4.0 (Sequenom) [18].
Statistical test for association
The statistical analyses were conducted using the PLINK tool set. Departure from HWE of rs61070260 frequency was assessed by the χ2 test in control subjects [19]. Genotype and allele frequencies of the cases and controls were assessed using Fisher's exact test under the allele, dominant and recessive models and using logistic regression analysis under an additive model. The sex and age of the patients contributing the samples were used as covariates. The odds ratio (OR) and 95% confidence interval (95% CI) were calculated, and P values less than 0.05 were considered to be statistically significant. Conditional logistic regression analyses were also performed for the associations of rs61070260 with the clinical characteristics of MM patients.
LD analysis
SNP genotype data of the HapMap CHB population were used to search for genomic regions in high LD (r2 > 0.8) with rs61070260 and the LD plot was constructed using the Haploview software.
Sequencing for causal variant(s)
Based on the results of the LD analysis, exons 26, 27, and 28 were sequenced in a new group of 178 patients with MM. Genomic DNA was extracted as previously mentioned and the primer sequences are listed in Table S1. Fragments were amplified and were subjected to Sanger sequencing. All sequencing chromatograms were compared with the reference sequence using Lasergene to detect the mutations.
The allele frequencies for the potential causal variants in current MM patients were compared to the Chinese Millionome Database (CMDB) and Genome Aggregation Database (gnomAD) (EAS) [20]. The effect of the causal variant on the protein function was estimated by SIFT [21] and PolyPhen [22].
Clinical features of subjects enrolled in this case-control study
| Characteristics | Classification | MM Patients | Controls |
|---|---|---|---|
| Age, y (mean ± SD) | 59.27 ± 10.11 | 47.83 ± 12.66 | |
| Sex, n (%) | Female | 324 (43.84) | 250 (42.23) |
| Male | 415 (56.16) | 342 (57.77) | |
| ISS stage, n (%) | I | 45 (6.09) | - |
| II | 105 (14.21) | - | |
| III | 184 (24.90) | - | |
| Isotype (Immunoglobulin), n (%) | IgM | 4 (0.54) | - |
| IgD | 21 (2.84) | - | |
| IgA | 144 (19.49) | - | |
| IgG | 312 (42.22) | - | |
| Light chain | 176 (23.82) | - | |
| Nonsecretory | 23 (3.11) | - | |
| NA | 59 (7.98) | - | |
| Isotype (Light chain), n (%) | κ | 312 (42.22) | - |
| λ | 345 (46.68) | - | |
| Nonsecretory | 23 (3.11) | - | |
| NA | 59 (7.98) | - | |
| Biochemical parameter (mean, range) | Ca (mmol/L) | 2.17 (1.13-4.41) | - |
| Crea (µmol/L) | 115.29 (5.30-857) | - | |
| β2-MG (mg/L) | 5.35 (0.73-38.5) | - | |
| Hb (g/L) | 115.01 (29.8-169) | - | |
| Alb (g/L) | 35.48 (2.70-51.00) | - |
Abbreviations: Alb, albumin; β2-MG, β2-microglobulin; Ca, calcium; Crea, creatinine; Hb, haemoglobin; κ, Kappa; λ, Lambda; NA, not applicable.
Results
Clinical Features of the Subjects
A total of 1331 participants, including 739 MM cases and 592 controls, were involved in this case-control study. Basic demographic and clinical information of the MM patients and controls are shown in Table 1. The average age was significantly lower for the 592 healthy controls than for the MM patients (P<0.001). This finding may be because MM is typical in elderly people, and over one-half of MM patients are older than 65 years at diagnosis [23, 24]; in contrast, healthy controls were mainly recruited during their physical examinations, and these were mostly young men.
Rs61070260 was strongly associated with MM susceptibility
No significant deviation from Hardy-Weinberg equilibrium (HWE) was observed for rs61070260 in the controls (P>0.05), and the case-control study showed a significant association between the SNP rs61070260 and MM risk, based on the result of the increased A allelic frequencies in MM patients (P=3.937×10-37) and clear differences in the genotype distribution in MM patients and healthy controls under the dominant model (AA+AG vs. GG: P=1.551×10-134), recessive model (AA vs. AG+GG: P=3.261×10-9), and additive model (AA vs. AG vs. GG: P=9.622×10-74) (Table 2). In the stratified analysis, no association was found between rs61070260 and any of the clinical features of MM patients (all P>0.05) (Table 3).
