Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(24):4611-4617. doi:10.7150/jca.26794 This issue Cite
Research Paper
Detection of CK19 mRNA Using One-step Nucleic Acid Amplification (OSNA) in Prostate Cancer: Preliminary Results
1. University Hospital for Urology, Klinikum Oldenburg, School of Medicine and Health Sciences, Carl von Ossietzky University Oldenburg, Rahel-Straus-Straße 10, 26133 Oldenburg, Germany
2. Institute of Pathology Oldenburg, Taubenstraße 28, 26122 Oldenburg, Germany
Received 2018-4-20; Accepted 2018-10-10; Published 2018-11-24
Abstract
Background: Accurate histopathological evaluation of lymph nodes (LNs) is essential for reliable staging in prostate cancer. In routine practice, conventional techniques only examine parts of the LN. Molecular nodal staging methods are limited by their high costs and extensive time requirement. One-step nucleic acid amplification (OSNA) determines the metastatic status of the complete LN and allows for rapid intraoperative detection of LN metastases. OSNA has been proposed for diagnosis of LN metastases from breast cancer by quantifying the CK19 mRNA copy number. To provide basic data for OSNA development for prostate cancer, we conducted an investigation of CK19 and OSNA in prostate cancer specimens.
Methods: OSNA is based on a short homogenization step and subsequent automated amplification of CK19 mRNA directly from the sample lysate, with results available in 30-40 min. A total of 20 prostate cancer specimens from consecutive patients with intermediate or high-risk prostate cancer (Gleason-Score ≥7) were investigated by both OSNA and conventional histopathology (H&E staining, CK19 immunohistochemistry). OSNA was performed on frozen samples using a ready-to-use amplification kit in an automated real-time detection system. Samples were defined as 'negative' or 'positive' according to mRNA copy number: >5000 copies/µl (++), 250-5000 copies/µl (+), and <250 copies/µl (-).
Results: Histopathological analysis confirmed prostate cancer in all samples: Gleason score 7 (n=11), Gleason score 8 (n=2), and Gleason score 9 (n=6). Gleason score could not be given for one patient who previously underwent hormonal treatment. OSNA analysis detected CK19 expression in 100% of the specimens and high numbers of CK19 mRNA copies in all cases (9 samples ++; 11 samples +). Immunohistochemistry confirmed CK19 expression in 19 of 20 cases. In the immunohistochemistry CK19-negative patient, a Gleason score 9 prostate cancer was diagnosed.
Conclusions: This is the first study using OSNA to detect CK19 expression in prostate cancer. Initial data indicate that this rapid method for molecular LN staging reliably identifies CK19 mRNA in prostate cancer. These results suggest that the OSNA assay may be suitable to improve (intraoperative) LN staging in prostate cancer. For further verification, OSNA analysis of LN specimens from prostate cancer patients is required.
Keywords: prostate cancer, CK19, OSNA, molecular lymph node staging, IDEAL-D
Background
Prostate cancer is the most common cancer and the third leading cause of cancer-related death among men in developed countries [1]. The lymph node (LN) status is a crucial prognostic factor in prostate cancer. Nodal metastases occur in 3-80% of men with clinically localized prostate cancer [2]. In addition, the presence of nodal metastases has a strong negative impact on survival [3, 4]. Furthermore, an accurate pathological assessment of LN status plays an important role in guiding the therapeutic management of prostate cancer patients. Multimodal treatments for LN-positive patients have been gaining interest in recent years, particularly in patients with high-stage diseases [5].
Despite the advances in imaging, the histological detection of LN metastases or pelvic lymphadenectomy is still the gold standard for LN staging in clinically localized prostate cancer [6]. The sensitivity of abdominal computed tomography (CT) and multiparametric magnetic resonance imaging (mpMRI) for detection of LN metastases is <40% [7]. New imaging methods, like 11C- or 18F- choline- and 68Ga-prostate-specific membrane antigen (PSMA) positron emission tomography (PET)/CT, enable the detection of small metastases, but the reliability of these procedures is limited by their spatial resolution (<5 mm) resulting in a low estimated sensitivity (49-66%) in the detection of LN (micro-)metastases before treatment of prostate cancer [8, 9]. Studies evaluating micrometastases (LN deposits <2 mm in diameter) identified occult LN metastases in 5-16% of patients [10, 11].
