Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(9):1623-1634. doi:10.7150/jca.24193 This issue Cite
Research Paper
Diagnostic and prognostic value of microRNA-628 for cancers
1. Department of Epidemiology, Binzhou Medical University, YanTai, ShanDong, 264003, P.R.China.
2. Department of Biochemistry and Molecular Biology, Binzhou Medical University, YanTai, ShanDong, 264003, P.R.China.
# These authors contributed equally to this work.
Received 2017-12-5; Accepted 2018-1-24; Published 2018-4-12
Abstract
Background: Many studies manifested miRNA-628 (miR-628) was deregulated in various cancers, indicating that miR-628 might serve as a novel biomarker of cancer diagnosis and prognosis, but it's role was still uncertain. This study aimed to evaluate the value of miR-628 in various cancers for diagnosis and prognosis, as well as its predictive power in combination biomarkers.
Materials and Methods: A literature search was performed using Medline (via PubMed), Embase, Web of Science databases, and Ovid platform up to November 2017. Meta-analysis was performed to provide summative outcomes. Quality assessment of each included study was performed.
Results: Twelve articles with 20 studies were included in our meta-analysis, including 8 articles with 15 studies for diagnostic meta-analysis and 4 articles with 5 studies for prognostic meta-analysis. For the diagnostic meta-analysis of miR-628 alone, the overall pooled results for sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and the area under the summary receiver operating characteristic (SROC) curve (AUC) were 0.81 (95% CI: 0.62-0.91), 0.72 (95% CI: 0.48-0.88), 2.90 (95% CI: 1.50-5.40), 0.27 (95% CI: 0.14-0.50), 11.0 (95% CI: 4.00-25.00), and 0.84 (95% CI: 0.80-0.87), respectively. For the diagnostic meta-analysis of miR-628-related combination biomarkers, the above six parameters were 0.89 (95% CI: 0.84-0.92), 0.93 (95% CI: 0.82-0.97), 12.30 (95% CI: 4.70-32.50), 0.12 (95% CI: 0.08-0.19), and 100.00 (95% CI: 28.00-354.00), 0.93 (95% CI: 0.90-0.95), respectively. For the prognostic meta-analysis, patients with lower miR-628 had significant shorter overall survival than high expression of miR-628 (HR = 1.553, 95% CI: 1.041-2.318, z = 2.16, P = 0.031).
Conclusions: This study confirms that miR-628 may be a promising biomarker for cancer diagnosis and prognosis. Expertly, microRNAs combination biomarkers could be a new alternative for clinical application.
Keywords: biomarker, miR-628, cancer, diagnosis, prognosis, meta-analysis.
Introduction
Cancer is currently the leading cause of human death worldwide, making it a public health issue [1-3]. The overall death rate increased due to the lack of appropriate techniques for cancer early detention and diagnosis [4-7]. For example, the average 5-year-surval rate of patients with lung cancer was only 14 % because of the delayed clinical manifestation and lack of early diagnosis [6]. However, the current diagnostic and prognostic markers with unsatisfactory sensitivity and specificity limit their utility. It is urgent to look for accurate and efficient diagnostic and prognostic biomarkers.
MicroRNAs (miRNAs), as endogenous and non-coding small RNAs (18-22 nucleotides in length), regulate a series of cellular functions, including cell growth, proliferation, differentiation, apoptosis and metabolism [8, 9]. Dysregulation of miRNAs play crucial roles in cancer development, progression, and response to therapy [10-12]. Emerging evidences suggest that some miRNAs may be predominant biomarkers for the diagnosis and prognosis of cancer, such as miR-361-5p, miR-4443 and miR-124-3p for breast cancer [13-15], miR-450b-5p for colorectal cancer [16, 17], and miR-148a as well as miR-375 for prostate cancer [18, 19].
MiR-628, located in a various cancer-related region at 15q21.3, commonly acts as a tumor suppressor involved in tumorigenesis. MiR-628 expression was associated with diagnosis and prognosis of various cancer types, including mesothelioma [20], lung cancer [21, 22], gastrointestinal stromal tumor (GIST) [23], renal cell carcinoma [2, 3], prostate cancer (PCa) [24], pancreatic cancer [25], hepatocellular carcinoma (HCC) [26], gastric carcinoma (GCR) [27], colorectal cancer(CRC) [28], and glioma [29]. However, the roles of miR-628 as a potential diagnostic and prognostic biomarker in cancers are inconsistent and needed further evaluated. Therefore, we conducted this meta-analysis to assess the potential roles of miR-628 in cancer diagnosis and prognosis.
Materials and methods
We followed the PRISMA (Preferred Reporting Items for Systematic Review and Meta-Analyses) statement [30].
Literature search
A systematical search was performed (updated to November 15, 2017) for obtaining potentially eligible publications using Medline (via PubMed), Embase, Web of Science databases, and Ovid platform. The literature search was carried using free-text words combined with Medical Subject Headings (MeSH), such as (“cancer” or “neoplasms” or “tumor” or “tumour” or “malignancy” or “neoplasia”) and (“microRNA-628” or “miRNA-628” or “miR-628” or “hsa-miR-628”). To ensure that the suitable studies were not missed, references cited in the retrieved articles were also searched manually.
Literature selection
All studies included in this study should be strictly agreed with the following criteria: (1) patients with any type of cancer; (2) the expression levels of miR-628 in blood, serum, plasma, cancer tissues, or buccal mucosa; and (3) sufficient data on the diagnosis and/or prognosis of cancer based on miR-628 expression levels. The exclusion criteria were as follows: (1) duplicate publications; (2) review papers, case reports, letters, or abstracts; (3) studies unrelated to our topic, without complete data or unqualified data; and (4) prognosis of outcome indicators is not overall survival (OS).
Data extraction and quality assessment
All included studies were carefully evaluated, and data were extracted by two independent investigators (Jing-Hua Li and Shan-Shan Sun) according to a standard protocol. Discrepancies were resolved through discussion with a third investigator (Chang-Jin Fu). After that, the following information were extracted from included studies, included (1) study features: surname of the first author, published year and country; (2) study population characteristics: ethnicity, sample size, cancer type, control type, miRNA type, and specimen; and (3) relevant data for meta-analysis. For diagnostic meta-analysis, sensitivity, specificity, value of AUC, and data of two-by-two tables including false negatives (FN), true negatives (TN), true positives (TP), and false positives (FP) were extracted. For prognostic meta-analysis, follow-up time, cut-off of miR-628 expression, hazard ratio (HR) of miR-628 for OS with corresponding 95% confidence intervals (CIs) and P value were extracted. If these statistical variables were not directly reported in some studies, we calculated survival data from survival curves using an Engauge Digitizer 4.1 software and the methods described by Tierney [31].