Genetic association analysis of SNP rs16070260 in patients with MM and controls
| SNP | Genetic model | Description | Cases/Controls | P value* |
|---|---|---|---|---|
| rs16070260 (G>A) | Allele model | A | 740/738 | 3.937×10-37 |
| G | 308/876 | |||
| Dominant model | AA+AG | 735/271 | 1.551×10-134 | |
| GG | 4/321 | |||
| Recessive model | AA | 5/37 | 3.261×10-9 | |
| AG+GG | 734/555 | |||
| Additive model | AA | 5/37 | 9.622×10-74 | |
| AG | 730/234 | |||
| GG | 4/321 |
* The additive model was analysed using regression analysis, and the other model was analysed by Fisher's exact test.
Association analysis of SNP rs61070260 with various clinical characteristics of MM patients
| Characters of MM | Phenotypes | Number (P/N) | rs61070260 (G>A) | |||
|---|---|---|---|---|---|---|
| F | χ2 | P value | OR | |||
| Immunoglobulin Isotype | Heavy chain paraprotein | 481/23 | G (0.4979/0.500) | 0.001188 | 0.9725 | 0.9917 |
| Light chain paraprotein | 176/23 | G (0.4915/0.500) | 0.01777 | 0.894 | 0.9665 | |
| Heavy vs. Light | 481/176 | G (0.4915/0.5021) | 0.1159 | 0.7336 | 0.9585 | |
| Biochemical parameter | Crea (µmol/L) | 148/554 | G (0.4966/0.4991) | 0.005728 | 0.9397 | 0.9901 |
| β2-MG (mg/L) | 420/151 | G (0.4975/0.4967) | 0.0005958 | 0.9805 | 1.003 | |
| Hb (g/L) | 402/313 | G (0.500/0.4984) | 0.003593 | 0.9522 | 1.006 | |
| Alb (g/L) | 294/408 | G (0.4983/0.4988) | 0.0003087 | 0.986 | 0.9981 | |
Abbreviations: N, patients negative for a certain phenotype; P, patients positive for a certain phenotype.
Candidate causal variation identified by Sanger sequencing
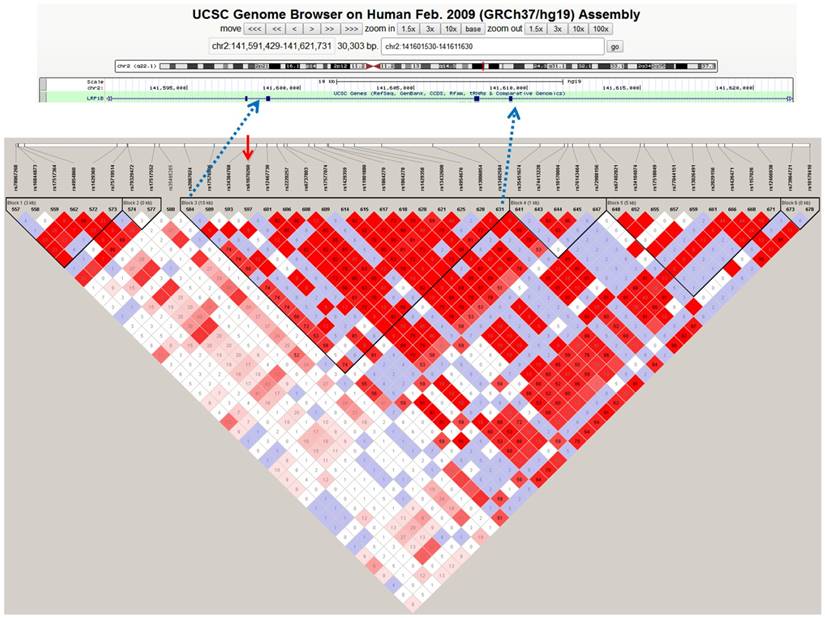
The LD analysis of rs61070260 revealed an LD block encompassing exons 26, 27 and 28 of the LRP1B gene (Figure 1). Therefore, Sanger sequencing of the three exons was conducted in another 178 MM patients to identify causal variations. We discovered three variant loci, rs113600691 (A>G) in exon 28 and rs762074421 (T>C) and rs756168629 (C>T) in exon 26 (Figure S1). Exon 27 was successfully sequenced, whereas no variant was found in this region. The SNPs rs113600691 and rs762074421 are located in the intronic region. The C allele frequency of rs762074421 and the G allele frequency of rs113600691 in MM patents were 0.0028 and 0.0112, respectively, which were lower than those in gnomAD (EAS) (Table 4). The SNP rs756168629 is a missense mutation located in the coding region (exon 26), which results in a substitution from arginine to histidine at position 1661 of the LRP1B protein. The frequency of the mutant T allele of this mutation in our MM patients was 0.0056. Particularly, according to the CMDB and gnomAD (EAS), this rare variant has not been found in Chinese populations to date. Furthermore, this variant is predicted to be deleterious or damaging by SIFT and PolyPhen. These findings provide original evidence of the role of the LRP1B gene in MM susceptibility and potentially its underlying pathogenesis.
