Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(1):211-222. doi:10.7150/jca.26444 This issue Cite
Research Paper
Deciphering Microenvironment of NSCLC based on CD8+ TIL Density and PD-1/PD-L1 Expression
1. Department of Respiratory Medicine, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China;
2. Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou, China.
*These authors have contributed equally to this work
Received 2018-4-2; Accepted 2018-8-6; Published 2019-1-1
Abstract
Purpose: To determine whether distinct tissue immune microenvironments differentially impact on clinical outcome in non-small cell lung cancer (NSCLC), an extended analysis of PD-1/PD-L1 and Tumor Infiltrating Lymphocytes (TILs) was performed.
Materials and Methods: 1016 NSCLC mRNA-sequence samples from The Genome Data Analysis Center (TCGA) and 275 NSCLC mRNA-microarray samples from Gene Expression Omnibus (GEO) were included as testing cohort and validation cohort respectively. Enrichment scores of CD8+ T cells' metagene were used for quantifying its infiltrating density. Based on the median values of CD8+ T cell density and PD-1/PD-L1 mRNA expression, the samples were classified into four Tumor Immune Microenvironment types (TIMTs). Overall survival, as well as clinicopathological features, mutational profiles, mismatch repair score etc. were compared across the four types.
Results: Neither PD-1 expression nor PD-L1 expression was associated with outcome in the overall NSCLC. Classification of TIMT based on PD-1/PD-L1 and CD8+ TIL could efficiently classify patients of different survival in ADC but not SCC, with the best overall survival achieved in TIMT3 (high CD8+ TIL and low PD-1/PD-L1), whereas TIMT2 (low CD8+ TIL and high PD-1/PD-L1) manifested the worst outcome. TIMT classification based on PD-1/ CD8+ TIL could better stratify patient of different prognosis than PD-L1/ CD8+ TIL based classification. EGFR wide type and IFNγ overexpression were associated with TIMT4 (high PD-1/PD-L1 and high CD8+ TIL), whereas tumor mutational burden (TMB) manifested no significant difference across four TIMTs.
Conclusion: The classification of tumors into four microenvironment subtypes based on PD-1/PD-L1 status and CD8+ TIL is an appropriate approach to stratify patients of different clinical outcome and better guide the practical use of immunotherapy.
Keywords: non-small-cell lung cancer (NSCLC), programmed cell death-1 (PD-1), programmed cell death-ligand 1 (PD-L1), tumor microenvironment, survival
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide, despite effective chemotherapeutic agents and driver mutation targeting agents, the prognosis remains dismal[1]. Immune checkpoint inhibitors (ICIs) have emerged as one of the main new therapeutic options for advanced non-small cell lung cancer (NSCLC) patients and may become a major treatment backbone in the next decades[2].
Programmed cell death-1 (PD-1), as a transmembrane immune regulatory molecule on T cells, is responsible for the negative regulation of T cell activation and peripheral tolerance through the engagement with its ligands PD-L1 and PD-L2. PD-L1 is constitutively expressed on antigen presenting cells (APCs) and tumor cells in numerous solid malignancies including NSCLC. Checkpoint inhibitors block PD-1/PD-L1 signaling pathway, thereby leading to an endogenous antitumor immune response[2, 3].
Even though ICIs demonstrated superiority towards standard chemotherapy in different disease settings, the response rates do not exceed 45% in highly molecularly selected patients[4-6]. This is related to known limitations of the available biomarkers, as well to the complex and dynamic nature of tumor microenvironment. PD-L1 expression was found to be correlated with treatment efficacy and used as a surrogate predictive marker for anti-PD-1/ PD-L1 therapy. Yet not all the patients with PD-L1 positive expression response well to immune checkpoint treatment, suggesting that some other microenvironment factors may also play an important role. Interestingly, the degree of tumor infiltrating lymphocyte (TIL) infiltration in the tumor microenvironment (TME) are also correlated with the clinical outcomes of anti-PD-1/PD-L1 therapies[7]. Further research found that higher tumor mutational burden (TMB), abundant neo-antigen, and micro-satellite (MSI)-high status were also associated with a good response to ICI[8].
Considering the complex nature of tumor immunity, a comprehensive immuno-genomic analysis of the tumor microenvironment based on the interaction between PD-1/PD-L1 and TILs is critical to deepen our understanding of the underlying mechanism and better guide us in tailoring optimal immunotherapeutic strategies for NSCLC.
Preliminary studies had proposed a classification of tumors into four categories based on the presence or absence of TILs and PD-L1 expression levels (type I: TILs+ and PD- L1+; type II: TILs- and PD-L1-; type III: TILs+ and PD- L1-; type IV: TILs- and PD-L1+) and investigated their difference from the perspective of bio-genomic features as well as clinical outcome[9-12]. But the yielded results were quite controversial because of the relative small studying population. Also, the previous categorization of tumor microenvironment didn't take the expression level of PD-1 into consideration, which also plays a major role in immune escape.
The tremendous transcriptome mRNA sequencing data together with the complete clinical information stored in the Cancer Genomic Atlas (TCGA, https://cancergenome.nih.gov) make it a suitable resource to investigate the interaction among immuno-genomic features as well as their survival relevance. Although TIL assessment in the tumor microenvironment remains a challenge, a novel and algorithmically-optimized method for scoring the enrichment of pre-defined gene sets of certain TIL subsets can be used to accurately measure the TILs density in a single sample[13].
Utilizing these resources and analytical tools, we classified a large set of NSCLC samples into four tumor immune microenvironment types base on CD8+ TIL density and mRNA expression levels of PD-L/PD-L1. The aim of the study is to determine the difference in clinicopathologic features, mutational burden, mismatch repair status, clinical outcome etc. among different microenvironment, which hopefully, could provide strategic information for the use of ICIs.
Materials and Methods
Sample and data collection
TCGA dataset: We retrospectively analyzed the gene mRNA sequence profiles of NSCLC frozen tumor tissue samples from TCGA (The Genome Data Analysis Center), which includes two datasets for NSCLC, LUAD (lung adenocarcinoma) and LUSC (lung squamous cell carcinoma). Level 3 TCGA mRNA-sequencing matrix plus clinical metadata and complete profiles of sequence-verified mutations for NSCLC were obtained with permission from the Cancer Genomics Hub (https://cghub. ucsc.edu). The compatibility for merging RNA-sequencing data of the LUAD and LUSC was assessed (details were shown in supplementary materials). To count the number of total somatic mutations, multiple somatic mutations including nonsynonymous mutations, insertion-deletion mutations, and silent mutations were each counted and summated, and germline mutations without somatic mutations were excluded.
