Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(2):539-546. doi:10.7150/jca.25507 This issue Cite
Research Paper
Inhibition of malignant human bladder cancer phenotypes through the down-regulation of the long non-coding RNA SNHG7
Hainan General Hospital, Haikou 570311, China
* First author
Received 2018-2-12; Accepted 2018-10-16; Published 2019-1-1
Abstract
There is abundant evidence that long non-coding RNAs play important roles in the development of tumors. In the present study, our main aim was to explore the relationship between lncRNA SNHG7 and human bladder cancer cells, thus finding a novel target for bladder cancer therapy and diagnosis. Expression of lncRNA SNHG7 was evaluated using real-time quantitative polymerase chain reaction in bladder tumor tissues and paired adjacent normal tissues from 72 patients diagnosed with urothelial bladder carcinoma. We analyzed the differences in expression according to grading and staging. Human bladder cancer cell lines UMUC, 5637, T24 and SW780 were transiently transfected with lncRNA SNHG7-specific siRNA and negative control siRNA. The changes in malignant phenotypes in transfected bladder cancer cells were determined using CCK-8 assay, wound-healing assay and ELISA. We found that lncRNA SNHG7 was correlated with human bladder cancer. lncRNA SNHG7 was overexpressed in bladder cancer tissues compared to paired normal tissues and expression of SNHG7 was higher in high-grade than low-grade tumors. The malignant phenotypes were significantly inhibited when we inhibited expression of lncRNA SNHG7 in several bladder cell lines. SNHG7 plays an oncogenic role in human bladder cancer and may be a potential novel therapeutic target for treating bladder cancer.
Introduction
Bladder cancer is one of the most frequent types of cancer worldwide in the male urinary system. The most common histopathological type of bladder cancer is urothelial carcinoma. Genetic and epigenetic changes play important roles in the development and progression of bladder cancer [1, 2]. At present, surgery, chemotherapy and radiotherapy are widely used to treat bladder cancer. However, such traditional therapeutic approaches also cause severe adverse effects as well as their tumoricidal effect in bladder cancer. In order to treat bladder cancer effectively and reduce the adverse effects, molecular-targeted therapy has aroused much attention from clinical scientists. Therefore, it is necessary to find a series of effective molecules that are highly correlated with the development of bladder cancer.
Long non-coding RNAs (lncRNAs) are a special kind of non-protein-coding RNA transcripts > 200 nt in length [3]. In recent years, cancer biomarker research has focused on ncRNAs, particularly on lncRNAs, which play an important role in regulating gene expression [4-7]. In addition, many studies have reported that lncRNAs are related to malignant phenotypes of cancer cells [8-11]. It has also been reported that expression of lncRNAs is correlated with cancer grading and staging. Therefore, lncRNAs are considered to special regulatory elements for the development of cancer. However, investigation of dysregulation and function of lncRNAs in cancer has just begun and more research is urgently needed for deeper understanding of the lncRNA regulatory network.
SNHG7 is a newly discovered lncRNA that is closely related to the development and carcinogenesis of several tumors, such as lung cancer [12], renal cell carcinoma [13] and hepatocellular carcinoma [14]. In our study, we found that SNHG7 was up-regulated in bladder cancer tissues compared to the paired adjacent normal tissues. We also found that expression of SNHG7 was higher in high-grade than low-grade tumor tissues. SNHG7 was more abundant in invasive than in non-invasive tumors. Therefore, we supposed that SNHG7 might be a potential effective biomarker in bladder cancer that could enable effective molecular-targeted therapy.
Materials and Methods
Patient samples
We collected 40 tumor specimens from the patients diagnosed with urothelial bladder carcinoma who were treated with partial or radical cystectomy. We preserved bladder cancer and paired adjacent normal tissues from patients by snap-freezing in liquid nitrogen immediately after surgical removal. Each patient agreed to donate their tumor tissues for our study and we obtained written informed consent from them. Our study was approved by the Institutional Review Board of our hospital.