Haplotype block of LRP1B containing rs61070260 based on 1000 genome CHB data. The standard Haploview LD colour scheme was based on D' and LOD (log of the likelihood odds ratio), and the number (divided by 100) in the small square represents the r2 value ranging from 0 (no linkage) to 1 (complete linkage). Haplotype blocks were defined using the confidence interval (CI) method. The red arrow indicates the position of the SNP rs61070260, which was strongly associated with MM risk.
Mutant, SIFT and PolyPhen results for LRP1B
| SNP ID | Localization | Exon | Number of patients (n/N) | Nucleotide change | Amino acid change | SIFT | PolyPhen | AF in MM patients | AF in gomAD (EAS) |
|---|---|---|---|---|---|---|---|---|---|
| rs762074421 | chr2: 141598463 | 26 | 1/178 | T>C | - | - | - | 0.0028 (1/356) | 0.0750 (3/40) |
| rs756168629 | chr2: 141598619 | 26 | 1/178 | C>T | R1661H | Deleterious | damaging | 0.0028 (1/356) | 0 (0/17220) |
| rs113600691 | chr2: 141609507 | 28 | 4/178 | A>G | - | - | 0.0112 (4/356) | 0.0220 (35/1622) |
Abbreviations: AF, allele frequency; gnomAD (EAS), Genome Aggregation Database (East Asians); n, number of patients with certain gene mutation; N, number of patients involved in the sequencing.
Discussion
The results presented in the current study imply the strong association of the SNP rs61070260 in LRP1B and MM risk in Chinese Han population. The role of LRP1B polymorphisms in predisposition to pancreas cancer as well as non-malignant diseases, such as Alzheimer's disease, increased ventricular volumes in psychosis, preeclampsia and cardiovascular disease has been reported [25-28]. A recent GWAS by Johnson et al. identified a suggestive association between rs61070260 and the risk of MM patients of European ancestry [7]. The present study confirmed this association in a Chinese Han population, but further stratified analysis according to various characteristics of our MM patients, including immunoglobulin isotype and biochemical parameters (Hb, Crea, Alb, and β2-MG), showed no correlation with the SNP rs61070260.
Rs61070260 is located in the intron region of LRP1B, and it is likely to be functional or in linkage disequilibrium with another nearby functional variant. To discover the potential causal variants in LD with rs61070260 in MM, we performed an LD analysis of LRP1B. The results suggest that rs61070260 was located in an LD block encompassing exons 26, 27 and 28. Our sequencing analysis of 178 MM patients in these three exons revealed a rare missense variant (rs756168629, 4982 C>T), which causes a substitution of arginine to histidine in position 1661 and is predicted to be deleterious and damaging by SIFT and PolyPhen. It is highly possible that this rare variant may cause genetic defects in the LRP1B gene, leading to an increased susceptibility to MM.
LRP1B is characterized as a candidate tumour suppressor gene because the homozygous deletion of individual exons is observed in non-small cell lung cancer cell lines [29, 30]. The protein encoded by the LRP1B gene is a multifunctional cell surface receptor that is a member of the LDL receptor family. The ability of the LRP1B protein to modulate the urokinase plasminogen system (uPA system) is thought to be a major underlying factor for the suppression of tumour invasion and metastasis. LRP1B displays a considerably slow rate of internalization of the uPA-PAI-1 complex, resulting in the subdued regeneration of unoccupied uPAR on the cell surface, which diminishes cell migration [31]. The down regulation of LRP1B in renal cell cancer tissues and cell lines is frequently observed, and the depletion of LRP1B increased cell migration and invasion in vitro and in vivo [14], which strongly validated that LRP1B may function as a tumour suppressor. Hitherto, point mutations, homozygous deletions, and DNA methylation of LRP1B have been detected in multiple primary cancers [9]. The variant that we detected in MM patients is damaging, indicating that it may contribute to the loss of LRP1B expression or be linked with specific clinical characteristics. Nevertheless, the prediction and deduction of the mutation remain to be verified.
