3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(7):1239-1247. doi:10.7150/jca.23808 This issue Cite
Research Paper
Association of WWOX rs9926344 polymorphism with poor prognosis of hepatocellular carcinoma
1. Department of Liver Surgery, Zhongshan Hospital, Liver Cancer Institute, Fudan University, Shanghai, 200032, China
2. Department of Surgery, Minhang Branch, Zhongshan Hospital, Fudan University, Shanghai, 201199, China.
3. Institute of Fudan-Minhang Academic Health System, Minhang Hospital, Zhongshan Hospital, Fudan University, Shanghai, 201199, China.
4. Institutes of Biomedical Sciences, Fudan University, Shanghai, 200032, China.
*These authors contributed equally to this study.
Received 2017-11-12; Accepted 2018-2-28; Published 2018-3-14
Abstract
Introduction: The WW domain-containing oxidoreductase (WWOX), widely expressed in human tissues, is considered as a tumor suppressor gene and plays an important role in the incidence and progression of human cancer, HCC included. This study was to investigate the correlation between single nucleotide polymorphisms (SNPs) of the WWOX gene and the prognosis of hepatocellular carcinoma (HCC) patients.
Materials and Methods: After a total of 152 HCC patients were recruited, 8 cases with tumor recurrence within 2-years after operation and 8 cases without recurrence were selected randomly for SNP genotyping and screening using Affymetrix Array 6.0. And then we confirmed candidate SNPs in the remaining 136 patients by time-of-flight mass spectrometry (TOF-MS).
Results: In total, 32 SNPs were screened and identified as candidate SNPs with one SNP in particular, (rs9926344), being further verified to be valuable. We found that AA+AG genotype and A allele of WWOX rs9926344 were significantly associated with recurrent risk of HCC (p=0.002 and p=0.001, respectively). The Kaplan-Meier curve showed that patients carrying rs9926344 AA +AG genotype had poor RFS (P=0.004) and OS (P=0.005) compared to those carrying GG genotypes. The multivariate COX regression analysis showed that the AA+AG genotype were an independent prognostic factor for tumor recurrence (HR 1.787, 95% CI 1.042-3.064, P=0.035). Furthermore, IHC analysis showed that the WWOX protein down-regulation is more frequent in patients with AG genotype compared to those with GG genotype (P=0.023).
Conclusion: Our findings indicate that WWOX rs9926344 polymorphism is positively correlated with tumor recurrence and can be used as an independent prognostic marker for HCC patients after operation.
Keywords: hepatocellular carcinoma, WWOX gene, single nucleotide polymorphism, recurrence, prognosis
Introduction
Hepatocellular carcinoma (HCC), which accounts for 70-90% of primary liver cancer, is one of the most common human cancers and considered to be the leading cause of cancer-related death worldwide [1, 2]. The global cancer statistics showed that a total of 782,500 new liver cancer cases and 745,500 deaths occurred during 2012, around half of those cases and deaths happen in China, mainly due to the high prevalence of hepatitis B virus (HBV) infection in its population [3]. Although its mortality decreased gradually due to the rapid development of therapeutic methods, including the technological progress of surgical operations, the long-term prognosis remains unsatisfactory. The main reason for the poor clinical outcome in HCC patients is postoperative recurrence. For example, recent study has pointed out that the recurrence rate after resection is approximate 50% within the first 2 year and 75% within the first 5 year [4]. However, no effective neoadjuvant or adjuvant therapies for reducing risk of recurrence are available. With the current lack of specific biomarkers for reliably predicting recurrence and long-term prognosis, the identification of new biomarkers for more effective prognosis prediction may contribute to the improvement of postoperative quality of life in HCC patients.