The methodological quality assessment of diagnostic studies was conducted by independent team members based on the guidelines of the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) criteria [32], which contains the following 4 categories (including 7 items): patient selection (2 items), index test (2 items), reference standard (2 items), and flow and timing (1 items) were used to judge the applicability and risk of bias. Each item will be assessed as 1 score (“yes” presents the low-risk bias), 0 score (“no” and “unclear” present the high-risk bias). Diagnostic studies with 4 or less points were regarded as low quality [33]. For prognostic studies, Newcastle-Ottawa Scale (NOS) was applied to assess the quality of prognostic studies [34]. It composes 3 key domains (selection, comparability, and outcome) and 8 items. Each item in selection and outcome parts receives 1 score, while each item in comparability part gains 2 scores [35]. Thus, the quality of study was determined on a scale ranged from 0 to 9 points. Studies with 7 or more points were regarded as high quality [36].
Statistical analysis
Meta-analysis was conducted using STATA 14.0 (Stata Corporation: College Station, TX, USA) and Review Manager 5.3 (Copenhagen: Nordic Cochrane Centre, the Cochrane Collaboration, 2014) software. For the diagnostic meta-analysis, considering that multiple miRNA assays and single miRNA assay may have different diagnostic efficacy for cancers, we collected data of individual miR-628 and miR-628-related combination markers from included studies, respectively. Therefore, we have calculated not only the pooled sensitivity, specificity, PLR, NLR, and DOR along with their 95% CIs of individual miR-628, but also the corresponding results of miR-628-related combination markers utilizing bivariate meta-analysis model [37]. In addition, we generated the corresponding AUC to assess the diagnostic power for cancers. Furthermore, Spearman correlation coefficient was used to examine the between-study heterogeneity caused by the threshold effects [38]. Heterogeneity among studies caused by the non-threshold effect was assessed by the Cochran's-Q and I-squared (I2) statistics index [39]. If the heterogeneity caused by non-threshold effects existed (P value < 0.10 or I2 > 50%), stratified analyses by variables (specimen types, control types, and miRNA types) were performed on each subgroup comprising of three or more studies. The Fagan's nomogram was conducted to explore the clinical diagnostic value of miR-628 in detection of cancers. Additionally, sensitivity analysis was performed in order to evaluate the robustness of our analysis. The Deek's funnel plot was used to check the probability of publication bias.
For the prognostic meta-analysis, HRs and their 95% CIs were used to assess the impact of miR-628 expression on survival of cancer patients. Cochran's-Q and I2 statistics index were used to assess the inter-study heterogeneity [39]. The random-effects model would be applied for P value less than 0.10 or I2 value more than 50%, otherwise, the fixed-effects model was used [40]. Sensitivity analysis was performed by excluding one study at a time and recalculating the risk effect. Moreover, the Begg's test and Egger's test were used to check the publication bias [41].
Results
A total of 144 potentially relevant articles for miR-628 and cancer were selected from electronic database and other sources, and 65 of them were excluded for duplications. After overlapping titles and abstracts, 45 articles were excluded due to unrelated to cancer or/and miR-628-3p/5p, or reviews and abstracts. Next, 34 articles were suitable for further assessment. After full-text assessment, 15 articles without sufficient data were excluded, 6 articles that were unrelated to the research topic were excluded, and 1 article that only investigated disease-free survival (DFS) not OS was excluded. Finally, 12 eligible articles with 20 studies were considered for this study (Figure 1). Among those articles, 1 article examined diagnostic accuracy of both individual miR-628 and miR-628-related combination markers. Thus, for the diagnostic meta-analysis, 6 articles with 8 studies were about diagnostic accuracy of individual miR-628, and 3 articles with 7 studies were about diagnostic accuracy of miR-628-related combination markers. Totally, 15 studies were included in diagnostic meta-analysis. For the prognostic meta-analysis, 4 articles with 5 studies were related to OS.
Diagnostic meta-analysis
Diagnostic meta-analysis of individual miR-628 for cancers
Study characteristics and quality assessment
Because Li' study [25] investigated the roles of miR-628 in three types of controls, we considered it as three independent studies in this analysis. Thus, 8 studies from 6 articles involving 245 cases and 172 controls, most of them were conducted in Caucasian or African American except Wang's study [42], investigated Asians. The cancer types included mesothelioma [20], lung cancer [21, 42], prostate cancer [24], renal cell carcinoma [3], and pancreatic cancer [25]. The specimen types contained serum (n = 4), plasma (n = 1), tissue (n = 1), blood (n = 1), and buccal mucosa sample (n = 1). The miR-628 types were classified as miR-628-3p (n = 5) and miR-628-5p (n = 3). Moreover, all studies were assessed by QUADAS-2 with scores > 4. The results are shown in Figure 2 and Table 1.
Diagnostic accuracy of individual miR-628 for cancers
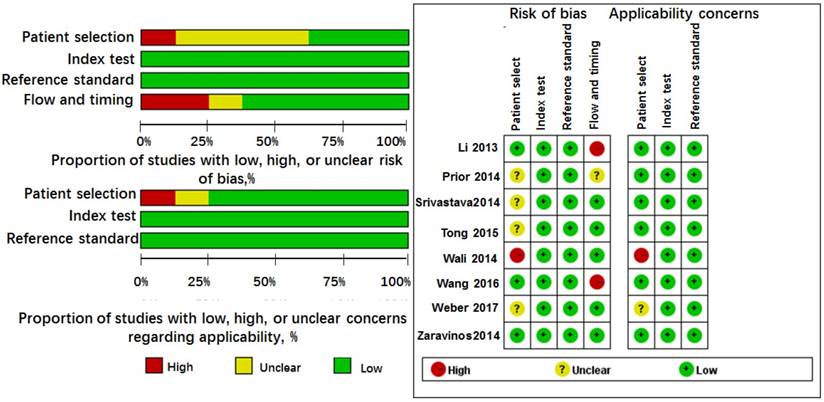
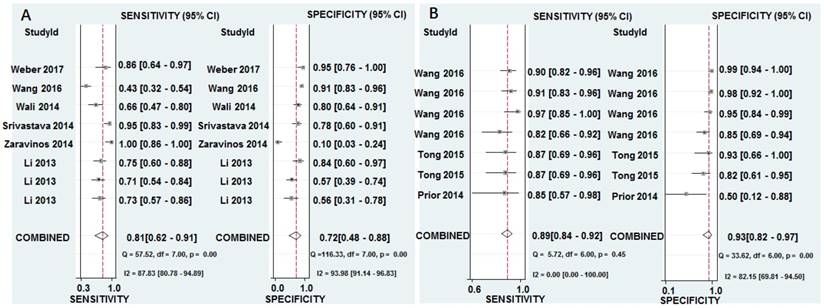
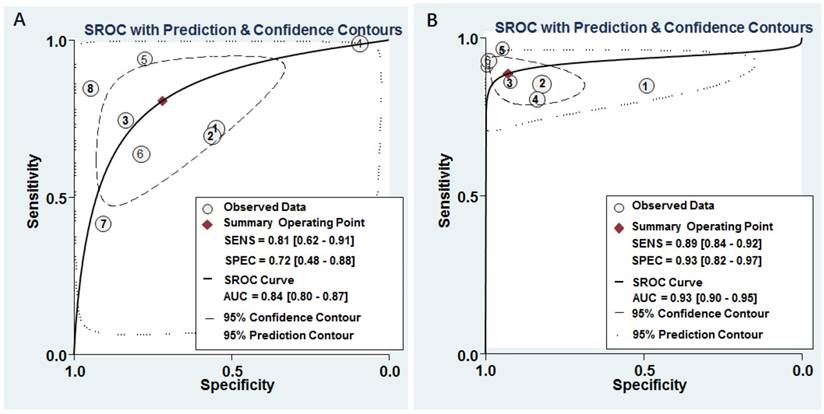
The pooled results for diagnostic accuracy of all included studies were as follows: sensitivity, 0.81 (95% CI: 0.62-0.91); specificity, 0.72 (95% CI: 0.48-0.88); PLR, 2.90 (95% CI: 1.50-5.40); NLR, 0.27 (95% CI: 0.14-0.50); and DOR, 11.0 (95% CI: 4.00-25.00), respectively (Figure 3A and Table 2). Diagnostic accuracy was further explored by plotting the SROC and calculating the AUC 0.84, (95% CI: 0.80-0.87) (Figure 4A).