Validation dataset: A public mRNA microarray dataset (GSE41271) containing sufficiently large numbers of NSCLC samples (n = 275) deposited in GEO (Gene Expression Omnibus) was used to constructed the validation cohort. Log-transformed and quantile-normalized mRNA expressing matrix plus clinical metadata was downloaded from GEO. For a gene represented by multiple probes, the mean value of those probes was calculated to obtain a unique expression value for that gene.
Sample enrichment scores (SES)
A novel and algorithmically-optimized method for scoring the enrichment of pre-defined genes set in single samples was applied in this study. The calculation was executed through Auto-compare SES software developed by Fournie and his colleague[13], the source for which are available at: https://sites.google.com/site/fredsoftwares/products/autocompare_ses. The RNA expression matrix prepared as txt files for each collapsed sample together with the gene sets prepared in txt file were used as input for Auto-compare SES software to calculate the enrichment of a gene set in a sample (SES) as previously described[13]. The characteristic gene set of CD8+ T lymphocyte were derived from previous published studies, the SES of which was used to represent the CD8+ T cell density. The functionally defined gene sets of “mismatch match repair”, “mitosis”, “inner cell mass proliferation” etc. were download from the Molecular Signatures Data base (MSigDB 3.0). The details of all the gene sets adopted in this study were shown in Table S1. SES for each of them were used to represent the activity of certain cellular function. Assessment analysis was performed to validate the feasibility of Auto-compare SES calculation (details were shown in supplementary materials).
Statistical analysis
As for sequence data from TCGA, Log 2-transformed values of RPKM (reads per kilobase of transcript per million reads mapped) for PD-L1/PD-1 were applied for further analysis. We transferred continual variables like CD8+ T cell infiltrating density, PD-1/PD-L1 mRNA expression level into categorical variables (high vs low) with median value as cutoff point. According to previous reports regarding the four types of tumor immune microenvironment we divided all of the NSCLC samples into four groups based on CD8+ T cell infiltrating level and PD-L1 mRNA expression as follows: type I, low CD8+ T cell density and low PD-L1 expression; type II, low CD8+ T cell density and high PD-L1 expression; type III, high CD8+ T cell density and low PD-L1 expression; and type IV, high CD8+ T cell density and highPD-L1 expression. Similar classification was also performed based on CD8+ T cell infiltrating level and PD-1 mRNA expression. The statistical significance of two continuous values, such as CD8+ T cell infiltrating density, PD-1/PD-L1 mRNA expression level, the number of mutations etc. was calculated by linear regression analysis. In the survival analysis section, we excluded patients who had received neoadjuvant therapy, or other pharmaceutical therapy owing to immune-modulating effects of some therapeutics before the surgery. We further exclude patients who have positive surgical margin, adjuvant therapy, radiation therapy or target therapy after the surgery and who have follow-up time or survival time less than 1 month when we do the survival analysis. The prognostic significance of categorical variables such as microenvironment subtypes, PD-1/PD-L1 expressing status, CD8+ T cell infiltrating status etc. was estimated using Kaplan-Meier plots (log-rank test) and Cox proportional hazards regression analysis. Survival times were calculated as months from initial pathological diagnosis to death, or the number of months from initial pathological diagnosis to the last time the patient was known to be alive. All statistical analyses and data presentations were performed in R language 3.1.3 (http://www. r-project.org) and the Statistical Package for the Social Sciences (SPSS, Chicago, IL, USA) version 15.0.
Results
Clinicopathological information and its association with PD-1/PD-L1 expression/CD8+ TIL infiltration
A total of 1013 resected NSCLC cases form TCGA were included in this study, with the demographic information demonstrated in Table S2. The median age at diagnosis was 67 (range: 33 - 90 years). 607 (60%) patients were male, and 774 (76.4%) patients had a history of smoking. The majority of the cases were of TNM early stage at the time of diagnosis, with 517(51.04%) being stage Ⅰ, 283 (27.94%) being stage Ⅱ, 168(16.58%) being stage Ⅲ, 33(32.57%) being stage Ⅳ. Adenocarcinoma (ADC, 513, 50.64%) and squamous cell carcinoma (SCC, 500, 49.36%) account for half of the cases respectively. The median follow-up time was 28.6 months; with 653 cases remain alive at the end of the follow-up.
As demonstrated in Table 1, enrichment score of CD8+ TIL was significant higher in tumor of female gender (P<0.001), TNM Ⅰ-Ⅱ (P=0.009) and adenocarcinoma (P=0.004). PD-1 mRNA expression level was higher in tumor of female gender (P<0.001), adenocarcinoma (P<0.001), tumor with mutant EGFR (P=0.007) and tumor with high mutation burden (P=0.010). While high PD-L1 mRNA expression level was associated with M0 stage (P=0.001) and squamous cell carcinoma (P=0.004). Of note, the analysis on driver mutation was performed only for adenocarcinoma, as all the mutant EGFR and 166 out of 171 mutant KRAS are of adenocarcinoma.
Association between PD-1/PD-L1 expression/CD8+ TIL density with clinical outcome
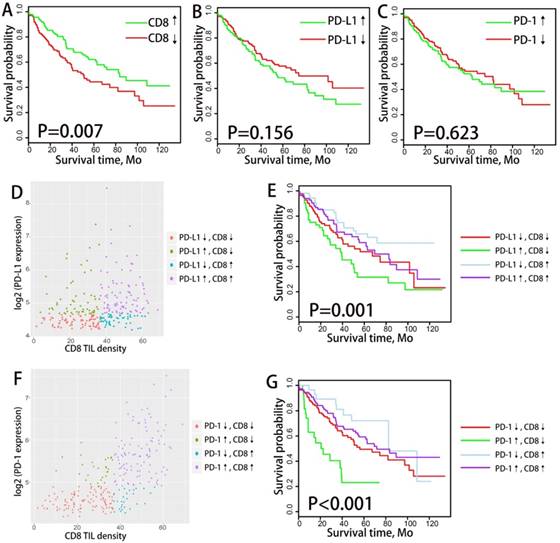
We divided the patients into subgroups of high CD8+ TIL infiltrating density and low CD8+ TIL infiltrating density according to the median CD8+ TIL enrichment score. Similarly, PD-1/PD-L1 expressing status was classified as high expression and low expression based on the mRNA expressing value, with the median as cutoff point. We analyzed the correlation between PD-1/PD-L1 expression/CD8+ TILs density and overall survival (OS) through Kaplan-Meier plots (log-rank test) and Cox proportional hazards regression analysis (as demonstrated in Fig.1). Neither the mRNA expression of PD-1 nor PD-L1 was associated with OS in the overall NSCLC population or in histological subtypes (ADC and SCC) population. CD8+ TIL infiltrating density manifested favorable prognostic significance in ADC population (P=0.01) but not in the overall NSCLC population (P=0.087) or SCC population (P=0.87).