The WW domain-containing oxidoreductase (WWOX) gene is located on chromosome 16q23.3-24.1 and spans the Common Chromosomal Fragile Site (CCFS) FRA16D. This gene is widely expressed in human tissues and encodes a protein that contains two N-terminal WW domains and a central short chain dehydrogenase/reductase domain (SDR) [5, 6]. Previous studies have implicated that WWOX is considered as a tumor suppressor [7, 8] and participates in a diversity of cellular activities, such as maintaining genomic stability, controlling CNS development and moderating the mitochondrial respiratory complex [9-11]. However, the exact molecular mechanisms of WWOX in the occurrence and progression of human cancer remains dismal and in need for further investigation. To date, a number of studies mainly focused on the correlations between aberrant expression of WWOX gene and human cancers [12-14]. Previous studies had also pointed out that breast, esophageal and non-small cell lung cancers show high loss of heterozygosity (LOH) rates of WWOX. The LOH of WWOX gene also occurred in hepatocellular carcinoma and HCC cell line [15, 16].
Considering the significant potential of WWOX in tumorigenesis and development, genetic variations within this gene are believed to affect the progression of tumor. But disappointedly, studies exploring the relationship between WWOX gene polymorphisms and tumor recurrence or HCC clinical outcome are very limited [17, 18]. To test this hypothesis, 8 HCC patients with early tumor recurrence and 8 patients without recurrence were selected randomly for genotype analysis in order to identify potential SNPs between these two groups and subsquently to determine whether those candidate SNPs may be associated with clinical outcome in another group of 136 patients after curative resection.
Materials and Methods
Study population
From January to July 2009, a total of 152 pathologically confirmed HCC patients were recruited from Department of Liver Surgery, Zhongshan Hospital, Fudan University. Samples were snap-frozen in liquid nitrogen and stored at -80℃. Clinical and pathological characteristics were obtained from the clinical, including age, HBV (hepatitis B virus) status, gender, tumor number and size, tumor thrombi, TMN staging and tumor differentiation. Among those patients, 8 cases with tumor recurrence within 2-years after operation and 8 cases without tumor recurrence were selected randomly for screening the candidate SNPs in WWOX gene by using the Affymetrix Genome-Wide Human SNP 6.0 platform. The remaining 136 patients were used to verify and determine the prognostic significance of the selected SNPs of human HCC. No patients had previous history of other cancers or received any anticancer treatment before curative resection. All patients have written informed consent, and the approval of the ethical committee of Zhongshan hospital was also obtained.
Follow-up
The follow-up data were obtained through medical chart review or direct calling. The follow-up period was calculated from the date of surgery to the date of death or the last follow-up time. The latest follow-up in this study was carried out in April 2015. For postoperative surveillance of tumor recurrence, AFP, ultrasonography and emission computed tomography were used every 3 months for the first 2 year, and semiannually thereafter. If recurrence was under suspicion, supplementary examinations such as Magnetic Resonance Imaging (MRI) were performed.
DNA extraction
After thawed, the cancerous tissues obtained from 16 HCC patients and the following 136 cases were used to extract the genomic DNA (gDNA) by using the Qiaamp DNA mini kit (Qiagen, Hilden, Germany) in strict accordance with the manufacturer's instructions. Thereafter, DNA qualities were assessed on 2% agarose gel and DNA concentration was measured by NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA).
SNP identification and verification
To identify candidate SNPs in WWOX gene, 16 patients, including 8 patients with tumor recurrence within 2-years after operation and 8 patents without tumor recurrence, were selected randomly for genotyping by Affymetrix Genome-Wide Human SNP 6.0 platform. Then we observed the similar distribution of genotypic and allelic frequencies of all detected SNPs in both recurrence group and non-recurrence group and calculated the difference between the two groups. Once the discrepancy rate was more than 50%, this SNP was defined as a candidate SNP and used for the following study.
To verify these candidate SNPs, the remaining 136 patients were also classified as recurrence group and non-recurrence group and then further genotyped by USA Sequenom MassARRAY (TOF-MS). We also observed the distribution of genotypic and allelic frequencies of these candidate SNPs and calculated the difference between the two groups.
Immunohistochemistry
Immunohistochemistry was performed as previously described, with minor modifications according to the previous study. Formalin-fixed paraffin sections were obtained from the Department of Pathology, Zhongshan hospital. The percentage of positive cells and the intensity of immunostaining were used to produce a weighted score for each case.