Due to the high proportion of small LN metastases or micrometastases in prostate cancer, the manner of histological LN evaluation has a considerable influence on the number of LN-positive patients. Wawroschek et al. found that the detection rate of LN metastases correlated with the number of sections examined [12]. Additionally, immunohistochemistry (IHC), e.g. pancytokeratin, and molecular analysis of the tissue (e.g., real time polymerase chain reaction (RT-PCR)) can help identify occult metastases or micrometastases [13, 14, 15]. Unlike in the case of other solid tumors, a highly specific marker for prostate cancer (prostate-specific antigen (PSA)) is available. PSA is not expressed in healthy LNs. The feasibility of molecular staging using PSA RT-PCR has been shown in various studies [16]. Schostak et al. demonstrated the additional value of molecular staging by PSA RT-PCR in a study examining 457 LNs from 70 patients with a 6-year follow-up [17]. However, in clinical routine, histopathological evaluation of LNs is usually not extensively carried out. Macroscopic identification of LNs without additional means of help and the preparation of at least one section (3 to 4 mm slices in case of larger LNs) in the longitudinal or transversal plane and staining with hematoxylin and eosin (H&E) is the standard in many centers [18]. Due to high expenditure of time and limitations in standardization and automatization, molecular methods for LN evaluation in prostate cancer including PSA RT-PCR are only applied in studies. For these reasons, RT-PCR is also not suitable for intraoperative use.
During the last few years, new biomolecular techniques have been used to assess LN metastases. In breast cancer, one-step nucleic acid amplification (OSNA) has recently been indicated as a fast-molecular intraoperative approach for the detection of LN metastases [19, 20, 21]. This procedure exploits that CK19 is normally not expressed in lymphatic tissue. The OSNA procedure has also shown promising results in other tumor types (e.g., colorectal cancer) and is already approved for in vitro LN diagnostics in other tumors [22]. OSNA is a new method that uses the reverse-transcription loop-mediated isothermal amplification (RT-LAMP) technique for gene amplification and quantifies the number of tissue CK19 mRNA copies [19, 23]. In conventional PCR, there is a risk of amplification of nonspecific genomic deoxyribonucleic acid (DNA). The high specific OSNA analysis eliminates this limitation by using 6 primers and an isothermal reaction. Immunohistochemical studies revealed that CK19 expression occurs frequently in basal as well as in luminal cells of normal, dysplastic and benign hyperplastic prostatic tissues and in prostate cancer [24]. However, data concerning CK19-RNA expression in prostatic tissue are still missing.
The aim of this study was to provide such data as a basis for the development of OSNA for LN staging in prostate cancer. Primary prostate cancer tissue samples were analyzed using OSNA and the findings were compared with standard histopathological techniques and IHC.
Methods
Patient population and inclusion criteria for analysis
This study was designed as an IDEAL-D Stage 2a study [25].
A total of 20 consecutive patients with intermediate or high-risk prostate cancer scheduled for open radical retropubic prostatectomy between June 2015 and February 2016 were included in this study (Research Registry UIN: researchregistry2705; http://www.researchregistry.com/browse-the-registry.html#home/registrationdetails/5956cf785c7b9761b99fcaa1/). Inclusion criteria were a Gleason score ≥7 and at least >50% cores affected with cancer on one side at the time when the diagnosis was made. Informed and written consent was obtained from all patients.