Classify TIMT based on combination of PD-L1 expression and CD8+ TIL density
TIMTs composition
All tumor samples were divided into four groups of tumor immune microenvironment type (TIMT) according to the median values of PD-L1 mRNA expression and CD8+ TIL enrichment score (as shown in Fig. 2A). The expression levels of PD-L1 and CD8+ TIL density were generally positively correlated regardless of histology, TNM stage, PD-L1 amplification status or driver mutation status (as shown in Supplementary Fig. S3). The proportion of TMIT samples was analyzed according to histological subtypes, TNM stages, EGFR mutation status and TMB level (Fig. 2B). The proportion of TIMT4 (high PD-L1 expression and high CD8+ TIL) is significantly lower in subgroup of TMN Ⅳ and subgroup of EGFR mutation.
Outcome analysis of TIMTs defined by PD-L1 and CD8+ TIL
We analyzed the difference in clinical outcome of the four TIMTs with Kaplan-Meier plots (log-rank test) and cox proportional hazards regression analysis were performed to decide the prognostic significance (Fig. 2C). Among the whole cohort, the best OS was achieved in TIMT3 (low PD-L1 expression/high CD8+ TIL infiltration), and the worst was in TIMT2 (high PD-L1 expression/low CD8+ TIL infiltration) (median OS 88.0 months & 37.2 months respectively, P=0.003). Similar tendency was observed in ADC cohort (P=0.004) (Fig. 2D) and SCC cohort (P=0.332) (Fig. 2E), though the later manifested no statistical significance.
TIMTs' correlation with Immune-genomic features
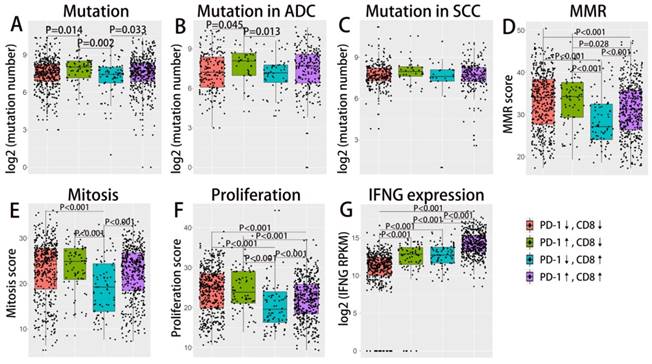
As demonstrated in previous studies, tumor mutation burden and tumor DNA mismatch repair status (MMR) could also reliably predict ICIs treatment response and may interact with the tumor microenvironment[14, 15]. IFN γ as an important cytokine in the immune microenvironment, has been closely associated with cytotoxic activity as well as the induction of PD-1/PD-L1 production[16]. We further investigate the correlation of these immune-genomic features with TIMTs (as demonstrated in Fig. 3). As mutation burden and MMR status may get affected by tumor proliferation index, the corresponding cellular function (mitosis and proliferation) were also compared across four TIMTs. MMR status as well as tumor mitosis and proliferation status was decided by the enrichment score of corresponding gene set, which was derived from MSigDB 3.0. Expression level of IFN γ was represent by the mRNA expressing level of its encoding gene, IFNG. Total somatic mutation number tend to be higher in tumor with high PD-L1 expression, which is even more obvious for ADC but not SCC (as shown in Fig. 3A,B,C). While TIMTs of high PD-L1 expression and low CD8+ TIL infiltration tend to higher have MMR score, mitosis score and proliferation score (Fig. 3D,E,F) (P<0.001). As expected, IFNG expression was significantly higher in TMIT4 tumors (high PD-L1 expression/high CD8+ TIL infiltration) (Fig. 3G).
Classify TIMT based on combination of PD-1 expression and CD8+ TIL density
TIMTs composition
Similarly, we divided the NSCLC tumor into four TIMTs based on the combination of PD-1 expression and CD8+ TIL density (as shown in Fig. 4A). PD-1 expression were evidently positively correlated with CD8+ TIL density regardless of histology, TNM stage, PD-L1 amplification status or driver mutation status (as shown in Supplementary Fig. S4). The distribution of TMIT sorted by histological subtypes, TNM stages, EGFR mutation status and TMB level were shown in Fig 4B. The proportion of TIMT4 (high PD-1 expression and high CD8+ TIL) is significantly lower in subgroup of TMN Ⅳ and subgroup of EGFR mutation.
Outcome analysis of TIMTs defined by PD-1 and CD8+ TIL
As for survival analysis, dramatic survival difference was observed among TIMTs. Similar to the results of TIMTs defined by PD-L1 and CD8+ TIL, the longest OS was achieved in TIMT3 (low PD-1 expression/high CD8+ TIL infiltration), while the shortest in TIMT2 (high PD-1 expression/low CD8+ TIL infiltration), the difference was statistically significant both for overall cohort (P=0.0043) and ADC cohort (P<0.001), but not for SCC cohort (P=0.53) (Fig. 4C,D,E). In particular, the difference in median OS across the four TIMTs defined by PD-1 and CD8+ TIL was much apparent than that defined by PD-L1 and CD8+ TIL, with median OS being 105.6 months for TIMT3 (low PD-1 expression/high CD8+ TIL infiltration) and 34.7 months for TIMT2 (high PD-1 expression/low CD8+ TIL infiltration).