Cell culture
The human bladder cancer cell lines SW780, T24, UMUC and 5637 were obtained from the Institute of Cell Research, Chinese Academy of Sciences, Shanghai, China. All the cells ware cultivated with minimal essential medium (Invitrogen, Carlsbad, CA, USA) and supplemented with 10% fetal bovine serum (FBS) (Invitrogen). The cell lines were grown in a 5% CO2 atmosphere.
Real-time quantitative polymerase chain reaction (qRT-PCR)
Total RNA of cancer cells was extracted from the tissue samples or cancer cell lines utilizing the TRIzol reagent (Invitrogen). cDNAs were synthesized from total RNA and extracted with the RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Hanover, MD, USA). The primer sequences of lncRNA SNHG7 were shown as follows: lncRNA-SNHG7 primers forward: 5'-GTCAGCCGCATCTTCTTTTG-3', reverse: 5'-GCGCCCAATACGACCAAATC-3'; GAPDH primers forward: 5'-CGCTCTCTGCTCCTCCTGTTC-3', reverse: 5'-ATCCGTTGACTCCGACCTTCAC-3'. Real-time PCR was performed in a total reaction volume of 20 μl including 0.4 μl forward primer, 0.4 μl reverse primer, 0.4 μl ROX Reference Dye, 1 μl First-Strand cDNA, 10 μl 2× All-in-OneTM qPCR Mix (GeneCopoiea, Rockville, MD, USA) and 7.8 μl double-distilled water. The reactions were performed and analyzed utilizing the ABI PRISM 7000 Fluorescent Quantitative PCR System (Applied Biosystems, Foster City, CA, USA). We performed the PCR in triplicate utilizing GAPDH as an internal control. The expression of lncRNA-SNHG7 was analyzed and evaluated relative to that of GAPDH mRNA. All results are shown as the mean ± SD of three independent experiments.
siRNA transfection
lncRNA-SNHG7 siRNA of was synthesized by Genepharma (Suzhou, China). The siRNA sequences: (sense) 5'-GCUGGAAUAAAGAGUAACAUU-3'; (antisense) 5'-AAUGUUACUCUUUAUUCCAGC-3'. We also purchased the negative control siRNA from Genepharma. We incubated the cells and transfected lncRNA-SNHG7 siRNA or negative control siRNA into cells when they were grown to 70%-80% confluence in tissue culture plates, using Lipofectamine 3000 Transfection Reagent (Invitrogen).
Cell proliferation assay
Cell proliferation was evaluated using Cell Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology, Shanghai, China). The cells were cultivated in 96-well plates for 24 h with normal medium. The cells were transfected with lncRNA-SNHG7 siRNA or negative control siRNA when the cells grew to 70%-80% confluence with Lipofectamine 3000. Ten microliters of CCK-8 (5 mg/ml) was added to each well after transfection at 0, 24, 48 and 72 h. The cells were incubated for 1 h and OD450 was measured using a microplate reader (Bio-Rad, Hercules, CA, USA). Finally, the OD450 values were converted to cell numbers by utilizing standard curves.
Apoptosis assay
Apoptosis was evaluated utilizing the Caspase-3 ELISA Kit (R&D, Minneapolis, MN, USA). The cells were cultivated in 96-well plates for 24 h with normal medium. The cells were transfected with lncRNA-SNHG7 siRNA or negative control siRNA when the cells grew to 70%-80% confluence with Lipofectamine 3000. At 48 h after transfection, apoptosis was evaluated by measuring and analyzing the activity of caspase-3. The microplate reader (Bio-Rad) was used to measure OD450. We calculated the ratios between the OD450 values of lncRNA-SNHG7 siRNA transfected cells and those of negative control siRNA transfected cells.
Wound healing assay
Bladder cancer cell lines were transfected with lncRNA-SNHG7 siRNA or negative control siRNA when the cells grew to 70%-80% confluence. The monolayer cells were scratched and then cultivated under normal conditions. We measured the migration distance at 0, 12 and 24 h after scratching for each bladder cancer cell line.