Statistical analysis
Clinicopathological data were expressed as the mean ± SD. The pearson's chi-squared test or Fisher's exact test were used for comparing clinicopathologic variables. Overall survival (OS) was defined as the interval from the date of radical surgery to the date of death. Recurrence-free survival (RFS) was defined as the interval from the date of radical surgery to the first date of recurrence. OS and RFS were calculated using the Kaplan-Meier method and the log-rank test was used for determining the significance. The univariate and multivariate Cox proportional hazards regression models were used for evaluating the effects of each clinical data, and WWOX genotypes on OS or RFS. All statistical analyses were performed using SPSS 19.0 for Windows (IBM, Armonk, NY, USA) and a P-value less than 0.05 was considered statistically significant.
Result
Clinical characteristics of the study population
A total of 152 patients enrolled in this study. The basic clinicopathologic data of the patients are summarized in Table 1. In the SNP array analysis, 16 patients, including 8 patients with tumor recurrence within 2-years after operation and 8 patients without tumor recurrence, were selected randomly. In the TOF-MS analysis, the remaining 136 patients, including 117 males and 19 females, were analyzed in the following study with a mean age of 53.1 ± 11.2 years, ranging from 26 to 78. 111 patients (81.6%) were HBV markers positive. 120 (88.2%) patients had liver cirrhosis and 48 (35.3%) patients had tumor thrombi, including macroscopic and microscopic tumor thrombi. 90 patients were TNM-StageⅠand Ⅱ based on the criterion of TNM staging system, established in 2010 by the the American Joint Committee on Cancer (AJCC) and the Union for International Cancer Control (UICC).
Distribution of 152 HCC patients' characteristics
| Variables | the SNP array analysis | the TOF-MS analysis | χ2 | P-value |
|---|---|---|---|---|
| All patients (n=16) | All patients (n=136) | |||
| Age, ≥ 50 years (%) | 7 (43.8) | 80 (58.8) | 1.328 | 0.249 |
| Gender, men (%) | 15 (93.8) | 117 (86.0) | 0.696* | |
| HbsAg, positive (%) | 16 (100) | 111 (81.6) | 0.075* | |
| AFP level, ≥ 20 ng/mL (%) | 9 (56.3) | 82 (60.3) | 0.097 | 0.755 |
| Liver cirrhosis, yes (%) | 16 (100) | 120 (88.2) | 0.221* | |
| Tumor number, single (%) | 10 (62.5) | 98 (72.1) | 0.560* | |
| Maximum tumor size, ≥ 5 cm (%) | 9 (56.3) | 67 (49.3) | 0.279 | 0.597 |
| Tumor thrombi, yes (%) | 9 (56.3) | 48 (35.3) | 2.682 | 0.101 |
| TNM Stage (I+II/III+IV) | 14/2 | 90/46 | 3.013 | 0.083 |
| Tumor differentiation (I+II/III+IV) | 11/5 | 93/43 | 0.001 | 0.976 |
* P-value was calculated by Fisher's exact test.
Abbreviation: AFP, alpha fetoprotein.
Identification of candidate SNPs within WWOX
Using the Affymetrix SNP array 6.0, 16 patients were successfully genotyped. Then we observed the distribution of genotypic and allelic frequencies of detected SNPs and selected the SNPs which allele frequency difference are more than 50% between recurrence and non- recurrence group, then selected the SNPs which show P-value less than 0.01 in the Cochran- Armitage trend test (weighted by the additive version, t= (0, 1, 2)). Finally there were 32 SNPs fitting the screen strategy and, therefore, selected as candidate SNPs (data were not shown)
Association analysis of candidate SNPs with HCC recurrence
Thereafter, we conducted genotyping experiments for these candidate SNPs in the remaining 136 HCC patients. In the early stage of our experiment, 58 cases have been diagnosed as tumor recurrence within 2-years after operation and 78 cases without tumor recurrence according to the early follow-up data after operation. The distribution of genotypic and allelic frequencies of candidate 32 SNPs were observed and the differences were also calculated.