The sample size of our study was limited, and large-scale population-based studies are warranted to validate our findings, especially the rare variant rs61070260 in MM patients. In addition, further functional studies that focus on delineating how variants of LRP1B identified in this study alter protein function and affect the development of MM are essential.
In summary, we confirmed the association between SNP rs61070260 in LRP1B and MM risk in a Chinese Han population and identified a rare mutation (p.R1661H) that may cause LRP1B protein defects. Further functional studies are required to fully understand the biological role of LRP1B in MM.
Abbreviations
MM: Multiple myeloma; GWAS: Genome-wide association studie; LDL: low density lipoprotein; HWE: Hardy-Weinberg equilibrium; OR: odds ratio; CI: confidence interval.
Supplementary Material
Supplementary figure and table.
Acknowledgements
This research was supported by grants from the National Natural Science Foundation of China Grants (81671618), the Capital Health Research and Development of Special (2014-1-4011), the CAMS Initiative for Innovative Medicine (2017-I2M-3-001), the CAMS Initiative for Innovative Medicine (2017-I2M-B&R-01), and the Scientific and Technological Project of Henan Province (172102310077).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bräutigam M, Biskup E. [CME: Multiple Myeloma - a Review]. Praxis (Bern 1994). 2018;107:749-54
2. Rand K, Song C, Dean E, Serie D, Curtin K, Sheng X. et al. A Meta-analysis of Multiple Myeloma Risk Regions in African and European Ancestry Populations Identifies Putatively Functional Loci. Cancer Epidemiol Biomarkers Prev. 2016;25:1609-18
3. Rio-Machin A, Ferreira B, Henry T, Gómez-López G, Agirre X, Alvarez S. et al. Downregulation of specific miRNAs in hyperdiploid multiple myeloma mimics the oncogenic effect of IgH translocations occurring in the non-hyperdiploid subtype. Leukemia. 2013;27:925-31
4. Bianchi G, Anderson KC. Understanding biology to tackle the disease: Multiple myeloma from bench to bedside, and back. CA: a cancer journal for clinicians. 2014;64:422-44
5. Soekojo C, de Mel S, Ooi M, Yan B, Chng W. Potential Clinical Application of Genomics in Multiple Myeloma. International journal of molecular sciences. 2018;19:1721
6. Li N, Johnson DC, Weinhold N, Kimber S, Dobbins SE, Mitchell JS. et al. Genetic Predisposition to Multiple Myeloma at 5q15 Is Mediated by an ELL2 Enhancer Polymorphism. Cell reports. 2017;20:2556-64
7. Johnson DC, Weinhold N, Mitchell JS, Chen B, Kaiser M, Begum DB. et al. Genome-wide association study identifies variation at 6q25.1 associated with survival in multiple myeloma. Nat Commun. 2016;7:10290
8. Liu CX, Musco S, Lisitsina NM, Yaklichkin SY, Lisitsyn NA. Genomic organization of a new candidate tumor suppressor gene, LRP1B. Genomics. 2000;69:271-4
9. Prazeres H, Salgado C, Duarte C, Soares P. LRP1B (low density lipoprotein receptor-related protein 1B). Springer New York. 2014
10. Liu CX, Li Y, Obermoeller-McCormick LM, Schwartz AL, Bu G. The putative tumor suppressor LRP1B, a novel member of the low density lipoprotein (LDL) receptor family, exhibits both overlapping and distinct properties with the LDL receptor-related protein. The Journal of biological chemistry. 2001;276:28889-96
11. Prazeres H, Torres J, Rodrigues F, Pinto M, Pastoriza MC, Gomes D. et al. Chromosomal, epigenetic and microRNA-mediated inactivation of LRP1B, a modulator of the extracellular environment of thyroid cancer cells. Oncogene. 2011;30:1302-17
12. Tabouret E, Labussiere M, Alentorn A, Schmitt Y, Marie Y, Sanson M. LRP1B deletion is associated with poor outcome for glioblastoma patients. Journal of the neurological sciences. 2015;358:440-3