Transwell assay
A total of 104 SW780, T24, UMUC and 5637 cells (~150 µl) were collected with serum-free medium and spread into the upper chamber. We filled the lower chamber with 600 μl of medium that contained 10% FBS. We pre-coated the membrane with Matrigel (Corning Inc., Corning, NY, USA) for 6 h at 37° C. The plates were incubated at 37° C and allowed to grow for 24 h. After 24 h, the membranes were fixed with pre-cooled methanol and stained with crystal violet.
Western blotting
We lysed the bladder cancer cells using RIPA protein extraction reagent (Beyotime, Beijing, China) supplemented with protease inhibitor cocktail and phenylmethylsulfonyl fluoride (Roche, Basel, Switzerland). The Bio-Rad Protein Assay Kit was utilized to measure the protein concentration. We performed western blotting according to the traditional western blot assay method [15]. GAPDH was utilized as an internal control. Antibodies (1:1000) against p21, Bax and E-cadherin were purchased from Cell Signaling Technology (Boston, MA, USA).
Statistical analysis
The differences in lncRNA-SNHG7 expression status between bladder cancer tissues and paired adjacent normal tissues were analyzed utilizing a paired samples t-test. The differences in lncRNA-SNHG7 expression between cancer subgroups were analyzed utilizing an independent samples t-test. The CCK-8 assay was analyzed at each time point using an independent samples t-test. Wound healing and apoptosis assays were also analyzed by an independent samples t-test. We performed all these statistical tests using SPSS version 21.0. P < 0.05 was considered statistically significant.
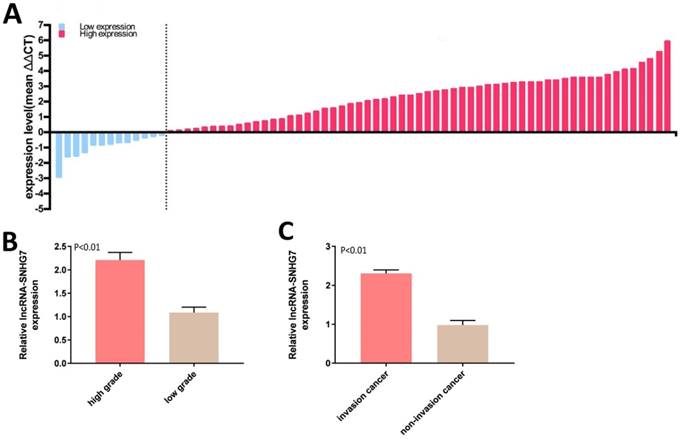
LncRNA-SNHG7 was overexpressed in bladder cancer. Relative lncRNA-SNHG7 expression was detected by utilizing qRT-PCR. A: The heights of the columns in the chart represent the log2-transformed fold changes (tumor/normal) in lncRNA-SNHG7 expression in 72 patients. B: Expression of lncRNA-SNHG7 was significantly higher in high-grade than low-grade cancer (P < 0.01). C: Expression of lncRNA-SNHG7 was higher in invasive than in non-invasive cancer. Data are shown as mean ± SEM.
Results
LncRNA-SNHG7 was overexpressed in bladder cancer tissues
Expression of lncRNA-SNHG7 was measured by qRT-PCR in 72 bladder cancer tissue samples. lncRNA-SNHG7 was overexpressed in bladder cancer compared to paired adjacent normal tissues. Expression status of lncRNA-SNHG7 in each sample (bladder cancer tissue/paired adjacent normal tissue) is shown in Figure 1A and the clinical characteristics of this set of patients are indicated in Table 1. We also analyzed the differences in lncRNA-SNHG7 expression according to grading and staging. lncRNA-SNHG7 was highly overexpressed (3.4-fold higher) in high-grade compared to low-grade tumors (Fig. 1B). We also found that lncRNA-SNHG7 expression was 2.45-fold higher in invasive tumors than in non-invasive tumors, (Fig. 1C).