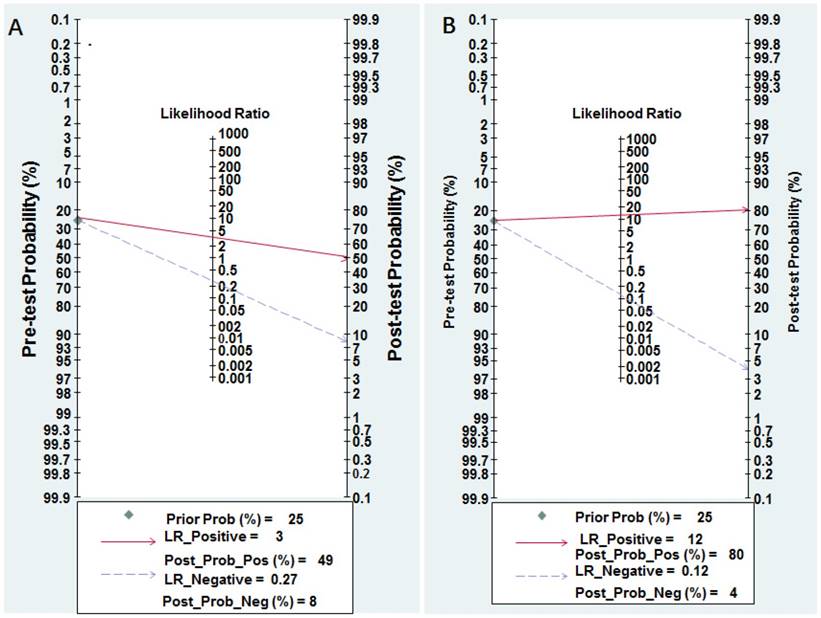
Nomogram of Fagan was regarded as a graphical tool for digging out the clinical diagnostic values of miR-628 in cancer detection. When 25% value was selected as the pre-test probability, the positive results of miR-628 showed the post-test probability of correctly diagnosing cancer would rise to 49%, while negative results of miR-628 indicated the post-test probability would drop to 8%, as demonstrated in the Fagan plot in Figure 5A.
Flow diagram of study selection process.
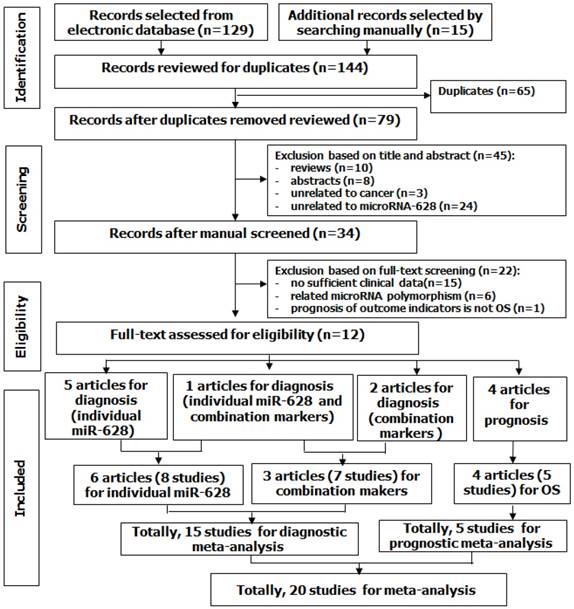
Quality assessment of diagnostic accuracy for the included studies.