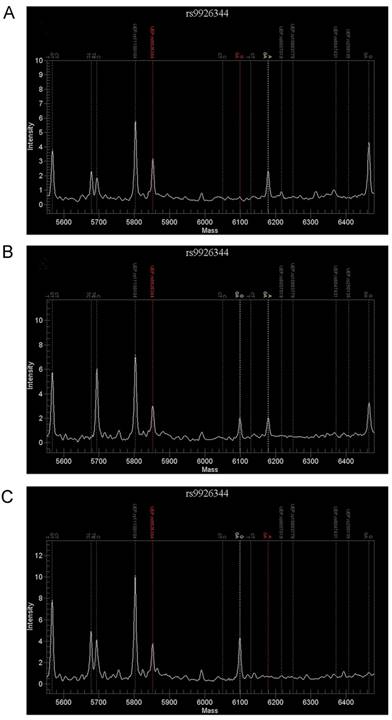
For rs9926344, 136 patients were genotyped successfully by the TOF-MS method and the GG genotype was detected in 108patients, AG in 24 patients and AA in 4 patients (Fig. 1). As is shown in Table 2, the recurrence group had a higher prevalence of AA+AG (31.1%) than the control subjects (10.6%, P = 0.002). For allele comparison, the recurrence group had also a higher A allele frequency (19.0%) than the recurrence group (6.4%, P = 0.001).
For the other candidate SNPs, the genotype and allele frequencies were not associated with the HCC recurrence after the adjustment with the aforementioned confounding factors.
The genotypes and allele frequencies of WWOX (rs9926344) in patients with and without recurrence
| SNPs | genetype | non-recurrence group (n=78) | recurrence group (n=58) | OR (95% CI) | P-value |
|---|---|---|---|---|---|
| rs9926344 | GG | 69 (81.2) | 39 (63.9) | 1(ref) | |
| AG | 8 (9.4) | 16 (26.2) | 3.538 (1.389-9.014) | 0.006 | |
| AA | 1 (1.2) | 3 (4.9) | 5.308 (0.534-52.780) | 0.147* | |
| AA+AG | 9 (10.6) | 19 (31.1) | 3.735 (1.542-9.050) | 0.002 | |
| G allele | 146 (93.6) | 94 (81.0) | 1(ref) | ||
| A allele | 10 (6.4) | 22 (19.0) | 3.417 (1.549-7.538) | 0.001 |
* P-value was calculated by Fisher's exact test.
Association of clinicopathological characteristics of patients carrying different genotypes of rs9926344
| Variables | Grading | All patients | rs9926344 | ||
|---|---|---|---|---|---|
| (n=136) | GG (n=108, %) | AA/AG (n=28, %) | P-value | ||
| Age, years | <50 | 56 | 46 (82.1) | 10 (17.9) | |
| ≥50 | 80 | 62 (77.5) | 18 (22.5) | 0.51 | |
| Gender | female | 19 | 17 (89.5) | 2 (10.5) | |
| male | 117 | 91 (77.8) | 26 (22.2) | 0.362* | |
| AFP level, ng/mL | <20 | 54 | 44 (81.5) | 10 (18.5) | |
| ≥20 | 82 | 64 (78.0) | 18 (22.0) | 0.628 | |
| HbsAg | negative | 25 | 20 (80.0) | 5 (20.0) | |
| positive | 111 | 88 (79.3) | 23 (20.7) | 0.936 | |
| Liver cirrhosis | no | 16 | 14 (87.5) | 2 (12.5) | |
| yes | 120 | 94 (78.3) | 26 (21.7) | 0.524* | |
| Tumor number | single | 98 | 78 (79.6) | 20 (20.4) | |
| multiple | 38 | 30 (78.9) | 8 (21.1) | 0.934 | |
| Tumor size, cm | <5 | 69 | 54 (78.3) | 15 (21.7) | |
| ≥5 | 67 | 54 (80.6) | 13 (19.4) | 0.736 | |
| Tumor thrombi | no | 88 | 73 (83.0) | 15 (27.0) | |
| yes | 48 | 35 (72.9) | 13 (27.1) | 0.167 | |
| TNM stage | I+II | 90 | 76 (84.4) | 14 (15.6) | |
| III+IV | 46 | 32 (69.6) | 14 (30.4) | 0.042 | |
| Tumor differentiation | I+II | 93 | 74 (79.6) | 19 (20.4) | |
| III+IV | 43 | 34 (79.1) | 9 (20.1) | 0.947 | |
* P-value was calculated by Fisher's exact test.