One-Step Nucleic Acid Amplification (OSNA) procedure
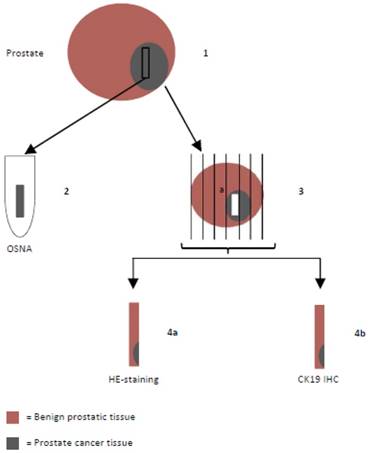
Prostates were harvested as native specimens immediately after surgery, put on ice and transported to the pathologist who extracted the tissue sample designated for OSNA (weight of the samples were 180-800 mg) after macroscopic and palpatory detection of the tumor areas. Directly adjacent sections were examined histopathologically to confirm tumorous tissue (instantaneous sections). In case of missing confirmation, another sample would have been taken. Because of including only patients with a high tumor volume (at least >50% cores affected with cancer on one side) or macroscopically well definable tumors this procedure was not necessary in any case. Samples were snap-frozen and stored at -80°C (Figure 1). For the OSNA analysis, samples were processed using a designated instrument (RD-100i) and reagent system (LYNOAMP and LYNORHAG) according to the instructions of the manufacturer (Sysmex Corporation, Kobe, Japan). Each prostate tissue sample was homogenized in 4 ml of lysing buffer LYNORHAG (0.2 M glycine-HCl pH 3.5, 5% Brij35 and 20% DMSO) for 60 s at 10,000 x g with a Polytron System PT1300D (Kinematica AG, Switzerland) and LYNOPREP blades to prepare homogeneous lysates. One milliliter of lysate was centrifuged to remove cell debris and then further diluted 1:10 and 1:100 with LYNORHAG. The diluted lysates were used directly for amplification without RNA extraction or purification. Isothermal amplification reactions were performed at 65°C using LYNOAMP amplification reagents. The rise time required for precipitation of magnesium pyrophosphate to reach a turbidity of 0.1 OD at 465 nm was obtained for each sample and the number of CK19 mRNA copies were determined using a calibration curve. According to previous studies, samples were defined as 'negative' or 'positive' according to mRNA copy number [19, 20, 22]. Positive OSNA results of >5000 copies/µl were designated as (++), 250-5000 copies/µl as (+) or (+)I (if tissue inhibition occurred during the amplification reaction and the result is based on the 1:100 diluted sample). Samples with less than 250 copies/µl were considered as a negative result (-).
Hematoxylin & eosin (H&E) staining
Prostate tissue directly adjacent to the sample that was prepared for OSNA was analyzed by conventional histopathology. Formalin-fixed tissue was embedded in paraffin and cut in 5 µm slices. After separation of paraffin and rehydration, slices were stained with H&E and CK19 (Figure 1).
Histopathological examination flow chart: Prostates were put on ice to avoid mRNA degradation and transported to the pathologist immediately after radical prostatectomy. (1) One tissue sample designated for OSNA was extracted, snap-frozen and stored at -80°C. (2) OSNA analysis. (3) Formalin-fixed prostate tissue was embedded in paraffin and conventionally sectioned. (4) Slices were stained with hematoxylin and eosin (H&E) (4a) and CK19 immunohistochemistry (IHC) was performed (4b).
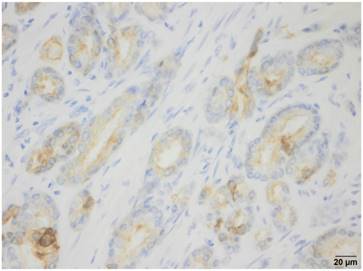
CK19 immunohistochemistry
CK19 staining was done automatically with the Dako Envision Dual Link System-HRP (Dako, Germany). After blocking of endogeneous peroxidase, slices were sequentially incubated with CK19 antibody (A53-B/A2.26, medac GmbH, Germany), labeled polymer and DAB, and counterstained afterwards with hematoxylin.
Results
A summary of patient and tumor characteristics is shown in Table 1. Conventional histopathological analysis confirmed prostate cancer in all samples: Gleason score 7 (n=11), Gleason score 8 (n=2), and Gleason score 9 (n=6). Gleason score could not be given for one patient who underwent hormonal treatment prior to radical retropubic prostatectomy.