Association between PD-1/PD-L1/ CD8+ TIL and clinicopathological features
| Variables | status | No. | CD8+ TIL density | log2 (PD-1 RPKM) | log2 (PD-L1 RPKM) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± sd | P | Mean ± sd | P | Mean ± sd | P | |||||
| age | >65 | 596 | 25.16±11.78 | 0.554 | 14.65±1.51 | 0.516 | 15.90±1.52 | 0.708 | ||
| <65 | 389 | 24.69±12.50 | 14.71±1.54 | 15.94±1.59 | ||||||
| gender | male | 607 | 23.74±12.05 | <0.001 | 14.50±1.57 | <0.001 | 15.89±1.62 | 0.354 | ||
| female | 406 | 26.87±11.90 | 14.95±1.40 | 15.98±1.45 | ||||||
| TNM | Ⅰ-Ⅱ | 517 | 25.46±12.25 | 0.009 | 14.70±1.53 | 0.289 | 15.93±1.52 | 0.586 | ||
| Ⅲ-Ⅳ | 283 | 22.96±11.28 | 14.57±1.50 | 15.86±1.65 | ||||||
| T stage | T1-T2 | 815 | 25.24±12.24 | 0.049 | 14.68±1.53 | 0.426 | 15.93±1.54 | 0.856 | ||
| T3-T4 | 155 | 23.14±11.56 | 14.57±1.52 | 15.90±1.67 | ||||||
| N stage | N0 | 618 | 25.21±12.31 | 0.201 | 14.64±1.52 | 0.839 | 15.86±1.52 | 0.164 | ||
| N1-N2 | 330 | 24.15±11.92 | 14.66±1.56 | 16.01±1.64 | ||||||
| M stage | M0 | 721 | 24.97±11.95 | 0.046 | 14.65±1.53 | 0.123 | 15.96±1.61 | 0.035 | ||
| M1 | 31 | 20.60±11.33 | 14.22±1.75 | 15.34±1.52 | ||||||
| Histology | ADC | 513 | 26.06±12.45 | 0.004 | 14.86±1.43 | <0.001 | 15.78±1.43 | 0.004 | ||
| SCC | 500 | 23.90±11.60 | 14.49±1.58 | 16.07±1.66 | ||||||
| Smoking history | yes | 774 | 24.71±12.03 | 0.421 | 14.64±1,53 | 0.722 | 15.95±1.59 | 0.245 | ||
| no | 239 | 25.80±13.02 | 14.70±1.57 | 15.74±1.32 | ||||||
| EGFR (ADC) | Mutant | 65 | 27.92±11.98 | 0.107 | 14.39±1.57 | 0.007 | 15.49±1.15 | 0. 051 | ||
| Wild type | 419 | 24.97±12.25 | 14.91±1.42 | 15.80±1.48 | ||||||
| KRAS (ADC) | Mutant | 166 | 26.94±12.73 | 0.093 | 14.84±1.27 | 0.982 | 15.79±1.43 | 0.848 | ||
| Wild type | 329 | 24.96±11.36 | 14.85±1.52 | 15.76±1.45 | ||||||
| Somatic mutation | >median | 507 | 24.83±12.08 | 0.891 | 14.78±1.54 | 0.010 | 15.83±1.48 | 0.092 | ||
| <median | 506 | 24.93±12.22 | 14.53±1.51 | 16.00±1.65 | ||||||
Abbreviations: PD-1, programmed cell death-1; PD-L1, programmed cell death ligand-1; ADC, adenocarcinoma; SCC, squamous cell carcinoma; EGFR, epidermal growth factor receptor; KRAS, Kirsten rat sarcoma viral oncogene; sd, standard deviation; RPKM, reads per kilobase of transcript per million reads mapped.
overall survival analysis of PD-1/PD-L1 and CD8+ TILs density among NSCLC patients. Kaplan-Meier plots of overall survival according to PD-1/PD-L1 expression level and CD8+ TILs density for total NSCLC patients (A,D,G), ADC patients (B,E,H) and SCC patients(C,F,I) are graphed. PD-1/PD-L1↓ and PD-1/PD-L1↑ refer to tumors with PD-1/PD-L1 mRNA expression value less or more than the median respectively. CD8↓ and CD8↑ refer to tumors with CD8+ TILs enrichment score more or less than the median respectively. Abbreviation: NSCLC, non-small cell lung cancer, ADC, adenocarcinoma, SCC, squamous cell carcinoma.
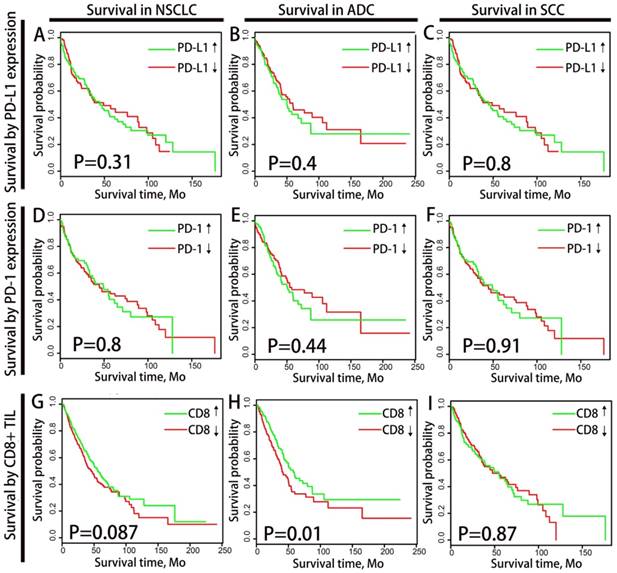
Distribution and overall survival analysis of TMIT defined by PD-L1/CD8+ TIL combination. A scatter plot of CD8+ TIL enrichment score and log 2-transformed values of RPKM of PD-L1 is shown (A). Tumors were classified into four TIMTs based on the combination of CD8+ TIL and PD-L1, with median values as cutoff points. The portion of TMIT [PD-L1↓/CD8↓, red (33%); PD-L1↑/CD8↓, green (17%); PD-L1↓/CD8↑, blue (17%); PD-L1↑/CD8↑; purple (15.4%)] (B) according to histology, TNM stage, EGFR status, KRAS status and TMB level are graphed. Kaplan-Meier plots of overall survival according to TMITs for total NSCLC patients (C), ADC patients (D) and SCC patients(E) are graphed. Abbreviation: TIMT, tumor immune microenvironment type; NSCLC, non-small cell lung cancer; ADC, adenocarcinoma; SCC, squamous cell carcinoma; RPKM, reads per kilobase of transcript per million reads mapped.
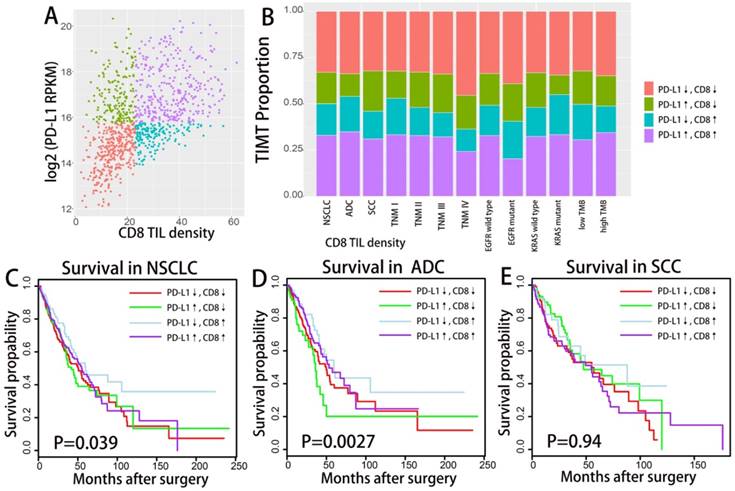
Difference in mutation burden, MMR, IFN γ expression etc. across TIMTs defined by PD-L1/CD8+ TIL combination. Box plot of the number of total somatic mutations (log 2-transformed) (A-C), MMR score (D), mitosis score (E), proliferation score (F), IFNG mRNA expression value (log 2-transformed) (G) according to TMITs (PD-L1↓/CD8↓, red; PD-L1↑/CD8↓, green; PD-L1↓/CD8↑, blue; PD-L1↑/CD8↑, purple) are plotted. Abbreviation: MMR, mismatch repair; RPKM, reads per kilobase of transcript per million reads mapped.