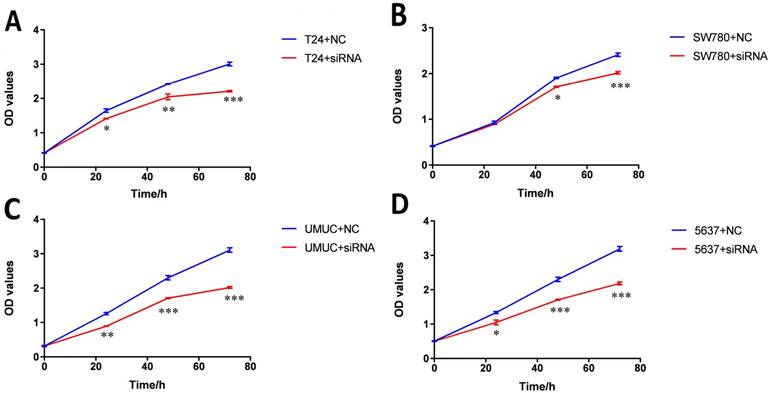
Knockdown of lncRNA-SNHG7 inhibited cell proliferation. Cell proliferation was evaluated by CCK-8 assay. After transfection of lncRNA-SNHG7 or negative control siRNA, we measured OD450 values and converted them to cell numbers. ANOVA was utilized to compare cell proliferation curves. A: Cell proliferation inhibition in bladder cancer cell line SW780. B: Cell proliferation inhibition in bladder cancer cell line T24. C: Cell proliferation inhibition in bladder cancer cell line UMUC. D: Cell proliferation inhibition in bladder cancer cell line 5637. Data are shown as mean ± SD. Each experiment in all cancer cell lines was performed in triplicate for three independent times. (*p<0.05, **p< 0.01, ***p< 0.001)
Correlation between lncRNA-SNHG7 expression and clinical characteristics of bladder cancer patients
| Parameters | Group | Total | lncRNA-SNHG7 | expression | P value |
|---|---|---|---|---|---|
| High | Low | ||||
| Age (years) | <60 | 29 | 25 | 4 | 0.440 |
| >60 | 43 | 34 | 9 | ||
| Gender | Male | 40 | 35 | 5 | 0.171 |
| Female | 32 | 24 | 8 | ||
| Tumor size (cm) | <3 cm | 42 | 31 | 11 | 0.034 |
| ≥3 cm | 30 | 28 | 2 | ||
| Histological grade | Low | 38 | 26 | 12 | 0.002 |
| High | 34 | 33 | 1 | ||
| TNM stage | I | 24 | 14 | 10 | 0.001 |
| II/III/IV | 48 | 45 | 3 | ||
| Lymphatic metastasis | NO | 31 | 20 | 11 | 0.001 |
| N1 or above | 41 | 39 | 2 |
Inhibition of cell proliferation by knockdown of lncRNA-SNHG7
Bladder cancer cell lines SW780, T24, UMUC and 5637 were transfected with lncRNA-SNHG7 siRNA or negative control siRNA. At 48 h after transfection, we analyzed lncRNA-SNHG7 expression using qRT-PCR. The inhibitory rate (lncRNA-SNHG7/negative control siRNA) was 83.22 ± 2.71 in SW780 cells, 79.45 ± 3.02 in T24 cells, 80.02 ± 2.91 in UMUC cells and 77.23 ± 1.98 in 5637 cells. We performed each experiment in triplicate for three independent times. We transfected the lncRNA-SNHG7 or negative control siRNA into SW780, T24, UMUC and 5637 cell lines. Changes in bladder cancer cell proliferation were determined using the CCK-8 assay. Cell proliferation arrest was observed in SW780 cells (Fig. 2A), T24 cells (Fig. 2B), UMUC cells (Fig. 2C) and 5637 cells (Fig. 2D).