OSNA analysis detected CK19 expression in 100% of the specimens. The OSNA assays revealed high numbers of CK19 mRNA copies in all cases (9 samples ++; 11 samples (+)I). IHC performed on prostate tissue directly adjacent to the sample used for OSNA detected CK19 expression in 19 of 20 cases. In the patient with negative CK19 staining by IHC, a Gleason score 9 prostate cancer was diagnosed by conventional histopathological examination. Figure 2 shows one example of CK19 IHC of a prostate cancer specimen.
Discussion
The assessment of the nodal status is a key factor in prostate cancer staging and therapy. The risk of progression can be calculated and the appropriate adjuvant therapy (e.g., hormonal treatment, radiotherapy) can be planned. Histopathological examination is currently the basic technique used to assess LNs in prostate cancer patients. However, consensus guidelines regarding the optimal approach are currently lacking [26]. The commonly used conventional histopathological analysis does not precisely evaluate the LN status. A certain proportion of small metastases or micrometastases can remain undetected by routine histopathology [12]. Usually, only several sections from each LN are examined. Serial sectioning is more sensitive, but many slides have to be examined [27]. Therefore, this procedure is not necessarily suitable for use as a standard procedure. Molecular methods allow the examination of the entire tissue of a LN, resulting in a higher sensitivity for the detection of minor tumor deposits in LNs compared with conventional histopathology. In prostate cancer, several studies showed that recurrence can occur in patients in which the routine histopathology from removed LNs was negative. In the study by Haas et al., four of 12 prostate cancer patients with biochemical relapse, but without (immuno)histologically detectable tumor cells, had LNs that were RT-PCR-positive for PSA [13].
In prostate cancer, molecular methods for the detection of LN invasion are not yet performed in routine clinical practice, due to the complexity and time-consuming nature. Moreover, they cannot be used as an intraoperative examination. Intraoperative LN frozen sections could, in principle, be used at radical prostatectomy. However, frozen section diagnosis of metastatic carcinoma in pelvic LNs before radical prostatectomy has a high false negative rate and is costly. Young et al. found a false-negative rate of 33%, which estimated the cost of metastatic cancer detection to be over $10,000 [28].
CK19 immunohistochemistry: Example of a CK19-positive prostate cancer specimen.
Patient and tumor characteristics and results of histopathological analysis, OSNA and CK19 immunohistochemistry
| Age range | Preoperative PSA [ng/ml] | Preoperative Gleason score | Histopathological analysis, pathologic stage | OSNA | IHC | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pT | Postoperative Gleason score | pN | CK19 mRNA[copies/ µl] | CK19 mRNA[copies/µl] per g | |||||||
| 1 | 72 | 10.09 | 8 (4+4) | 3b | 7 (4+3) | 1 | 5,200 | 8,661 | (++) | + | |
| 2 | 73 | 49.68 | 9 (4+5) | 4 | 9 (4+5) | 1 | 1,100 | 1,454 | (+)I | + | |
| 3* | 59 | 160.00 | 8 (4+4) | ypT3b | n/a | 1 | 39,000 | 82,750 | (++) | + | |
| 4 | 65 | 52.84 | 7 (4+3) | 3a | 7 (3+4) | 1 | 12,000 | 17,544 | (++) | + | |
| 5 | 66 | 13.17 | 8 (4+4) | 3b | 8 (4+4) | 0 | 5,200 | 18,446 | (+)I | + | |
| 6 | 59 | 18.00 | 7 (4+3) | 3b | 7 (3+4) | 0 | 5,200 | 16,169 | (+)I | + | |
| 7 | 68 | 5.00 | 7 (3+4) | 2c | 7 (3+4) | 0 | 7,800 | 14,734 | (+)I | + | |
| 8 | 66 | 7.18 | 7 (4+3) | 3a | 9 (4+5) | 1 | 39,000 | 136,555 | (++) | + | |
| 9 | 68 | 19.40 | 8 (4+4) | 3a | 8 (4+4) | 1 | 8,800 | 39,747 | (+)I | + | |
| 10 | 55 | 12.54 | 7(3+4) | 2c | 7 (3+4) | 0 | 47,000 | 263,789 | (++) | + | |