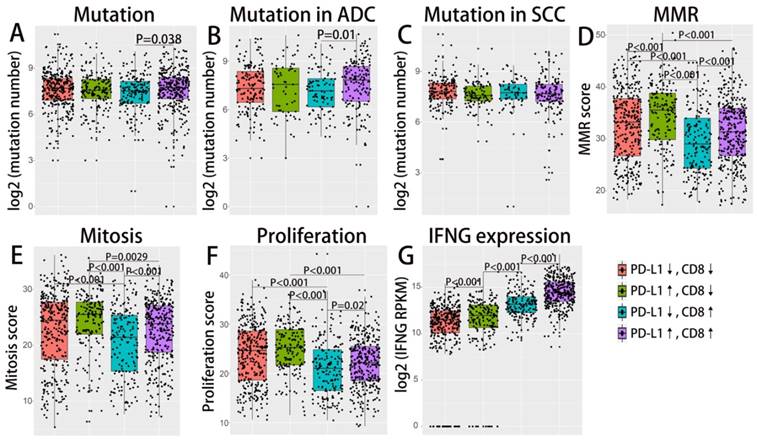
Distribution and overall survival analysis of TMIT defined by PD-1/CD8+ TIL combination. A scatter plot of CD8+ TIL enrichment score and log 2-transformed values of RPKM of PD-1 is shown (A). Tumors were classified into four TIMTs based on the combination of CD8+ TIL and PD-1, with median values as cutoff points. The portion of TMITs [PD-1↓/CD8↓, red (42.5%); PD-1↑/CD8↓, green (7.5%); PD-1↓/CD8↑, blue (7.7%); PD-1↑/CD8↑; purple (42.3%)] (B) according to histology, TNM stage, EGFR status, KRAS status and TMB level are graphed. Kaplan-Meier plots of overall survival according to TMITs of total NSCLC patients (C), ADC patients (D) and SCC patients(E) are graphed. Abbreviation: TIMT, tumor immune microenvironment type; NSCLC, non-small cell lung cancer; ADC, adenocarcinoma; SCC, squamous cell carcinoma; RPKM, reads per kilobase of transcript per million reads mapped.
TIMTs' correlation with Immune-genomic features
Association between immune-genomic features and TIMTs was analyzed as previously descried. Tumor with high PD-1 and low CD8+ TIL) tend to have higher somatic number, which is even more obvious for ADC but not SCC (as shown in Fig. 5A,B,C). Similar to the results of TIMTs defined by PD-L1 and low CD8+ TIL, TIMTs of high PD-1 expression and low CD8+ TIL infiltration tend to higher have MMR score, mitosis score and proliferation score (Fig. 5D,E,F) (P<0.001). Also, IFNG expression was significantly higher in TMIT4 tumors (high PD-1 expression/high CD8+ TIL infiltration) (Fig. 5G).
Summary of clinicopathologic features correlated with TMIT4
We further perform logistic regression analysis to identify clinicalpathologic features including age, gender, TNM staging, driver mutation status, TMB etc. that associating with TMIT4 (as shown in Table 2). Only EGFR wild type and IFNγ were significantly associated with TMIT 4 (tumor with high PD-1/ PD-L1 expression and CD8+ TIL density.
Logistic regression analysis for predicting TIMT4 according to clinicopathological characteristics
| High PD-L1/High CD8+ TIL | High PD-L1/High CD8+ TIL | ||||
|---|---|---|---|---|---|
| OR | P | OR | P | ||
| age | >65y | 1.32 (1.007-1.729) | 0.044 | 1.172 (0.907-1.513) | 0.224 |
| gender | female | 1.45 (1.112-1.891) | 0.006 | 1.549 (1.201-1.998) | 0.001 |
| histology | SCC | 0.838 (0.645-1.090) | 0.188 | 0.897 (0.699-1.151) | 0.391 |
| smoking history | smoker | 0.809 (0.514-1.273) | 0.35 | 0.846 (0.545-1.313) | 0.455 |
| EGFR | mutant | 0.52 (0.285-0.951) | 0.031 | 0.45 (0.256-0.790) | 0.005 |
| KRAS | mutant | 1.045 (0.737-1.486) | 0.801 | 1.094 (0.780-1.528) | 0.596 |
| T stage | T3-4 | 0.883 (0.609-1.282) | 0.514 | 0.815 (0.572-1.160) | 0.256 |
| N stage | N1-2 | 0.996 (0.748-1.326) | 0.98 | 0.93 (0.708-1.220) | 0.599 |
| M stage | M1-2 | 0.522 (0.211-1.291) | 0.159 | 0.466 (0.206-1.057) | 0.068 |
| TMB | >median | 1.191 (0.911-1.558) | 0.202 | 1.122 (0.869-1.447) | 0.377 |
| IFNγ expression | >median | 13.581 (9.542-19.33) | <0.001 | 13.296 (9.752-18.128) | <0.001 |
OR, odds ratio; PD-1, programmed cell death-1; PD-L1, programmed cell death ligand-1; SCC, squamous cell carcinoma; EGFR, epidermal growth factor receptor; KRAS, Kirsten rat sarcoma viral oncogene.
Validation in public dataset
To further validate the survival relevance of CD8+ TIL and PD-1/PD-L1 expression, we performed similar survival analysis at a public mRNA expression dataset (GSE41271) containing sufficiently large numbers of NSCLC samples (n = 275) deposited in GEO. The clinicopathological information of GSE41271 was summarized in table S3.
Difference in mutation burden, MMR, IFN γ expression etc. across TIMTs defined by PD-1/CD8+ TIL combination. Box plot of the number of total somatic mutations (log 2-transformed) (A-C), MMR score (D), mitosis score (E), proliferation score (F), IFNG mRNA expression value (log 2-transformed) (G) according to TMITs (PD-1↓/CD8↓, red; PD-1↑/CD8↓, green; PD-1↓/CD8↑, blue; PD-1↑/CD8↑, purple) are plotted. Abbreviation: MMR, mismatch repair; RPKM, reads per kilobase of transcript per million reads mapped.
As mentioned before, CD8+ TIL density and PD-1/PD-L1 mRNA expression level were transferred into categorical variables (high vs low) with median value as cutoff point. Similar with the results of TCGA cohort, expression level of PD-1/PD-L1 manifested no survival significance in validation cohort (Fig. 6). High CD8+ T cell infiltration was significantly associated with favorable survival in ADC but not SCC (Fig. S5). We further classified the samples of validation cohort into four TIMTs based on the combination of CD8+ TIL and PD-1/PD-L1 as previously described. Consistent with previous finding, the best outcome was achieved in TIMT3 (low PD-1/ PD-L1 expression/high CD8+ TIL infiltration) while the worst in TIMT2 (high PD-1/ PD-L1 expression/low CD8+ TIL infiltration) (Fig. 6). Subgroup analysis according to histologic subtype indicated similar tendency for ADC but not SCC, which may due to the modest sample size in SCC subgroup (Fig. S5).