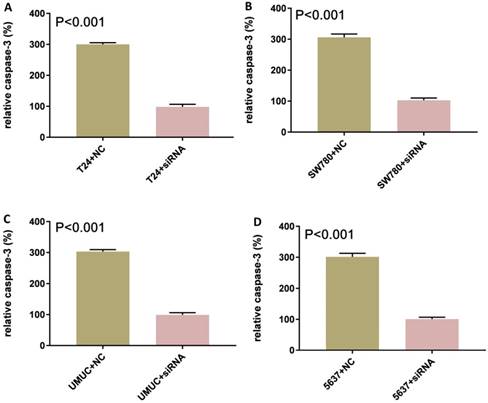
Knockdown of lncRNA-SNHG7-induced apoptosis
Bladder cancer cell lines SW780, T24, UMUC and 5637 were cultured and transfected with lncRNA-SNHG7 or negative control siRNA. After transfection, we cultured these cells for 48 h. Changes in apoptosis in SW780, T24, UMUC and 5637 cell lines were determined by ELISA (Fig. 3). We concluded that knock down of lncRNA-SNHG7 in the bladder cell lines induced apoptosis.
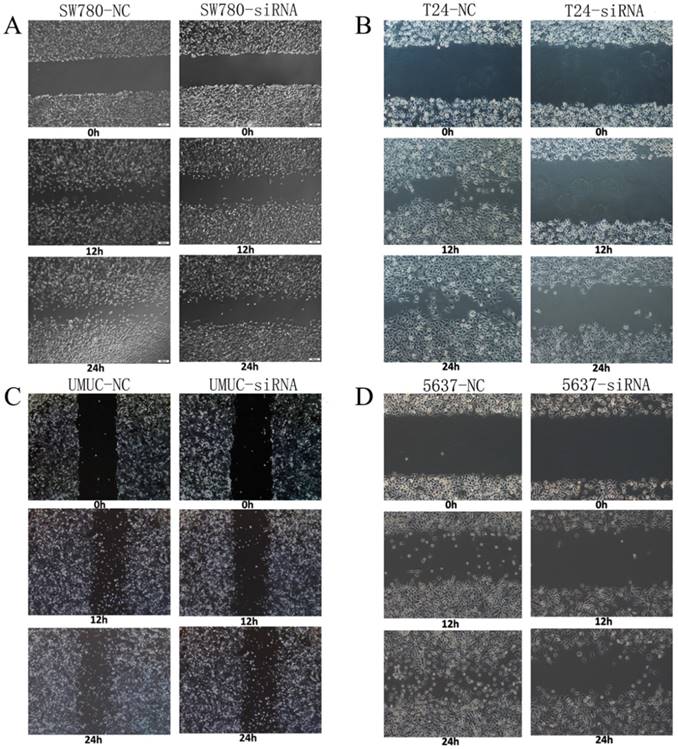
Motility changes induced by knockdown of lncRNA-SNHG7
Changes in motility of bladder cancer cell lines SW780, T24, UMUC and 5637 were detected and analyzed by wound healing assay. We transfected lncRNA-SNHG7 and negative control siRNA into all the bladder cell lines and cultured them for 48 h. We performed the wound healing assay in monolayer culture and then cultured the cells under normal cell culture conditions. Decreased cell motility was observed in SW780, T24, UMUC and 5637 cell lines (Fig. 4). We concluded that silencing lncRNA-SNHG7 repressed the motility of cancer cells.