| 11 | 55 | 29.00 | 7 (4+3) | 3b | 7 (4+3) | 0 | 250,000 | 980,392 | (++) | + | |
| 12 | 77 | 8.58 | 9 (5+4) | 3b | 9 (5+4) | 1 | 20,000 | 212,670 | (++) | - | |
| 13 | 75 | 6.76 | 9 (5+4) | 3b | 9 (5+4) | 1 | 320 | 627 | (+)I | + | |
| 14 | 66 | 9.61 | 7 (4+3) | 2c | 9 (4+5) | 0 | 32,000 | 60,264 | (+)I | + | |
| 15 | 69 | 25.57 | 7 (4+3) | 3a | 7 (4+3) | 0 | 6,900 | 12,591 | (+)I | + | |
| 16 | 60 | 10.29 | 7 (3+4) | 3a | 7 (4+3) | 0 | 6,900 | 229,358 | (++) | + | |
| 17 | 65 | 5.81 | 8 (4+4) | 3b | 9 (4+5) | 1 | 20,000 | 46,948 | (+)I | + | |
| 18 | 59 | 7.32 | 7 (3+4) | 3b | 7 (3+4) | 1 | 170,000 | 213,836 | (+)I | + | |
| 19 | 75 | 14.00 | 7 (4+3) | 3b | 7 (4+3) | 1 | 71,000 | 387,978 | (+)I | + | |
| 20 | 66 | 4.90 | 7 (3+4) | 2c | 7 (3+4) | 0 | 110,000 | 223,810 | (++) | + | |
| Median | 66 | 11.42 | |||||||||
The OSNA procedure could represent a possible alternative for intraoperative detection of LN metastases in prostate cancer patients. This method is clinically used for the evaluation of LN metastases in breast cancer and has already been used in colorectal cancer, gastric cancer and head and neck cancer [22, 29-32]. OSNA quantitatively measures the levels of CK19 mRNA, which is not expressed in normal lymphatic tissue. Previous studies and results from the current study have demonstrated that CK19 expression occurs frequently in prostatic tissue or prostate cancer, suggesting that OSNA could also be suitable for the detection of LN metastases in prostate cancer [24]. OSNA offers the advantage that the supernatant of a homogenized LN solution can be directly analyzed without the mRNA purification process that is usually required in PCR. The isothermally performed reaction can detect CK19 mRNA without the interference of pseudogenes. The OSNA assay can also differentiate contamination of a few benign epithelial cells by using a verified cut off value. The use of an automated gene amplification machine permits a standardized and rapid procedure. A single examination takes 30-40 min. Besides, the procedure is accompanied with reasonable costs. For example, in prostate cancer, the cost would be around 80 euros per node.
In the present study, OSNA was used for the first time to detect CK19 expression in prostate cancer patients. Our results showed that a high number of CK19 mRNA copies could be detected by this automated amplification approach in all prostate cancer specimens examined regardless of the degree of tissue differentiation (Gleason score). Thus, our findings help to provide basic data for OSNA development for prostate cancer.
The OSNA assay, for example, combined with sentinel node biopsy, could be a tool that gives the possibility to the surgeon to choose intraoperatively whether to perform an additional extended pelvic lymphadenectomy in prostate cancer patients. A systematic literature review showed a high diagnostic accuracy for sentinel node biopsy in detecting positive LNs in prostate cancer [33]. In a recent consensus panel meeting, consensus was obtained on the statement that sentinel guided LN dissection should be combined with an extended lymphadenectomy, at least in patients with intermediate and high-risk prostate cancer, to resect additional LN metastases in non-sentinel nodes [34]. However, the rate of complications rises along with the number of LNs removed [35, 36]. Accordingly, one possible procedure might be to perform a sentinel LN dissection only as a first step. During the removal of the prostate the sentinel nodes could be analyzed using the OSNA approach. In case of positive sentinel nodes, an extended lymphadenectomy could follow after the prostatectomy. In the case of negative OSNA results, patients could be spared from unnecessary morbidity caused by extended lymphadenectomy.