Discussion
Accumulating evidence suggests that PD-1/ PD-L1 antibodies are effective for treating many types of human cancer including NSCLC[4-6, 17, 18]. But the screening of treatment responding patients remains the challenge for immunotherapy. Both PD-1/PD-L1 and TIL infiltrating status had been associated with treatment response. The interaction of PD-1/PD-L1 and TIL in the immune microenvironment and how they affect the clinical outcome as a whole is yet to be clarified. Utilizing a large-scale TCGA dataset, we analyzed the immune microenvironment of NSCLC from the perspective of PD-L1/ CD8+ TIL combination as well as PD-1/CD8+ TIL combination. Our studies suggest that classifying the immune microenvironment based on PD-1/PD-L1 and CD8+ TIL combination could better stratify patients of different outcome for ADC but not for SCC in NSCLC. The interpretation of tumor microenvironment should take PD-1/PD-L1 expression and TIL infiltrating into consideration at the same time, which could bring more prognostic information and better guide us in tailoring optimal immunotherapeutic strategies for NSCLC.
PD-1/PD-L1 is well known as a biomarker for immune escape in the tumor microenvironment and supposed to be associated with worse prognosis for malignant tumor in theory. But its actual survival relevance remains in dispute, with some studies claiming it to be an unfavorable prognostic marker, while others indicating a better survival relevance [19-22]. Similar controversy occurs for CD8+ TIL, which by theory is associated with better outcome because of its tumor eradicating function, whereas many clinical studies didn't observe prominent survival significance for CD8+ TIL[23, 24]. The major drawback of the previous studies was neglecting the interaction between PD-1/PD-L1 and TIL, both of which exert influence on the immune microenvironment as a whole. Since the propose of classifying tumor microenvironment base on TIL and PD-L1[9], quite a few study set out to unravel the clinical significance of such classification[10, 11, 25, 26]. But the yielded results were inconsistent with one another, which may owe to the modest sample size or the diverse detecting methods as well as various cutoff values adopted in the PD-L1 detection[27, 28]. At the same time, such classification fails to take PD-1 expression status into consideration, which plays an equal important role in immune evasion. Our study was carried out based on the transcriptome sequencing data, which could avoid the bias caused by immunohistochemistry detecting to some extent. No survival significance was observed for PD-1 mRNA expression or PD-L1 mRNA expression in the overall population. CD8+ TIL density, measured by enrichment score of CD8+ TIL's characteristic gene set, was associated with favorable survival for ADC but not SCC. Even for ADC, the median overall survival of subgroups classified by CD8+ TIL was quite close with each other, which indicates that CD8+ TIL along could not stratify patients of different prognosis efficiently. Many studies had suggested that CD8+ TILs could produce IFN-γ and induce PD-L1 expression in different solid tumors, which indicates a co-evolvement of immune activity and tumor immune escape. The survival significance of each of them is neutralized by the co-existence of the counterpart. That's why classifying the immune microenvironment based on PD-1/PD-L1 and CD8+ TIL combination could better stratify patients of different outcome. The best survival was achieved in patients with high CD8+ TIL and low PD-1/PD-L1 expression. Patients with high CD8+ TIL and high PD-1/PD-L1 expression have similar survival with patients of low CD8+ TIL, which indicates the prominent immune suppressing function of PD-1/ PD-L1 in high TIL infiltrating microenvironment. Also, Patients with high CD8+ TIL and high PD-1/PD-L1 expression may benefit the most form anti-PD-1/PD-L1 therapy, as in which case the immune escape status of TIL could get reversed to the maximum. It is worth noting that PD-1/CD8+ TIL combination could better stratified patients of different prognosis than PD-L1/ CD8+ TIL combination in our study. A recent study also indicates that low PD-1 expression in cytotoxic CD8+ Tumor infiltrating lymphocytes confers an Immune privileged microenvironment in NSCLC[29]. Overall, these findings underline the need to define the expressing status of PD-1 on TIL to better assess immune status and tailor regimen of ICIs treatment.
Survival analysis in validation cohort. Kaplan-Meier plots of overall survival according to CD8+ TILs density and PD-1/PD-L1 expression level are graphed (A, B, C). PD-1/PD-L1↓ and PD-1/PD-L1↑ refer to tumors with PD-1/PD-L1 mRNA expression value less or more than the median respectively. CD8↓ and CD8↑ refer to tumors with CD8+ TILs enrichment score more or less than the median respectively. Scatter plots of CD8+ TIL enrichment score and log 2-transformed values of PD-1/ PD-L1 mRNA expression is shown (D, F). Tumors were classified into four TIMTs based on the combination of CD8+ TIL and PD-1/PD-L1, with median values as cutoff points. Kaplan-Meier plots of overall survival according to TIMTs are graphed (E, G).
Despite the overall efficacy demonstrated by the immune checkpoint inhibitors in NSCLC, the use of these drugs in practice remains imprecise, with a limited ability to identify patients who will benefit from treatment. Tumor mutational burden (TMB) and tumor DNA mismatch repair (MMR) deficiency are newly proposed as promising predictive biomarkers, which could work as a supplement to PD-1/PD-L1 and TIL[14, 30]. The combination of TMB and PD-L1 could better identify patients with good response to anti-PD-1/PD-L1 immunotherapy[31]. Tumors with higher TMB have been hypothesized to have more neoantigens that can be recognized by the immune system in response to checkpoint inhibition[32]. Also, Mutational epitopes had been associated with higher TIL infiltration and elevation of CD8A and PD-1 gene expression[33-36], which indicates the interaction between the tumor neoantigens and immune microenvironment. In our study, total somatic mutation number was slightly elevated in subgroups with high PD-1/PD-L1 expression but no significance changes in subgroups of different CD8+ TIL level. As not all the somatic mutations could bring about immunogenic neoantigens, further study exemplify it's the neoantigen number rather the total mutation number that positively correlated with immune cytotoxic activity and PD-1/PD-L1 expression[34]. Deficiency in MMR could leads to accelerated accumulation of genetic errors and neoantigens burden, and thus were linked to ICIs treatment response. Usually, MMR status is detected through immunohistochemistry for the presence of the four MMR proteins[37]. In our study, we decided MMR status with enrichment score of MMR related genes. Interestingly, MMR score tend to be lower in tumor with higher CD8+ TIL and lower PD-1/PD-L1 expression, which group also manifested the lowest mutation burden and mitosis/proliferation score. We assume that MMR score is positively correlated with cellular mitosis and proliferation status, whereas MMR deficiency not necessarily indicates higher mutation burden in NSCLC. A recent study also suggested that MMR deficiency status was irrelevant to TIL and PD-1/PD-L1 expression and may not be a useful marker for immune checkpoint inhibitor treatment in NSCLC[38].