Characteristics and quality assessment of diagnostic clinical the studies included in the meta-analysis
| Author | Year | Country | Ethnicity | Cancer type | Control type | miRNA type | Case/control | Specimen | AUC | SEN | SPE | TP | FP | FN | TN | QUADAS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Weber | 2017 | Germany | Caucasian | mesothelioma | asbestos-exposed | miR-628-5p | 21/21 | blood | NM | 0.860 | 0.950 | 18 | 1 | 3 | 20 | 5 |
| Wang | 2016 | China | Asian | lung cancer | healthy | miR-628-3p | 82/91 | plasma | 0.73 | 0.427 | 0.912 | 35 | 8 | 47 | 35 | 6 |
| Wali | 2014 | UAS | Caucasian | lung cancer | non-lung cancer | miR-628-5p | 37/39 | buccal* | >0.800 | 0.660 | 0.800 | 24 | 8 | 13 | 31 | 5 |
| Srivastava | 2014 | USA | Mixed* | PCa | healthy | miR-628-5p | 40/32 | serum | 0.940 | 0.948 | 0.777 | 38 | 7 | 2 | 25 | 6 |
| Zaravinos | 2014 | Greece | Caucasian | ccRCC | healthy | miR-628-3p | 24/40 | tissue | 0.868 | 1.000 | 0.100 | 24 | 36 | 0 | 4 | 7 |
| Li | 2013 | UAS | Caucasian | pancreatic cancer | healthy | miR-628-3p | 41/19 | serum | 0.820 | 0.750 | 0.840 | 31 | 3 | 10 | 16 | 6 |
| cancer | CP | miR-628-3p | 41/35 | serum | 0.690 | 0.710 | 0.570 | 29 | 15 | 12 | 20 | 6 | ||||
| PNET | miR-628-3p | 41/18 | serum | 0.680 | 0.730 | 0.560 | 30 | 8 | 11 | 10 | 6 | |||||
| Wang | 2016 | China | Asian | lung cancer | healthy | miR-628-3pa | 82/91 | plasma | 0.976 | 0.902 | 0.989 | 74 | 1 | 8 | 74 | 6 |
| healthy | miR-628-3pb | 82/91 | plasma | 0.974 | 0.915 | 0.978 | 75 | 2 | 7 | 75 | 6 | |||||
| healthy | miR-628-3pb | 36/43 | plasma | NM | 0.972 | 0.953 | 35 | 2 | 1 | 35 | 6 | |||||
| healthy | miR-628-3pb | 38/39 | plasma | NM | 0.816 | 0.846 | 31 | 6 | 7 | 31 | 6 | |||||
| Tong | 2015 | China | Asian | GIST | BO | miR-628-5pc | 30/14 | tissue | 0.960 | 0.870 | 0.930 | 26 | 1 | 4 | 13 | 6 |
| BB | miR-628-5pd | 30/23 | tissue | 0.906 | 0.870 | 0.820 | 26 | 4 | 4 | 19 | 6 | |||||
| Prior | 2014 | Spain | Caucasian | MRCC | resistant | miR-628-5pe | 14/6 | tissue | 0.619 | 0.850 | 0.500 | 12 | 3 | 2 | 3 | 5 |
Mixed*, Caucasian and African American; GIST, gastrointestinal stromal tumor; MRCC, metastatic renal-cell-carcinoma; Pca, prostate cancer; ccRCC, clear cell renal cell carcinoma; BO, borderline GIST; BB, benign or borderline GIST; CP, chronic pancreatitis; miR-628-3pa, miR-628-3p, 425-3p, 532, 339-3p; miR-628-3pb, miR-628-3p, 425-3p, 532; miR-628-5pc, miR6285p, 29b2#, let7c, 891b, 218, 204, 2043p, 744, 29c#, 625; miR-628-5pd, miR6285p, let7c, 218, 2043p, 204, 891b, 488#, 145,891a; miR-628-5pe, miR-628-5p, 942, 133a, 484; AUC, area under ROC curve; buccal*, buccal mucosa samples; NM, not mentioned; TP, true positive; FP, false positive; FN, false negative, TN, true negative; SEN, sensitivity; SPE, specificity; QUADAS; Quality Assessment of Diagnostic Accuracy Studies.
Forest plots of sensitivity and specificity from test accuracy studies of miR-628 in the diagnosis of cancer. A Individual miR-628. B MiR-628-related combination markers.
Summary results for diagnostic accuracy of miR-628 for cancers
| Subgroup | N | SEN(95% CI) | SPE (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) | AUC (95% CI) | |
|---|---|---|---|---|---|---|---|---|
| Individual | Overall | 8 | 0.81(0.62-0.91) | 0.72(0.48-0.88) | 2.90(1.50-5.40) | 0.27(0.14-0.50) | 11.00(4.00-25.00) * | 0.84(0.80-0.87) * |
| Specimen types | ||||||||
| blood | 6 | 0.76(0.59-0.87) | 0.80(0.65-0.90) | 3.80(2.00-7.20) | 0.30(0.17-0.54) | 13.00(5.00-36.00) | 0.85(0.82-0.88) | |
| Control types | ||||||||
| healthy | 4 | 0.95(0.33-1.00) | 0.69(0.27-0.93) | 3.00(1.00-8.80) | 0.07(0.00-1.53) | 45.00(4.00-509.00) | 0.88(0.85-0.91) * | |
| non-healthy | 4 | 0.73(0.64-0.80) | 0.75(0.53-0.89) | 2.90 (1.30-6.40) | 0.36(0.23-0.57) | 8.00(2.00-26.00) | 0.76(0.72-0.80) * | |
| miRNA types | ||||||||
| miR-628-3p | 5 | 0.77(0.53-0.91) | 0.60(0.27-0.85) | 1.90(1.00-3.50) | 0.39(0.25-0.60) | 5.00(3.00-10.00) | 0.76(0.72-0.79) * | |
| miR-628-5p | 3 | 0.85(0.64-0.95) | 0.83(0.73-0.89) | 4.90(3.00-8.00) | 0.18(0.07-0.49) | 27.00(7.00-99.00) | 0.84(0.80-0.87) * | |
| Combination | Overall | 7 | 0.89(0.84-0.92) | 0.93(0.82-0.97) | 12.30(4.70-32.50) | 0.12(0.08-0.19) | 100.00(28.00-354.00) * | 0.93(0.90-0.95) * |
N, number of studies; SEN, sensitivity; 95 % CI, 95% confidence interval; SPE, specificity; PLR, positive likelihood ratio; NLR, negative likelihood ratio; DOR, diagnostic odds ratio; AUC, area under ROC curve; *statistically significant results.
SROC curves of miR-628 for the diagnosis of cancers. A Individual miR-628. B MiR-628-related combination markers.
Pre-test probability of miR-628 in cancer detection. A Individual miR-628. B MiR-628-related combination markers.
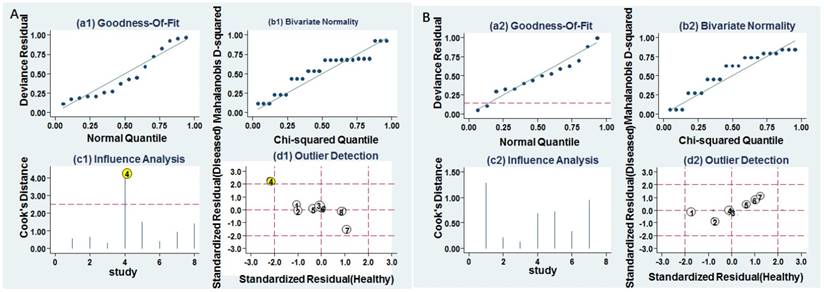
Influence analysis and outlier detection. A Individual miR-628. B MiR-628-related combination markers. (a) goodness of fit, (b) bivariate normality, (c) influence analysis, and (d) outlier detection.
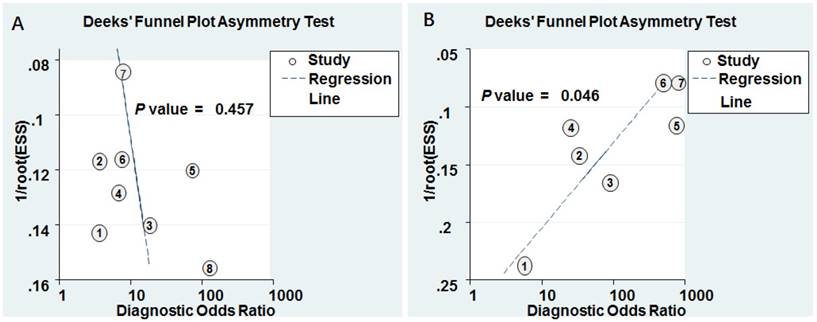
Deeks' funnel plots for the assessment of potential diagnosis bias in meta-analysis for diagnosis. A Individual miR-628. B MiR-628-related combination markers.