13. Wang Z, Sun P, Gao C, Chen J, Li J, Chen Z. et al. Down-regulation of LRP1B in colon cancer promoted the growth and migration of cancer cells. Experimental cell research. 2017;357:1-8
14. Ni S, Hu J, Duan Y, Shi S, Li R, Wu H. et al. Down expression of LRP1B promotes cell migration via RhoA/Cdc42 pathway and actin cytoskeleton remodeling in renal cell cancer. Cancer Sci. 2013;104:817-25
15. Beaudoin M, Goyette P, Boucher G, Lo K, Rivas M, Stevens C. et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 2013;9:e1003723
16. Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9362-7
17. Kyle RA, Rajkumar SV. Criteria for diagnosis, staging, risk stratification and response assessment of multiple myeloma. Leukemia. 2009;23:3-9
18. Peng M, Zhao G, Yang F, Cheng G, Huang J, Qin X. et al. NCOA1 is a novel susceptibility gene for multiple myeloma in the Chinese population: A case-control study. PloS one. 2017;12:e0173298
19. Li B, Shi X, Yuan Y, Peng M, Jin H, Qin D. ERCC1 rs11615 polymorphism increases susceptibility to breast cancer: a meta-analysis of 4547 individuals. Bioscience reports. 2018;38(3):BSR20180440
20. Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285-91
21. Kumar P, Henikoff S, Ng P. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073-81
22. Adzhubei I, Jordan D, Sunyaev S. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;07:Unit7.20
23. Larocca A, Palumbo A. Frail Patients with Newly Diagnosed Multiple Myeloma. Neoplastic Diseases of the Blood, 6th ed. Cambridge: Cambridge University Press. 2018:539-549
24. Salvini M, D'Agostino M, Bonello F, Boccadoro M, Bringhen S. Determining treatment intensity in elderly patients with multiple myeloma. Expert review of anticancer therapy. 2018;18:917-30
25. Cotterchio M, Lowcock E, Bider-Canfield Z, Lemire M, Greenwood C, Gallinger S. et al. Association between Variants in Atopy-Related Immunologic Candidate Genes and Pancreatic Cancer Risk. PloS one. 2015;10:e0125273
26. Alliey-Rodriguez N, Grey TA, Shafee R, Padmanabhan J, Tandon N, Klinger M. et al. Common variants of NRXN1, LRP1B and RORA are associated with increased ventricular volumes in psychosis - GWAS findings from the B-SNIP deep phenotyping study. bioRxiv. 2017 doi:10.1101/175489
27. Shang Z, Lv H, Zhang M, Duan L, Wang S, Li J. et al. Genome-wide haplotype association study identify TNFRSF1A, CASP7, LRP1B, CDH1 and TG genes associated with Alzheimer's disease in Caribbean Hispanic individuals. Oncotarget. 2015;6:42504-14
28. Løset M, Johnson M, Melton P, Ang W, Huang R, Mori T. et al. Preeclampsia and cardiovascular disease share genetic risk factors on chromosome 2q22. Pregnancy hypertension. 2014;4:178-85
29. Liu CX, Musco S, Lisitsina NM, Forgacs E, Minna JD, Lisitsyn NA. LRP-DIT, a putative endocytic receptor gene, is frequently inactivated in non-small cell lung cancer cell lines. Cancer research. 2000;60:1961-7
30. Liu CX, Musco S, Lisitsina NM, Yaklichkin SY, Lisitsyn NA. Genomic Organization of a New Candidate Tumor Suppressor Gene, LRP1B ☆. Genomics. 2000;69:271-4
31. Li Y, Knisely J, Lu W, McCormick L, Wang J, Henkin J. et al. Low density lipoprotein (LDL) receptor-related protein 1B impairs urokinase receptor regeneration on the cell surface and inhibits cell migration. J Biol Chem. 2002;277:42366-71
Author contact
![]() Corresponding authors: Dongchun Qin, Email: qindongchunedu.cn; or Yongzhe Li, Email: yongzhelipumchcom.
Corresponding authors: Dongchun Qin, Email: qindongchunedu.cn; or Yongzhe Li, Email: yongzhelipumchcom.

 Global reach, higher impact
Global reach, higher impact