Results of MassARRAY detection for rs9926344. Using the MassARRAY system, 3 genetypes were detected in rs9926344, which included AA genetype (A), AG genetype (B) and GG genetype (C).
Association of different rs9926344 genotypes with clinicopathological variables
Then, we analyzed the association between rs9926344 genotypes and the clinicopathological features in 136 HCC patients. Compared to the rs9926344 GG genotype, the rs9926344 AA+AG genotypes appeared in high frequency in advanced TNM-stage (P = 0.042). No significant differences were found between the two groups for other preoperative and postoperative characteristics (Table 3).
WWOX rs9926344 polymorphisms was associated with the poor prognosis of HCC patients
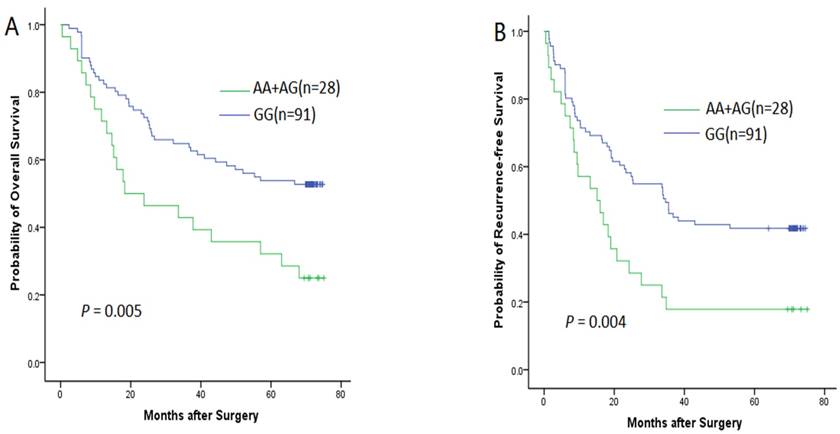
The follow-up data were obtained through medical chart review or direct calling and the latest follow-up in this study was carried out in April 2015. However, 17 patient lost follow-up before the deadline. Therefore, we got the remaining 119 HCC patients' follow-up data for prognosis evaluation. Out of 119 patients, 76 cases were diagnosed as tumor recurrence and 64 cases died of the tumor. Thereafter, we evaluated the association between the WWOX SNP and the prognosis of HCC patients after surgical resection. A significant relationship was found between rs9926344 and RFS or OS in Kaplan-Meier analysis (Fig. 2). The patients who carried the AA+AG genotype had poorer prognosis than those with GG genotype. The Kaplan-Meier curve showed that the median OS time in patients carrying AA+AG genotype was 35.6 months, which was apparently lower than that patients carrying GG genotype (the median OS time was 50.4 months), and the difference is statistically significant (P = 0.005); the median RFS time in patients carrying AA+AG genotype was 24.2 months, which was also lower than that patients carrying GG genotype (the median RFS time was 40.9 months), and the difference is also statistically significant (P = 0.004).
To further evaluate the prognostic value of the polymorphisms of rs9926344 in HCC patients, univariate and multivariate Cox regression analysis were performed. Using the univariate analysis, we found that patients with the AA+AG genotype had a significantly decreased RFS [unadjusted hazard ratio (HR) 2.021, 95% confidence interval (CI) 1.233-3.312, P = 0.005] and OS (unadjusted HR 2.089, 95% CI 1.237-3.526, P = 0.006) compared with those with the GG genotype (Table 4). Besides, we also found that the AFP level, tumor number and size, tumor thrombi, TNM stage and poor tumor differentiation may have significant influence on the prognosis of patients with HCC. Adjusted by these predictive indicators above, multivariate Cox proportional hazards regression model was next performed to evaluate the independent predictive value of rs9926344 polymorphisms on RFS and OS (Table 5). The results showed that, compared to patients carrying the GG genotype, patients with the AA+AG genotype had a higher risk of relapse and death (adjusted HR 1.787, 95% CI 1.042-3.064, P = 0.035 and adjusted HR 1.621, 95% CI 0.917- 2.816, P = 0.097 for RFS and OS, respectively), especially in predicting the tumor recurrence. Taken together, these results imply that rs9926344 AA+AG genotypes could be used as an independent prognostic marker for HCC after operation.