Investigations of heterogeneity
The heterogeneity may be related to threshold effect and/or non-threshold effect. Spearman correlation coefficient in 8 studies was 0.72, with a P value of 0.52, suggesting that the heterogeneity was not caused by the threshold effect.
Heterogeneity in sensitivity and specificity were observed by I-squared statistics index (I2 = 87.83% and I2 = 93.98%), which indicated a significant heterogeneity appeared among the included studies. To further analyze the heterogeneity of the included studies, subgroup analyses based on specimen (blood sample vs. non-blood sample), miRNA types (miR-628-3p vs. miR-628-5p), and control types (healthy control vs. non-healthy control) were performed on those comparing of three or more studies (Table 2). For non-blood sample with only 2 studies, the results were not shown in Table 2. The subgroup analysis based on miR-628 types indicated miR-628-5p single is a more promising biomarker for cancer detection than miR-628-3p single with AUC of 0.84 versus 0.76. Meanwhile, healthy control may be a better diagnostic accuracy than non-healthy control in cancer detection, and the corresponding AUC values were 0.88 versus 0.76.
Sensitivity analysis and publication bias
Goodness of fit and bivariate normality analyses were performed to assess the stability of our study by using the random effects bivariate model. Besides that, influence analysis and outlier detection identified one outlier study (Figure 6A). After exclusion, the pooled sensitivity, specificity, PLR, NLR, DOR and AUC were 0.74 (95% CI: 0.60-0.85), 0.80 (95% CI: 0.67-0.88), 3.70 (95% CI: 2.20-6.20), 0.32 (95% CI: 0.20-0.52), 11.00 (95% CI: 5.00-27.00), and 0.84 (95% CI: 0.81-0.85), respectively, which had only minimal changes compared with previous results. Sensitivity results indicated that minimal changes did not significantly affect the overall estimates. Furthermore, Deeks' funnel plot indicated no significant publication bias (P = 0.457) in our study (Figure 7A).
Characteristics and quality assessment of prognostic clinical studies included in the meta-analysis
| Author | Year | Country | Ethnicity | Cancer type | N | Specimen | miRNA type | Results | Cut off | Follow-up (month) | P | HR | LL | UL | NOS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Liu | 2017 | China | Mixed* | HCC | 322 | tissue | miR-628 | OS | median | 96(max) | 0.031 | 1.560 | 1.041 | 2.337 | 9 |
| Gao | 2016 | China | Asian | GCA | 50 | tissue | miR-628-3p | OS | median | NM | 0.291 | 0.756 | 0.282 | 2.028 | 7 |
| Schou | 2015 | Dasnish | Caucasian | CRC | 138 | blood | miR-628-5p | OS | median | 76(max) | >0.050 | 1.049 | 0.779 | 1.412 | 9 |
| Li | 2013 | China | Asian | glioma | 80 | tissue | miR-628-5p | OS | median | 22.84(mean) | <0.050 | 1.883 | 1.435 | 2.475 | 7 |
| 80 | tissue | miR-628-5p | OS | median | 22.49(mean) | <0.050 | 4.380 | 1.810 | 10.600 | 7 |
Mixed*, Asian, White, Black, American Indian, and unknown races.; HCC, hepatocellular carcinoma; GCA, gastric cardia adenocarcinoma; CRC, colorectal cancer; N, number; OS, overall survival; DFS, disease-free survival; NM, not mentioned; HR, hazard ratio; LL, Lower limit of 95% confidence interval; UL, Upper limit of 95% confidence interval; NOS, Newcastle-Ottawa Scale.
Diagnostic meta-analysis of miR-628-related combination markers for cancers
Study characteristics and quality assessment
Among these articles, Wang's study [42] investigated the roles of two kinds of miR-628-3p panels in three independent populations (one panel in one population, anther panel in three populations). Tong's study [23] also examined two different types of miR-628-5p panels in one case population. Thus, 7 studies with 200 cases and 216 controls assessed the accuracy of miR-628-related combination markers for the diagnosis of cancer. One study was in Europe region, and 6 studies were in Asia region. The control types were classified as healthy controls (n = 4) and healthy controls (n = 4). Three studies used tissue samples, and 4 studies used plasma samples. The miR-628 types included miR-628-3p (n = 4) and miR-628-5p (n = 3). The QUADAS scores ranged from 5 to 6 (Table 1 and Figure 2).
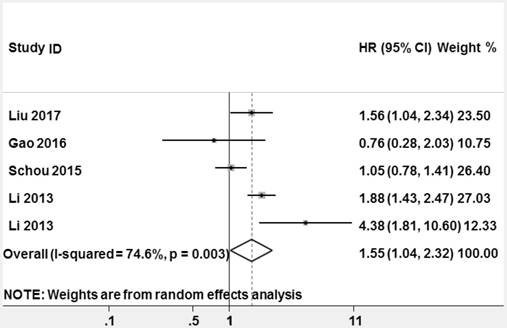
Forest plots of studies evaluating hazard ratios (HRs) of miR-628 for overall survival.
Diagnostic accuracy of miR-628-related combination markers for cancer
The pooled sensitivity, specificity, PLR, NLR, and DOR were 0.89 (95% CI: 0.84-0.92), 0.93 (95% CI: 0.82-0.97), 12.30 (95% CI: 4.70-32.50), 0.12 (95% CI: 0.08-0.19), and 100.00 (95% CI: 28.00-354.00), respectively (Figure 3B and Table 2). Diagnostic accuracy was further explored by plotting the SROC and calculating the AUC, which was 0.93 (95% CI = 0.90 - 0.95) (Figure 4B). Additionally, Figure 5B showed that the post-test probability of cancer using miR-628-related combination markers for a positive test result was 80%, while the probability of a negative test result was 4%, with a pre-test probability of 25% (Figure 5B).
Sensitivity analysis and publication bias
Goodness of fit and bivariate normality analyses were used to study the robustness of the pooled estimates. No study was identified by influence analysis and outlier detection (Figure 6B). Additionally, The P value of Deek's test was 0.046, indicating that there may exist publication bias (Figure 7B).
Prognostic meta-analysis
Study characteristics and quality assessment
Five studies with 670 cancer patients were included for OS. The ethnicity of study included Asian (n = 3), Caucasian (n = 1), and mixed races (n = 1) (Asian, White, Black, American, Indian, and unknown races). Four studies used tissue samples and 1 used blood sample. Median were selected as cut-off value in all studies. Additionally, the mean length of follow-up time was greater than 22 months. The quality of the included studies was assessed by NOS, and the quality scores varied from 7 to 9 (Supplementary Figure 1 and Table 3).
Correlation between miR-628 expression and survival outcomes
High heterogeneity among the studies appeared (P = 0.003, I2 = 74.6%), thus a random-effect model was used to pool HR. The pooled HR and its 95% CI were statistically significant (HR = 1.553, 95% CI: 1.041-2.318, z = 2.16, P = 0.031, Figure 8).