Knockdown of lncRNA-SNHG7 inhibited cell invasion
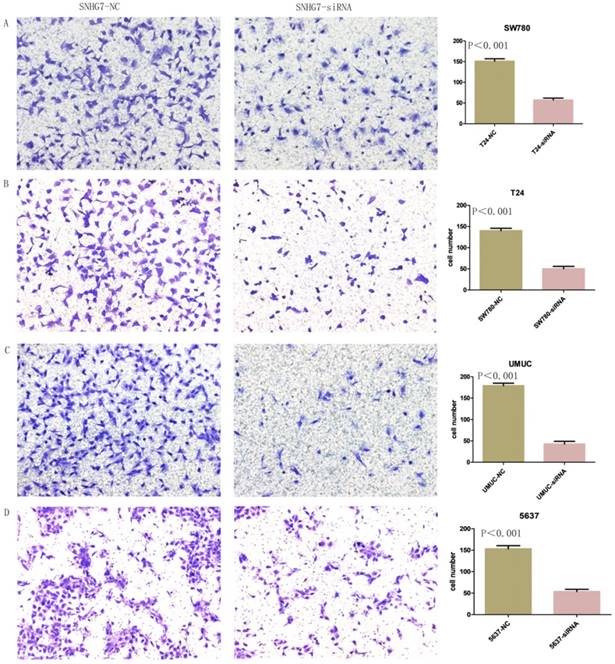
Changes in invasiveness of SW780, T24, UMUC and 5637 cell lines were measured by Transwell assay. We transfected lncRNA-SNHG7 and negative control siRNA into all the bladder cell lines and cultured them for 48 h. We collected the cells and used them for the Transwell assay, which showed that suppression of lncRNA-SNHG7 repressed the motility of cancer cells (Fig. 5).
Knockdown of lncRNA-SNHG7 increased expression levels of Bax, p21 and E-cadherin protein
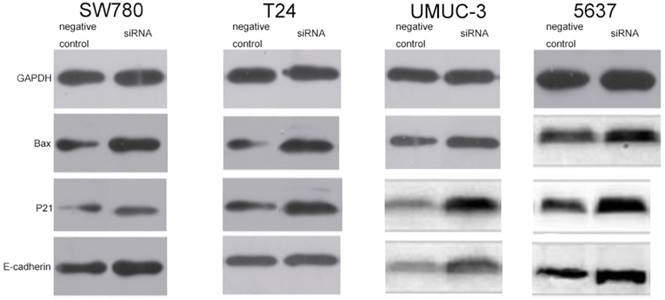
In order to explore the potential downstream targets that induced the above phenotypic changes after inhibition of lncRNA-SNHG7, we utilized western blotting to measure the protein expression levels of Bax, p21 and E-cadherin, which are known to be involved in the development of bladder cancer. As previously reported, Bax protein plays an important role in inducing apoptosis and is inhibited in a variety of tumors [16]. To demonstrate the relationship between lncRNA-SNHG7 and Bax protein, we used western blotting to detect the expression level of Bax after knockdown of lncRNA-SNHG7. Compared to the negative control group, the cells transfected with lncRNA-SNHG7 siRNA showed up-regulation of Bax (Fig. 6). p21 is a hallmark of cell cycle regulation and acts as an inhibitor of cell growth [17]. Western blotting showed that expression of p21 was significantly increased when lncRNA-SNHG7 was suppressed by siRNA (Fig. 6). As shown in a previous study, E-cadherin serves as a strong suppressor of cell migration [18]. We also found that expression of E-cadherin was obviously increased after silencing lncRNA-SNHG7 (Fig. 6). These data suggested that Bax, p21 and E-cadherin genes might serve as potential downstream targets of lncRNA-SNHG7, and that lncRNA-SNHG7 plays a role in regulating bladder cancer cell phenotypes by down-regulating expression of Bax, p21 and E-cadherin protein.
Discussion
At present, cancer therapy is a major challenge worldwide. In order to decrease the severe adverse effects of traditional cancer therapy, molecular-targeted therapy has aroused widespread attention. Therefore, exploring tumor-related targets is essential for the development of molecular-targeted therapy in cancer. Recent studies have indicated that lncRNAs can be key players in the development of cancer formation. lncRNAs are highly correlated with several human diseases through regulation of DNA methylation, transcription and chromatin remodeling [5, 19, 20]. Furthermore, increasing evidence shows that lncRNAs are correlated with the expression of some genes [21, 22]. Therefore, it has been confirmed that lncRNAs have significant clinical potential to be a group of cancer biomarkers and effective therapeutic targets [23, 24].