Some limitations of our study include the small sample size and that only primary specimens with a relatively high tumor load were examined. For further verification, OSNA analysis of LN specimens from prostate cancer patients are required. However, in order to assure highest transparency of results and to avoid redundant replications of similar study protocols we have registered this study at the newly built online-based registry platform [37, 38]. The IDEAL recommendations have primarily been developed for surgical techniques, however, the recommendations are shifting and being elaborated in order to cover devices, non-invasive techniques or diagnostic tools and innovations as is the case in this current study. Although, the OSNA technique has been used in other cancer types, our study represents the first of its kind in prostate cancer and merits the respective classification in this particular indication. Protocol- and number wise the current study mostly fits the characteristics of an IDEAL-D device protocol [25]. Therefore it has been labeled Stage 2a accordingly. In addition, IDEAL clearly provides specific strategies and settings for further studies to pursue OSNA analysis of LN specimens from prostate cancer patients.
Conclusions
This is the first study using OSNA to detect CK19 expression in prostate cancer specimens. Initial data indicate that this new rapid method for molecular LN staging reliably identifies CK19 mRNA in prostate cancer. Therefore, OSNA analysis may be suitable to determine the metastatic status of the complete LN and could improve (intraoperative) LN staging in prostate cancer. To verify this promising opportunity, OSNA analysis of LN specimens from prostate cancer patients are required.
Abbreviations
CT: computed tomography; CK19: cytokeratin 19; H&E: hematoxylin and eosin; IHC: immunohistochemistry; LN: lymph node; mpMRI: multiparametric magnetic resonance imaging; OSNA: one-step nucleic acid amplification; PET: positron emission tomography; PSA: prostate-specific antigen; PSMA: prostate-specific membrane antigen; RT-LAMP: reverse transcription loop-mediated isothermal amplification.
Acknowledgements
We thank Gabrielle White Wolf, PhD, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Authors' contributions
AW participated in the conception of the study and acquisition and interpretation of data, and drafted the manuscript. SE performed the analysis, reviewed the literature, and was involved in drafting the manuscript. PG participated in the acquisition and analysis of data. MCS participated in the acquisition and analysis of data. RPH carried out histological examinations and was involved in critically revising the manuscript for important intellectual content. HG participated in the conception of the study, acquisition and interpretation of data, and was involved in critically revising the manuscript for important intellectual content. FW participated in the conception and design of the study and interpretation of data, and was involved in critically revising the manuscript for important intellectual content. All authors have read and approved the final manuscript.
Funding
The cost to publish as open access was financed by the DFG (Deutsche Forschungsgemeinschaft).
Ethics approval and consent to participate
The study protocol was approved by the ethics committee of the Hannover Medical School, Germany (No. 2697-2015). Informed and written consent was obtained from all patients.
Availability of data and materials
All the data and materials supporting the conclusions were included in the main paper. Further data were available from the corresponding author on reasonable request.
Competing Interests
The OSNA assay was supported by Sysmex Europe, Norderstedt, Germany. Otherwise the authors declare that they have no competing interests.