Sensitivity analysis and publication bias
A low sensitivity will be found in the results of the Begg's test when the number of eligible studies is < 10 [43], so it was not used for this study. Egger's test was applied to assess the publication bias. Its value was 0.821, suggesting there is no obvious publication bias in prognostic meta-analysis.
Discussion
The early diagnosis, treatment and prognosis of cancer remain clinically challenging over the past decades [44, 45]. It is estimated that new cancer cases increased by 6 % per year worldwide [4-7]. Unfortunately, most patients with cancer had missed the best period of treatment, for they were diagnosed with cancer at the final stage. Therefore, it is urgent to find a new convenient, inexpensive, non-invasive, and sensitive detection methods or markers so as to ensure early diagnosis of cancer and improve prognosis of cancer [46].
Recently, due to their roles in tumorigenesis and their correlation with clinical characteristics, miRNAs have attracted wide attention. Further, accumulating evidence supports that their abnormal expression levels are associated with various diseases, such as lung cancer [47, 48], prostate cancer [22, 49, 50], and cervical cancer [51, 52]. More importantly, miRNAs present in various biological materials, like serum, plasma, and tissue, are extremely stable, even in extreme thermal environment [53, 54]. Besides, miRNAs are easy to be measured by mean of qRT-PCR, which is the evidence of their reproducibility [55, 56]. Therefore, miRNAs may be useful biomarkers for diagnosis and prognosis of cancers.
MiR-628 is a diagnostic biomarker for cancers
Numerous studies have supported that miRNAs obtained from body may serve as ideal tools for detection cancer [57-59]. To date, miR-628, one of recently studied miRNAs, was down-regulated in various cancers [2, 3, 20, 21, 23, 24, 29, 42]. The expression of miR-628 appears to be tissue, plasma, serum, or type-specific, which is useful for classifying human cancers [60], distinguishing tumor subtypes [42], and correlated with prognosis [26]. MiR-628-3p regulated ATRX, SLC45A2, and TNRC6B expression in lung cancer and exhibited aberrantly expression between lung cancer patients and healthy controls [42]. MiRNA profiling of tissue fractions revealed that miR-628-3p/5p were capable to elucidate the characteristics of cancer subtypes [23, 42].
The above mentioned studies indicated that roles of miR-628 for cancer diagnosis presented inconsistent or inverse [20, 21, 23, 29, 42]. Thus, we conducted this meta-analysis to evaluate the roles of miR-628 alone in detecting cancer. The pooled sensitivity and specificity were 0.81 and 0.72, respectively, indicating that miR-628 single has a moderate level of accuracy to detect cancer. The DOR, ranging from 0 to infinity, is a single test accuracy index that combined sensitivity and specificity into a single number. The higher DOR indicates better diagnostic performance [61]. In our study, the DOR value was 11.00, indicating that miR-628 might be a promising biomarker for cancer diagnosis. Furthermore, the AUC represents an analytical summary of test performance. The AUC value close to 1.0 indicates that diagnostic test has almost perfect discrimination [62, 63]. The AUC of this meta-analysis was 0.84, suggesting a moderate level of diagnostic accuracy. Generally, biomarkers with a PLR > 5 and an NLR < 0.1 are served as excellent clinical value indices. The results of our study (PLR < 5, NLR > 0.1) indicated that the level of overall accuracy is not high enough for clinical application. Thus, serving application of miR-628 single as a clinical diagnostic marker for distinguishing patients with cancer from healthy people or no-cancer patients would take a long time.
Diagnostic accuracy of miR-628 single varies from different specimens, control types, and miR-628 types. Here, we only demonstrated that miR-628 single had precisely diagnostic performance for detecting cancer by using blood samples not tissue. Similarly, Zhou's study showed the miRNA in blood sample had a relatively high level of diagnostic accuracy, indicating noninvasive and convenient nature of miR-628 may promote its application for cancer diagnosis in the future [64]. In addition, the control type may influence the diagnostic accuracy. Healthy controls seem to be superior to those of controls of patients with other disease. Similarly, Li's study [29] reported that the diagnostic accuracy of healthy controls was found to be significantly higher than that of other non-health controls. Additionally, we found that miR-628-5p had more credible diagnostic compared to miR-628-3p. A possible explanation for the conclusion is their different stability.
From the perspective of cancer development, an individual biomarker is unlikely to dominate the complex development of cancer. Based on the hypothesis that combination biomarkers will help to explain the underlying mechanisms of cancer developments and the external factors affecting it. Furthermore, a meta-analysis for miR-628-related combination markers for cancers was performed to test whether it was better than individual miR-628 for cancer. The pooled sensitivity, specificity, DOR, and AUC were 0.89, 0.93, 100.00, and 0.93, respectively, indicating that combination diagnostic power was relatively high. The combined PLR and NLR were 12.30 and 0.12, respectively, which further suggested that miR-628-related combination markers has sufficient power to confirm or exclude cancer.
MiR-628 is a prognostic biomarker for cancers
MiR-628 is important for the prognosis of cancer patients. It has been served as a potentially useful predictor survival in cancers. Liu et al [26] demonstrated that overall survival of HCC patients group with higher miR-628 was significantly longer than low-expression group (P = 0.03). Li and his colleagues [29] found that the expression between miR-628-5p and miR-524-5p was significantly positively correlated in glioma (R = 0.41, P < 0.001), which was related to glioma progression. Schou's study [28] revealed that high levels of miR-628-5p were associated with short OS. The latest findings reveled that miR-628-5p served as a potential biomarker for the prognosis of patients with ovarian cancer [65]. The increased miR-628-5p was also associated with prolonged MRCC patients' survival [66]. The altered miR-628 expression may further mediate the progression of ductal carcinoma in situ to invasive breast cancer and lympho-vascular invasion [67].
The abovementioned studies indicated that increased miR-628 expression may predict a better OS [25, 26, 66]. However, Gao's study [27] showed that there was no association of miR-628-3p expression with OS of GCA. Therefore, we further evaluated the roles of miR-628 in cancer prognosis and found that higher miR-628 expression is a prognostic factor for better OS, with the pooled HR of 1.553, 95% CI: 1.041-2.318, and this association was statistically significant (z = 2.16, P = 0.031). Our results were supported by some studies, which reported that miR-628 might be a promising biomarker for predicting the OS of patients with cancer [26, 29].
Strengths and limitations
Our study had several strengths. Frist, we conducted quantitative analyses for estimating the diagnostic and prognostic roles of miR-628 in cancers for the first time. Next, the diagnostic value of miR-628 was detected in different miR-628 types, including miR-628-3p and miR-628-5p. Moreover, different samples sources were measured to find the most suitable one for clinical application. We also performed a meta-analysis for miR-628-related combination markers in cancers to check whether they better than miR-628 alone in diagnosis of cancers for the first time.