Knockdown of lncRNA-SNHG7 induced apoptosis. Transfection of lncRNA-SNHG7 or negative control siRNA into bladder cancer cells for 48 h. We determined the changes in apoptosis using ELISA. Apoptosis induction was observed in lncRNA-SNHG7 siRNA-transfected bladder cancer cell lines SW780 (A), T24 (B), UMUC (C) and 5637 (D) using ELISA (P < 0.01). Data are shown as mean ± SD. We performed each experiment in all cell lines in triplicate for three independent times.
Knockdown of lncRNA-SNHG7 decreased cell motility. After transfection of lncRNA-SNHG7 or negative control siRNA, we used wound healing assay to evaluate motility changes in bladder cancer cells. A: Representative images of wound healing assay in SW780 cell lines, and decreased motility was observed in SW780 cell lines (P < 0.01). B: Representative images of wound healing assay in T24 cell lines, and decreased motility was observed in T24 cell lines (P < 0.01). C: Representative images of wound healing assay in UMUC cell lines, and decreased motility was observed in UMUC cell lines (P < 0.01). D: Representative images of wound healing assay in 5637 cell lines, and decreased motility was observed in 5637 cell lines (P < 0.01).
Several studies have characterized the oncogenic properties of lncRNA-SNHG7. lncRNA-SNHG7 affects cancer formation by interacting with miR-5095 [25], miR-193b [26], FAIM2 [12], p15 and p16 [27]. lncRNA-SNHG7 produces an important effect on cancer cells. In our study, we investigated the potential relationship between lncRNA-SNHG7 and the downstream proteins Bax, p21 and E-cadherin, which contribute to cancer development in some way. Our study also explained the possible mechanism by which lncRNA-SNHG7 affects the malignant phenotypes of human bladder cancer. Therefore, we suppose that lncRNA-SNHG7 plays an important role in the development of bladder cancer, and silencing SNHG7 may inhibit the malignant phenotypes of bladder cancer cells. The important conclusions of our study were that lncRNA-SNHG7 was more highly expressed in bladder cancer tissues compared with paired adjacent normal tissues. We also found that expression of lncRNA-SNHG7 was higher in high-grade than in low-grade cancer and that lncRNA-SNHG7 expression was higher in invasive than in non-invasive bladder cancer. We silenced lncRNA-SNHG7 by siRNA in the bladder cancer cell lines SW780, T24, UMUC and 5637 and found that the malignant phenotypes of these bladder cancer cells were inhibited.
Knockdown of lncRNA-SNHG7 decreased cell invasion. After transfection of lncRNA-SNHG7 or negative control siRNA, Transwell assay was used to detect invasiveness of bladder cancer cells. A. Representative images of Transwell assay in T24 cells. B. Representative images of Transwell assay in SW780 cells. C. Representative images of Transwell assay in UMUC cells. D. Representative images of Transwell assay in 5637 cells.
Knockdown of FAL1 increased expression of p21, Bax and E-cadherin protein. After transfection of lncRNA-SNHG7 or negative control siRNA, western blotting was used to detect changes in expression of p21, Bax and E-cadherin in bladder cancer cells. A. Representative images of western blotting in T24 cells. B. Representative images of western blotting in SW780 cells. C. Representative images of western blotting in UMUC cells. D. Representative images of western blotting in 5637 cells.