References
1. Haas GP, Delongchamps N, Brawley OW, Wang CY, de la RG. The worldwide epidemiology of prostate cancer: perspectives from autopsy studies. Can J Urol. 2008;15:3866-71
2. Winter A, Kneib TH, Wasylow C. et al. Updated nomogram incorporating percentage of positive cores to predict probability of lymph node invasion in prostate cancer patients undergoing sentinel lymph node dissection. J Cancer. 2017;8:2692-98
3. Eggener SE, Scardino PT, Walsh PC. et al. Predicting 15-year prostate cancer specific mortality after radical prostatectomy. J Urol. 2011;185:869-75
4. Joniau S, Briganti A, Gontero P, et al.; European Multicenter Prostate Cancer Clinical, Translational Research Group (EMPaCT). Stratification of high-risk prostate cancer into prognostic categories: a European multi-institutional study. Eur Urol. 2015;67:157-64
5. Gandaglia G, Karnes RJ, Cozzarini C, Montorsi F, Briganti A. Node-positive prostate cancer: a call for level 1 evidence. Oncology. 2015;29:118 125,141
6. Mottet N, Bellmunt J, Briers E. et al. EAU - ESTRO - ESUR - SIOG guidelines on prostate cancer 2017. Available from: http://uroweb.org/guideline/prostate-cancer/. Accessed October. 2017
7. Heidenreich A, Bastian PJ, Bellmunt J. et al. European Association of Urology. EAU guidelines on prostate cancer. part 1: screening, diagnosis, and local treatment with curative intent — update 2013. Eur Urol. 2014;65:124-37
8. Evangelista L, Guttilla A, Zattoni F, Muzzio PC, Zattoni F. Utility of choline positron emission tomography/computed tomography for lymph node involvement identification in intermediate- to high-risk prostate cancer: a systematic literature review and metaanalysis. Eur Urol. 2013;63:1040-8
9. Maurer T, Gschwend JE, Rauscher I. et al. Diagnostic efficacy of 68gallium-PSMA positron emission tomography compared to conventional imaging for lymph node staging of 130 consecutive patients with intermediate to high risk prostate cancer. J Urol. 2016;195:1436-43
10. Clobes H, Fossâ SD, Waehre H, Jocham D, Berner A. The immunohistochemical assessment of occult regional lymph node metastases in patients with T3pN0M0 prostate cancer before definitive radiotherapy. BJU Int. 2000;85:270-5
11. Gomella LG, White JL, McCue PA, Byrne DS, Mulholland SG. Screening for occult nodal metastasis in localized carcinoma of the prostate. J Urol. 1993;149:776-8
12. Wawroschek F, Wagner T, Hamm M. et al. The influence of serial sections, immunohistochemistry, and extension of pelvic lymph node dissection on the lymph node status in clinically localized prostate cancer. Eur Urol. 2003;43:132-6
13. Haas CJ, Wagner T, Wawroschek F, Arnholdt H. Combined application of RT-PCR and immunohistochemistry on paraffin embedded sentinel lymph nodes of prostate cancer patients. Pathol Res Pract. 2005;200:763-70
14. Terakawa T, Miyake H, Kurahashi T, Furukawa J, Takenaka A, Fujisawa M. Improved sensitivity for detecting micrometastases in pelvic lymph nodes by real-time reverse transcriptase polymerase chain reaction (RT-PCR) compared with conventional RT-PCR in patients with clinically localized prostate cancer undergoing radical prostatectomy. BJU Int. 2009;103:1074-8
15. Heck MM, Retz M, Bandur M. et al. Topography of lymph node metastases in prostate cancer patients undergoing radical prostatectomy and extended lymphadenectomy: results of a combined molecular and histopathologic mapping study. Eur Urol. 2014;66:222-9
16. Kollermann J, Müller M, Goessl C. et al. Methylation-specific PCR for DNA-based detection of occult tumor cells in lymph nodes of prostate cancer patients. Eur Urol. 2003;44:533-8
17. Schostak M, Krause H, Miller K, Schrader M, Kempkensteffen C, Kollermann J. Does the molecular staging in pelvic lymph nodes improve the detection of relevant prostate cancer metastases? An assessment after 6 years. BJU Int. 2007;99:1409-14
18. Lawrence WD; Association of Directors of Anatomic and Surgical Pathology. ADASP recommendations for processing and reporting of lymph node specimens submitted for evaluation of metastatic disease. Virchows Arch. 2001;439:601-3