Consistent with the previous findings[39, 40], EGFR driver mutation is associated with lower PD-1 expression (P=0.007). Also, Patients with EGFR mutation tend to have lower PD-L1expression and higher CD8 + TIL infiltrating though the difference is not statistically significant. Quite a few have found that NSCLCs harboring EGFR mutations are associated with low overall response rate to PD-1/PD-L1 inhibitors[41], which is partly explained by its negative association with PD-1/PD-L1 expression and uninflamed tumor microenvironment[42]. A careful evaluation of EGFR mutation status will be required to better tailor immunotherapy regimen for NSCLC. At variance with current literature, we don't find any association between PD-1/PD-L1/TIL and KRAS mutation. Unlike EGFR, KRAS mutation had been associated with superior efficacy in the OAK and Checkmate 057 trials[5, 43]. Subsequently, quite a few studies implied that KRAS mutation was associated higher PD-L1 expression [44-46]. Noteworthy, our study carried out the analysis based on mRNA expressing level of PD-1/PD-L1, whereas the previous studies analyzed PD-1/PD-L1 from the perspective of protein expression. A newly published study[47] indicated that RAS regulates PD-L1 through post-transcriptional mechanism, which explain the negative finding in our study. Our study also indicates that both CD8+ TIL and PD-1 is positively associated with female gender and adenocarcinoma histology subtype whereas PD-L1 expression level tend to be higher in squamous cell carcinoma, which is consistent with the previous report[11, 48].
Several limitations were attached with our study. TILs are not randomly distributed but are located in specific areas, like core of tumor or invasive margin. Yet the gene signature based quantification of CD8+ TIL fails to classify TILs of different localization, which might reduce the precision of TIL quantification and constrain further analysis. Also, our analysis was carried out based on transcriptional level of PD-1/PD-L1, which may not actually reflect the protein expression status as the traditional detecting method do. But at the same time, it could actually become an advantage as it could get rid of the congenital drawbacks like the differences in the choice of primary IHC antibody, cut-off value of positivity etc. that associate with immunohistochemistry. Nevertheless, the clinical validation of this approach is definitely warranted. Although our study has some limitations, including that the cutoff values of PD-L1 and PD-1 need clinical validation, the fidelity of gene set enrichment scoring needs further confirmation, it is nevertheless valuable because we exemplify the clinical significance of tumor microenvironment classification based on PD-1/PD-L1 and TIL combination on a large scale of NSCLC cases. Further efforts to integrate PD-1/PD-L1, TIL and many other associated biomarkers to better unravel tumor immune microenvironment status are warranted.
In conclusion, the classification of tumors into four microenvironment subtypes based on PD-1/ PD-L1 status and CD8+ TIL is an appropriate approach to stratify patients of different clinical outcome and better guide the application of immunotherapy in the clinical practice.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work is supported by grants from the National Natural Science foundation of China (81570008, Yanbin Zhou) and the Natural Science Foundation of Guangdong Province of China (2014A030313052, Yanbin Zhou).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferrara R, Mezquita L, Besse B. Progress in the Management of Advanced Thoracic Malignancies in 2017. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2018
2. Mathew M, Safyan RA, Shu CA. PD-L1 as a biomarker in NSCLC: challenges and future directions. Annals of translational medicine. 2017;5:375
3. Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS. et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563-7
4. Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE. et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. The New England journal of medicine. 2015;373:1627-39
5. Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J. et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet (London, England). 2017;389:255-65
6. Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J. et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet (London, England). 2016;387:1837-46
7. Wu SP, Liao RQ, Tu HY, Wang WJ, Dong ZY, Huang SM. et al. Stromal PD-L1+regulatory T cells and PD-1+CD8+ T cells define the response of different subsets of non-small-cell lung cancer to PD-1/PD-L1 blockade immunotherapy. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2017
8. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (New York, NY). 2015;348:124-8
9. Teng MW, Ngiow SF, Ribas A, Smyth MJ. Classifying Cancers Based on T-cell Infiltration and PD-L1. Cancer research. 2015;75:2139-45
10. Cui S, Dong L, Qian J, Ye L, Jiang L. Classifying Non-Small Cell Lung Cancer by Status of Programmed Cell Death Ligand 1 and Tumor-Infiltrating Lymphocytes on Tumor Cells. Journal of Cancer. 2018;9:129-34
11. Yang H, Shi J, Lin D, Li X, Zhao C, Wang Q. et al. Prognostic value of PD-L1 expression in combination with CD8(+) TILs density in patients with surgically resected non-small cell lung cancer. Cancer medicine. 2018;7:32-45
12. Ock CY, Keam B, Kim S, Lee JS, Kim M, Kim TM. et al. Pan-Cancer Immunogenomic Perspective on the Tumor Microenvironment Based on PD-L1 and CD8 T-Cell Infiltration. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016;22:2261-70
13. Tosolini M, Algans C, Pont F, Ycart B, Fournie JJ. Large-scale microarray profiling reveals four stages of immune escape in non-Hodgkin lymphomas. Oncoimmunology. 2016;5:e1188246