In conclusion, our study demonstrated that lncRNA-SNHG7 serves as an oncogene that can promote migration, proliferation and invasion and inhibit apoptosis of bladder cancer cells. The malignant phenotypes of bladder cancer cells were inhibited by suppression of expression of lncRNA-SNHG7 by utilizing siRNA. This finding shows that lncRNA-SNHG7 is an important factor for bladder cancer progression, thus providing a potentially useful target for treating bladder cancer.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chen LM, Chang M, Dai Y, Chai KX, Dyrskjot L, Sanchez-Carbayo M. et al. External validation of a multiplex urinary protein panel for the detection of bladder cancer in a multicenter cohort. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2014;23:1804-12
2. Wang J, Zhang X, Wang L, Dong Z, Du L, Yang Y. et al. Downregulation of urinary cell-free microRNA-214 as a diagnostic and prognostic biomarker in bladder cancer. Journal of surgical oncology. 2015;111:992-9
3. Gloss BS, Dinger ME. The specificity of long noncoding RNA expression. Biochimica et biophysica acta. 2016;1859:16-22
4. Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH. et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nature genetics. 2010;42:1113-7
5. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature reviews Genetics. 2009;10:155-9
6. Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178-81
7. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Molecular cell. 2011;43:904-14
8. Saxena A, Carninci P. Long non-coding RNA modifies chromatin: epigenetic silencing by long non-coding RNAs. BioEssays: news and reviews in molecular, cellular and developmental biology. 2011;33:830-9
9. Paralkar VR, Weiss MJ. A new 'Linc' between noncoding RNAs and blood development. Genes & development. 2011;25:2555-8
10. Lakhotia SC. Long non-coding RNAs coordinate cellular responses to stress. Wiley interdisciplinary reviews RNA. 2012;3:779-96
11. Liu X, Li D, Zhang W, Guo M, Zhan Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. The EMBO journal. 2012;31:4415-27
12. She K, Huang J, Zhou H, Huang T, Chen G, He J. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncology reports. 2016;36:2673-80
13. He HT, Xu M, Kuang Y, Han XY, Wang MQ, Yang Q. Biomarker and competing endogenous RNA potential of tumor-specific long noncoding RNA in chromophobe renal cell carcinoma. OncoTargets and therapy. 2016;9:6399-406
14. Cui H, Zhang Y, Zhang Q, Chen W, Zhao H, Liang J. A comprehensive genome-wide analysis of long noncoding RNA expression profile in hepatocellular carcinoma. Cancer medicine. 2017;6:2932-41
15. Hnasko TS, Hnasko RM. The Western Blot. Methods in molecular biology (Clifton, NJ). 2015;1318:87-96
16. Gu J, Zhang L, Huang X, Lin T, Yin M, Xu K. et al. A novel single tetracycline-regulative adenoviral vector for tumor-specific Bax gene expression and cell killing in vitro and in vivo. Oncogene. 2002;21:4757-64
17. Cherrier T, Suzanne S, Redel L, Calao M, Marban C, Samah B. et al. p21(WAF1) gene promoter is epigenetically silenced by CTIP2 and SUV39H1. Oncogene. 2009;28:3380-9
18. Junxia W, Ping G, Yuan H, Lijun Z, Jihong R, Fang L. et al. Double strand RNA-guided endogeneous E-cadherin up-regulation induces the apoptosis and inhibits proliferation of breast carcinoma cells in vitro and in vivo. Cancer science. 2010;101:1790-6
19. Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nature reviews Molecular cell biology. 2013;14:699-712
20. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nature structural & molecular biology. 2013;20:300-7
21. Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Molecular biology and evolution. 2001;18:1611-30
22. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11667-72
23. Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocrine reviews. 2015;36:25-64
24. Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends in cancer. 2015;1:93-109
25. Ren J, Yang Y, Xue J, Xi Z, Hu L, Pan SJ. et al. Long noncoding RNA SNHG7 promotes the progression and growth of glioblastoma via inhibition of miR-5095. Biochemical and biophysical research communications. 2018
26. She K, Yan H, Huang J, Zhou H, He J. miR-193b availability is antagonized by LncRNA-SNHG7 for FAIM2-induced tumour progression in non-small cell lung cancer. Cell proliferation. 2018:51
27. Wang MW, Liu J, Liu Q, Xu QH, Li TF, Jin S. et al. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. European review for medical and pharmacological sciences. 2017;21:4613-22
Author contact
![]() Corresponding author: Xinli Kang (2151220207szu.edu.cn)
Corresponding author: Xinli Kang (2151220207szu.edu.cn)

 Global reach, higher impact
Global reach, higher impact