Limitations do exist despite important discoveries revealed by the meta-analysis. First, only 12 articles are involved in our meta-analysis. Our conclusion needs to be reconfirmed by more prospective studies. Second, the blind design was not used in some studies, which limits the diagnostic accuracy. Third, heterogeneity exists among these studies. Fourth, due to limited studies, the prognostic value of miR-628 was only merged OS.
Conclusions
This study demonstrated that miR-628 might be used as a novel biomarker for diagnosis and prognosis of cancer. MiR-628-related combination markers have also emerged as new beneficial alternatives for clinical application. However, our findings require further evaluation in future large-scale prospective studies.
Supplementary Material
Supplementary figure.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No.81772281, 31371321), the Shandong Science and Technology Committee (No. 2017GSF221011, 2015GSF118073, ZR2016CL09), Shandong province Taishan Scholar Program, and National Training Program of Innovation and Entrepreneurship for Undergraduates (201710440009, 201710440139).
Authorship contributions
PYW, JHL, SYX: study conception and design, data analysis, and manuscript preparation. JHL, SSS, CJF, AQZ: data collection, data analysis. CW, RX: data analysis. All authors approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67:7-30
2. Prior C, Perez-Gracia JL, Garcia-Donas J, Rodriguez-Antona C, Guruceaga E, Esteban E. et al. Identification of tissue microRNAs predictive of sunitinib activity in patients with metastatic renal cell carcinoma. PloS one. 2014;9:e86263
3. Zaravinos A, Lambrou GI, Mourmouras N, Katafygiotis P, Papagregoriou G, Giannikou K. et al. New miRNA profiles accurately distinguish renal cell carcinomas and upper tract urothelial carcinomas from the normal kidney. PloS one. 2014;9:e91646
4. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: a cancer journal for clinicians. 2012;62:10-29
5. Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA: a cancer journal for clinicians. 2011;61:212-36
6. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA: a cancer journal for clinicians. 2013;63:11-30
7. Shen J, Stass SA, Jiang F. MicroRNAs as potential biomarkers in human solid tumors. Cancer letters. 2013;329:125-36
8. Visone R, Croce CM. MiRNAs and cancer. The American journal of pathology. 2009;174:1131-8
9. Fang YX, Gao WQ. Roles of microRNAs during prostatic tumorigenesis and tumor progression. Oncogene. 2014;33:135-47
10. Berardi E, Pues M, Thorrez L, Sampaolesi M. miRNAs in ESC differentiation. American journal of physiology Heart and circulatory physiology. 2012;303:H931-9
11. Zheng B, Liang L, Huang S, Zha R, Liu L, Jia D. et al. MicroRNA-409 suppresses tumour cell invasion and metastasis by directly targeting radixin in gastric cancers. Oncogene. 2012;31:4509-16
12. Zheng B, Liang L, Wang C, Huang S, Cao X, Zha R. et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:7574-83
13. Cao ZG, Huang YN, Yao L, Liu YR, Hu X, Hou YF. et al. Positive expression of miR-361-5p indicates better prognosis for breast cancer patients. Journal of thoracic disease. 2016;8:1772-9
14. Chen X, Zhong SL, Lu P, Wang DD, Zhou SY, Yang SJ. et al. miR-4443 Participates in the Malignancy of Breast Cancer. PloS one. 2016;11:e0160780
15. Zhang F, Wang B, Long H, Yu J, Li F, Hou H. et al. Decreased miR-124-3p Expression Prompted Breast Cancer Cell Progression Mainly by Targeting Beclin-1. Clinical laboratory. 2016;62:1139-45
16. Ye YP, Wu P, Gu CC, Deng DL, Jiao HL, Li TT. et al. miR-450b-5p induced by oncogenic KRAS is required for colorectal cancer progression. Oncotarget. 2016;7:61312
17. Zhao M, Liu Y, Huang F, Qu H. A gene browser of colorectal cancer with literature evidence and pre-computed regulatory information to identify key tumor suppressors and oncogenes. Scientific reports. 2016;6:30624
18. Metcalf GA, Shibakawa A, Patel H, Sita-Lumsden A, Zivi A, Rama N. et al. Amplification-Free Detection of Circulating microRNA Biomarkers from Body Fluids Based on Fluorogenic Oligonucleotide-Templated Reaction between Engineered Peptide Nucleic Acid Probes: Application to Prostate Cancer Diagnosis. Analytical chemistry. 2016;88:8091-8
19. Stuopelyte K, Daniunaite K, Bakavicius A, Lazutka JR, Jankevicius F, Jarmalaite S. The utility of urine-circulating miRNAs for detection of prostate cancer. British journal of cancer. 2016;115:707
20. Weber DG, Gawrych K, Casjens S, Brik A, Lehnert M, Taeger D. et al. Circulating miR-132-3p as a Candidate Diagnostic Biomarker for Malignant Mesothelioma. Disease markers. 2017;2017:9280170
21. Wali RK, Hensing TA, Ray DW, Dela Cruz M, Tiwari AK, Radosevich A. et al. Buccal microRNA dysregulation in lung field carcinogenesis: gender-specific implications. International journal of oncology. 2014;45:1209-15
22. Wang Y, Lieberman R, Pan J, Zhang Q, Du M, Zhang P. et al. miR-375 induces docetaxel resistance in prostate cancer by targeting SEC23A and YAP1. Molecular cancer. 2016;15:70
23. Tong HX, Zhou YH, Hou YY, Zhang Y, Huang Y, Xie B. et al. Expression profile of microRNAs in gastrointestinal stromal tumors revealed by high throughput quantitative RT-PCR microarray. World journal of gastroenterology. 2015;21:5843-55
24. Srivastava A, Goldberger H, Dimtchev A, Marian C, Soldin O, Li X. et al. Circulatory miR-628-5p is downregulated in prostate cancer patients. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:4867-73
25. Li A, Yu J, Kim H, Wolfgang CL, Canto MI, Hruban RH. et al. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:3600-10
26. Liu G, Wang H, Fu JD, Liu JY, Yan AG, Guan YY. A five-miRNA expression signature predicts survival in hepatocellular carcinoma. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica. 2017;125:614-22
27. Gao S, Zhou F, Zhao C, Ma Z, Jia R, Liang S. et al. Gastric cardia adenocarcinoma microRNA profiling in Chinese patients. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:9411-22
28. Schou JV, Rossi S, Jensen BV, Nielsen DL, Pfeiffer P, Hogdall E. et al. miR-345 in metastatic colorectal cancer: a non-invasive biomarker for clinical outcome in non-KRAS mutant patients treated with 3rd line cetuximab and irinotecan. PloS one. 2015;9:e99886
29. Li Y, Xu J, Chen H, Bai J, Li S, Zhao Z. et al. Comprehensive analysis of the functional microRNA-mRNA regulatory network identifies miRNA signatures associated with glioma malignant progression. Nucleic acids research. 2013;41:e203
30. Moher D, Liberati A, Tetzlaff J, Altman DG. Reprint-preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Physical therapy. 2009;89:873-80
31. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16
32. Wade R, Corbett M, Eastwood A. Quality assessment of comparative diagnostic accuracy studies: our experience using a modified version of the QUADAS-2 tool. Research synthesis methods. 2013;4:280-6
33. Ren J, Kuang TH, Chen J, Yang JW, Liu YX. The diagnostic and prognostic values of microRNA-21 in patients with gastric cancer: a meta-analysis. European review for medical and pharmacological sciences. 2017;21:120-30
34. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European journal of epidemiology. 2010;25:603-5
35. Wang F, Zhou J, Zhang Y, Wang Y, Cheng L, Bai Y. et al. The Value of MicroRNA-155 as a Prognostic Factor for Survival in Non-Small Cell Lung Cancer: A Meta-Analysis. PloS one. 2015;10:e0136889
36. Margulis AV, Pladevall M, Riera-Guardia N, Varas-Lorenzo C, Hazell L, Berkman ND. et al. Quality assessment of observational studies in a drug-safety systematic review, comparison of two tools: the Newcastle-Ottawa Scale and the RTI item bank. Clinical epidemiology. 2014;6:359-68
37. Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. Journal of clinical epidemiology. 2005;58:982-90
38. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Statistics in medicine. 2002;21:1539-58
39. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ (Clinical research ed). 2003;327:557-60
40. Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Annals of internal medicine. 1997;127:820-6
41. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clinical research ed). 1997;315:629-34
42. Wang Y, Zhao H, Gao X, Wei F, Zhang X, Su Y. et al. Identification of a three-miRNA signature as a blood-borne diagnostic marker for early diagnosis of lung adenocarcinoma. Oncotarget. 2016;7:26070-86
43. Ioannidis JP, Trikalinos TA. The appropriateness of asymmetry tests for publication bias in meta-analyses: a large survey. CMAJ: Canadian Medical Association journal = journal de l'Association medicale canadienne. 2007;176:1091-6
44. Fendler A, Jung K. MicroRNAs as new diagnostic and prognostic biomarkers in urological tumors. Critical reviews in oncogenesis. 2013;18:289-302
45. DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA: a cancer journal for clinicians. 2014;64:52-62
46. Gao Y, Zhao H, Lu Y, Li H, Yan G. MicroRNAs as potential diagnostic biomarkers in renal cell carcinoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:11041-50
47. Sun CC, Li SJ, Zhang F, Zhang YD, Zuo ZY, Xi YY. et al. The Novel miR-9600 Suppresses Tumor Progression and Promotes Paclitaxel Sensitivity in Non-small-cell Lung Cancer Through Altering STAT3 Expression. Molecular therapy Nucleic acids. 2016;5:e387
48. Gao X, Wu Y, Yu W, Li H. Identification of a seven-miRNA signature as prognostic biomarker for lung squamous cell carcinoma. Oncotarget. 2016;7:81670
49. Yates C, Long MD, Campbell MJ, Sucheston-Campbell L. miRNAs as drivers of TMPRSS2-ERG negative prostate tumors in African American men. Frontiers in bioscience (Landmark edition). 2017;22:212-29
50. Filella X, Foj L. Prostate Cancer Detection and Prognosis: From Prostate Specific Antigen (PSA) to Exosomal Biomarkers. International journal of molecular sciences. 2016;17:E1784
51. Virtanen E, Pietila T, Nieminen P, Qian K, Auvinen E. Low expression levels of putative HPV encoded microRNAs in cervical samples. SpringerPlus. 2016;5:1856
52. Li J, Liu Q, Clark LH, Qiu H, Bae-Jump VL, Zhou C. Deregulated miRNAs in human cervical cancer: functional importance and potential clinical use. Future oncology (London, England). 2017;13:743
53. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10513-8
54. Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ. et al. The microRNA spectrum in 12 body fluids. Clinical chemistry. 2010;56:1733-41
55. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K. et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell research. 2008;18:997-1006
56. Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic acids research. 2011;39:7223-33
57. Meyer-Rochow GY, Jackson NE, Conaglen JV, Whittle DE, Kunnimalaiyaan M, Chen H. et al. MicroRNA profiling of benign and malignant pheochromocytomas identifies novel diagnostic and therapeutic targets. Endocrine-related cancer. 2010;17:835-46
58. Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH. et al. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic acids research. 2009;37:1672-81
59. Iguchi T, Todoroki R, Yamaguchi S, Takasugi N. Changes in the uterus and vagina of mice treated neonatally with antiestrogens. Acta anatomica. 1989;136:146-54
60. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D. et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-8
61. Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM. The diagnostic odds ratio: a single indicator of test performance. Journal of clinical epidemiology. 2003;56:1129-35
62. Jones CM, Athanasiou T. Summary receiver operating characteristic curve analysis techniques in the evaluation of diagnostic tests. The Annals of thoracic surgery. 2005;79:16-20
63. Rutter CM, Gatsonis CA. A hierarchical regression approach to meta-analysis of diagnostic test accuracy evaluations. Statistics in medicine. 2001;20:2865-84
64. Zhou XJ, Dong ZG, Yang YM, Du LT, Zhang X, Wang CX. Limited diagnostic value of microRNAs for detecting colorectal cancer: a meta-analysis. Asian Pacific journal of cancer prevention: APJCP. 2013;14:4699-704
65. Li M, Qian Z, Ma X, Lin X, You Y, Li Y. et al. MiR-628-5p decreases the tumorigenicity of epithelial ovarian cancer cells by targeting at FGFR2. Biochem Biophys Res Commun. 2017 pii: S0006-291X(17)32440-3
66. Puente J, Lainez N, Duenas M, Mendez-Vidal MJ, Esteban E, Castellano D. et al. Novel potential predictive markers of sunitinib outcomes in long-term responders versus primary refractory patients with metastatic clear-cell renal cell carcinoma. Oncotarget. 2017;8:30410-21
67. Soon PS, Provan PJ, Kim E, Pathmanathan N, Graham D, Clarke CL. et al. Profiling differential microRNA expression between in situ, infiltrative and lympho-vascular space invasive breast cancer: a pilot study. Clin Exp Metastasis. 2017
Author contact
![]() Corresponding author: Ping-Yu Wang. Binzhou Medical University, YanTai, ShanDong, 264003, P.R.China. Tel: +86 535 6913217. E-mail address: wpingyugirlcom.
Corresponding author: Ping-Yu Wang. Binzhou Medical University, YanTai, ShanDong, 264003, P.R.China. Tel: +86 535 6913217. E-mail address: wpingyugirlcom.

 Global reach, higher impact
Global reach, higher impact