19. Tsujimoto M, Nakabayashi K, Yoshidome K. et al. One-step nucleic acid amplification for intraoperative detection of lymph node metastasis in breast cancer patients. Clin Cancer Res. 2007;13:4807-16
20. Visser M, Jiwa M, Horstman A. et al. Intra-operative rapid diagnostic method based on CK19 mRNA expression for the detection of lymph node metastases in breast cancer. Int J Cancer. 2008;122:2562-7
21. Di Filippo F, Di Filippo S, Ferrari AM. et al. Elaboration of a nomogram to predict nonsentinel node status in breast cancer patients with positive sentinel node, intraoperatively assessed with one step nucleic amplification: Retrospective and validation phase. J Exp Clin Cancer Res. 2016;35:193
22. Croner RS, Schellerer V, Demund H. et al. One step nucleic acid amplification (OSNA) - a new method for lymph node staging in colorectal carcinomas. J Transl Med. 2010;8:83
23. Notomi T, Okayama H, Masubuchi H. et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63
24. Peehl DM, Sellers RG, McNeal JE. Keratin 19 in the adult human prostate: tissue and cell culture studies. Cell Tissue Res. 1996;285:171-6
25. Sedrakyan A, Campbell B, Merino JG, Kuntz R, Hirst A, McCulloch P. IDEAL-D: a rational framework for evaluating and regulating the use of medical devices. BMJ. 2016;353:i2372
26. Prendeville S, van der Kwast TH. Lymph node staging in prostate cancer: perspective for the pathologist. Clin Pathol. 2016;69:1039-1045
27. Schiavina R, Capizzi E, Borghesi M. et al. Nodal occult metastases in intermediate- and high-risk prostate cancer patients detected using serial section, immunohistochemistry, and real-time reverse transcriptase polymerase chain reaction: prospective evaluation with matched-pair analysis. Clin Genitourin Cancer. 2015;13:e55-64
28. Young MP, Kirby RS, O'Donoghue EP, Parkinson MC. Accuracy and cost of intraoperative lymph node frozen sections at radical prostatectomy. J Clin Pathol. 1999;52:925-7
29. Yamamoto H, Sekimoto M, Oya M. et al. OSNA-based novel molecular testing for lymph node metastases in colorectal cancer patients: results from a multicenter clinical performance study in Japan. Ann Surg Oncol. 2011;18:1891-1898
30. Yaguchi Y, Sugasawa H, Tsujimoto H. et al. One-step nucleic acid amplification (OSNA) for the application of sentinel node concept in gastric cancer. Ann Surg Oncol. 2011;18:2289-2296
31. Goda H, Nakashiro K, Oka R. et al. One-step nucleic acid amplification for detecting lymph node metastasis of head and neck squamous cell carcinoma. Oral Oncol. 2012;48:958-63
32. Matsuzuka T, Takahashi K, Kawakita D. et al. Intraoperative Molecular Assessment for Lymph Node Metastasis in Head and Neck Squamous Cell Carcinoma Using One-Step Nucleic Acid Amplification (OSNA) Assay. Ann Surg Oncol. 2012;19:3865-3870
33. Wit EM, Acar C, Grivas N. et al. Sentinel Node Procedure in Prostate Cancer: A Systematic Review to Assess Diagnostic Accuracy. Eur Urol. 2017;71:596-605
34. van der Poel HG, Wit EM, Acar C, van den Berg NS, et al.; Sentinel Node Prostate Cancer Consensus Panel Group members. Sentinel node biopsy for prostate cancer: report from a consensus panel meeting. BJU Int. 2017;120:204-211
35. Briganti A, Chun FK, Salonia A. et al. Complications and other surgical outcomes associated with extended pelvic lymphadenectomy in men with localized prostate cancer. Eur Urol. 2006;50:1006-13
36. Winter A, Vogt C, Weckermann D, Wawroschek F. Complications of pelvic lymphadenectomy in clinically localised prostate cancer: different techniques in comparison and dependency on the number of removed lymph nodes. Aktuelle Urol. 2011;42:179-83
37. Agha R, Fowler AJ, Limb C. et al. The First 500 Registrations to the Research Registry®: Advancing Registration of Under-Registered Study Types. Front Surg. 2016;3:50
38. Agha RA, Jafree DJ, Vella-Baldacchino M. et al. Surveying opinions of 149 registrants to the Research Registry: Awareness of and attitudes towards research registration. Int J Surg. 2017;39:182-187
Author contact
![]() Corresponding author: AW: winter.alexanderde
Corresponding author: AW: winter.alexanderde

 Global reach, higher impact
Global reach, higher impact