14. Mutation Burden Predicts Anti-PD-1 Response. Cancer discovery. 2018.
15. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science (New York, NY). 2017;357:409-13
16. Abiko K, Matsumura N, Hamanishi J, Horikawa N, Murakami R, Yamaguchi K. et al. IFN-gamma from lymphocytes induces PD-L1 expression and promotes progression of ovarian cancer. British journal of cancer. 2015;112:1501-9
17. Muller M, Schouten RD, De Gooijer CJ, Baas P. Pembrolizumab for the treatment of non-small cell lung cancer. Expert review of anticancer therapy. 2017;17:399-409
18. Horn L, Spigel DR, Vokes EE, Holgado E, Ready N, Steins M. et al. Nivolumab Versus Docetaxel in Previously Treated Patients With Advanced Non-Small-Cell Lung Cancer: Two-Year Outcomes From Two Randomized, Open-Label, Phase III Trials (CheckMate 017 and CheckMate 057). Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2017 Jco2017743062
19. Boxberg M, Steiger K, Lenze U, Rechl H, von Eisenhart-Rothe R, Wortler K. et al. PD-L1 and PD-1 and characterization of tumor-infiltrating lymphocytes in high grade sarcomas of soft tissue - prognostic implications and rationale for immunotherapy. Oncoimmunology. 2018;7:e1389366
20. Takada K, Toyokawa G, Shoji F, Okamoto T, Maehara Y. The Significance of the PD-L1 Expression in Non-Small-Cell Lung Cancer: Trenchant Double Swords as Predictive and Prognostic Markers. Clinical lung cancer. 2018;19:120-9
21. Vrankar M, Zwitter M, Kern I, Stanic K. PD-L1 expression can be regarded as prognostic factor for survival of non-small cell lung cancer patients after chemoradiotherapy. Neoplasma. 2018;65:140-6
22. Rashed HE, Abdelrahman AE, Abdelgawad M, Balata S, Shabrawy ME. Prognostic Significance of Programmed Cell Death Ligand 1 (PD-L1), CD8+ Tumor-Infiltrating Lymphocytes and p53 in Non-Small Cell Lung Cancer: An Immunohistochemical Study. Turk patoloji dergisi. 2017;1:211-22
23. Wakabayashi O, Yamazaki K, Oizumi S, Hommura F, Kinoshita I, Ogura S. et al. CD4+ T cells in cancer stroma, not CD8+ T cells in cancer cell nests, are associated with favorable prognosis in human non-small cell lung cancers. Cancer science. 2003;94:1003-9
24. Mori M, Ohtani H, Naito Y, Sagawa M, Sato M, Fujimura S. et al. Infiltration of CD8+ T cells in non-small cell lung cancer is associated with dedifferentiation of cancer cells, but not with prognosis. The Tohoku journal of experimental medicine. 2000;191:113-8
25. Lin G, Fan X, Zhu W, Huang C, Zhuang W, Xu H. et al. Prognostic significance of PD-L1 expression and tumor infiltrating lymphocyte in surgically resectable non-small cell lung cancer. Oncotarget. 2017;8:83986-94
26. Tokito T, Azuma K, Kawahara A, Ishii H, Yamada K, Matsuo N. et al. Predictive relevance of PD-L1 expression combined with CD8+ TIL density in stage III non-small cell lung cancer patients receiving concurrent chemoradiotherapy. European journal of cancer (Oxford, England: 1990). 2016;55:7-14
27. Hofman P. PD-L1 immunohistochemistry for non-small cell lung carcinoma: which strategy should be adopted?. Expert review of molecular diagnostics. 2017:1-12
28. Troncone G, Gridelli C. The reproducibility of PD-L1 scoring in lung cancer: can the pathologists do better? Translational lung cancer research. 2017;6:S74-s7
29. Mazzaschi G, Madeddu D, Falco A, Bocchialini G, Goldoni M, Sogni F. et al. Low PD-1 expression in Cytotoxic CD8+ Tumor infiltrating Lymphocytes Confers an Immune Privileged Tissue Microenvironment in NSCLC with a Prognostic and Predictive Value. Clinical cancer research: an official journal of the American Association for Cancer Research. 2017
30. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD. et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. The New England journal of medicine. 2015;372:2509-20
31. Rizvi H, Sanchez-Vega F, La K, Chatila W, Jonsson P, Halpenny D. et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand (PD-L)-Ligand 1 Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2018 Jco2017753384
32. McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK. et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science (New York, NY). 2016;351:1463-9
33. Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH. et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome research. 2014;24:743-50
34. Chen YP, Zhang Y, Lv JW, Li YQ, Wang YQ, He QM. et al. Genomic Analysis of Tumor Microenvironment Immune Types across 14 Solid Cancer Types: Immunotherapeutic Implications. Theranostics. 2017;7:3585-94
35. Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S. et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165:35-44
36. Danilova L, Wang H, Sunshine J, Kaunitz GJ, Cottrell TR, Xu H. et al. Association of PD-1/PD-L axis expression with cytolytic activity, mutational load, and prognosis in melanoma and other solid tumors. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E7769-e77
37. Lee V, Murphy A, Le DT, Diaz LA Jr. Mismatch Repair Deficiency and Response to Immune Checkpoint Blockade. The oncologist. 2016;21:1200-11
38. Takamochi K, Takahashi F, Suehara Y, Sato E, Kohsaka S, Hayashi T. et al. DNA mismatch repair deficiency in surgically resected lung adenocarcinoma: Microsatellite instability analysis using the Promega panel. Lung cancer (Amsterdam, Netherlands). 2017;110:26-31
39. Zhang M, Li G, Wang Y, Wang Y, Zhao S, Haihong P. et al. PD-L1 expression in lung cancer and its correlation with driver mutations: a meta-analysis. Scientific reports. 2017;7:10255
40. Haratake N, Toyokawa G, Takada K, Kozuma Y, Matsubara T, Takamori S. et al. Programmed Death-Ligand 1 Expression and EGFR Mutations in Multifocal Lung Cancer. The Annals of thoracic surgery. 2017
41. Gainor JF, Shaw AT, Sequist LV, Fu X, Azzoli CG, Piotrowska Z. et al. EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD-1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016;22:4585-93
42. Soo RA, Lim SM, Syn NL, Teng R, Soong R, Mok TSK. et al. Immune checkpoint inhibitors in epidermal growth factor receptor mutant non-small cell lung cancer: Current controversies and future directions. Lung cancer (Amsterdam, Netherlands). 2018;115:12-20
43. Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A. et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. The New England journal of medicine. 2016;375:1823-33
44. Parra ER, Villalobos P, Zhang J, Behrens C, Mino B, Swisher S. et al. Immunohistochemical and Image Analysis-Based Study Shows That Several Immune Checkpoints are Co-expressed in Non-Small Cell Lung Carcinoma Tumors. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2018;13:779-91
45. Chen N, Fang W, Lin Z, Peng P, Wang J, Zhan J. et al. KRAS mutation-induced upregulation of PD-L1 mediates immune escape in human lung adenocarcinoma. Cancer immunology, immunotherapy: CII. 2017;66:1175-87
46. Falk AT, Yazbeck N, Guibert N, Chamorey E, Paquet A, Ribeyre L. et al. Effect of mutant variants of the KRAS gene on PD-L1 expression and on the immune microenvironment and association with clinical outcome in lung adenocarcinoma patients. Lung cancer (Amsterdam, Netherlands). 2018;121:70-5
47. Coelho MA, de Carne Trecesson S, Rana S, Zecchin D, Moore C, Molina-Arcas M. et al. Oncogenic RAS Signaling Promotes Tumor Immunoresistance by Stabilizing PD-L1 mRNA. Immunity. 2017;47:1083-99.e6
48. Janzic U, Kern I, Janzic A, Cavka L, Cufer T. PD-L1 Expression in Squamous-cell Carcinoma and Adenocarcinoma of the Lung. Radiology and oncology. 2017;51:357-62
Author contact
![]() Corresponding author: Professor Yanbin Zhou; E-mail: sysuzybcom; Department of Respiratory Medicine, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China; Tel: +86-13925188388
Corresponding author: Professor Yanbin Zhou; E-mail: sysuzybcom; Department of Respiratory Medicine, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China; Tel: +86-13925188388

 Global reach, higher impact
Global reach, higher impact