The Kaplan-Meier survival curves for overall survival (A) and recurrence-free survival (B) of the HCC patients stratified by WWOX rs9926344 genotypes.
Univariate analyses of factors associated with OS and RFS of HCC (n=119)
| Variables | Patients | OS | RFS | ||||
|---|---|---|---|---|---|---|---|
| n =119 (%) | Death n (%) | HR (95%CI) | P-value | Recurrence n (%) | HR (95%CI) | P-value | |
| Gender | |||||||
| female | 16(13.4) | 6(9.4) | Ref. | 7(9.2) | Ref. | ||
| male | 103(86.6) | 58(90.6) | 1.674(0.722-3.884) | 0.230 | 69(90.8) | 1.914(0.879-4.168) | 0.102 |
| Age, years | |||||||
| <50 | 46(38.7) | 25(39.1) | Ref. | 30(39.5) | Ref. | ||
| ≥50 | 73(61.3) | 39(60.9) | 0.932(0.564-1.540) | 0.784 | 46(60.5) | 0.869(0.549-1.377) | 0.550 |
| AFP, ng/mL | |||||||
| <20 | 49(41.2) | 20(31.3) | Ref. | 26(34.2) | Ref. | ||
| ≥20 | 70(58.8) | 44(68.7) | 1.923(1.132-3.267) | 0.016 | 50(65.8) | 1.867(1.160-3.004) | 0.010 |
| HbsAg | |||||||
| negative | 21(17.6) | 15(23.4) | Ref. | 16(21.1%) | Ref. | ||
| positive | 98(82.4) | 49(76.6) | 0.606(0.340-1.081) | 0.090 | 60(78.9) | 0.725(0.417-1.258) | 0.253 |
| Liver cirrhosis | |||||||
| no | 15(12.6) | 5(7.8) | Ref. | 9(11.8) | Ref. | ||
| yes | 104(87.4) | 59(92.2) | 2.024(0.812-5.046) | 0.130 | 67(88.2) | 1.209(0.603-2.424) | 0.593 |
| Tumor number | |||||||
| single | 83(69.7) | 38(59.4) | Ref. | 46(60.5) | Ref. | ||
| multiple | 36(30.3) | 26(40.6) | 1.902(1.154-3.137) | 0.012 | 30(39.5) | 2.128(1.339-3.382) | 0.001 |
| Tumor size | |||||||
| <5cm | 58(48.7) | 25(39.1) | Ref. | 32(42.1) | Ref. | ||
| ≥5cm | 61(51.3) | 39(60.9) | 2.061(1.245-3.411) | 0.005 | 44(57.9) | 1.933(1.224-3.051) | 0.005 |
| Tumor thrombi | |||||||
| no | 78(65.5) | 33(51.6) | Ref. | 42(55.3) | Ref. | ||
| yes | 41(34.5) | 31(48.4) | 2.647(1.615-4.339) | <0.001 | 34(44.7) | 2.490(1.577-3.932) | <0.001 |
| TNM stage | |||||||
| I+II | 77(64.7) | 31(48.4) | Ref. | 41(53.9) | Ref. | ||
| II+IV | 42(35.3) | 33(51.6) | 3.260(1.986-5.352) | <0.001 | 35(46.1) | 2.907(1.841-4.589) | <0.001 |
| Tumor differentiation | |||||||
| I+II | 83(69.7) | 40(62.5) | Ref. | 48(63.2) | Ref. | ||
| II+IV | 36(30.3) | 24(37.5) | 1.848(1.112-3.070) | 0.018 | 28(36.8) | 2.866(1.168-2.981) | 0.009 |
| rs9926344 | |||||||
| GG | 91 (70.5) | 43 (63.2) | Ref. | 53 (66.3) | Ref. | ||
| AA+AG | 28(21.7) | 21 (30.9) | 2.089(1.237-3.526) | 0.006 | 23 (28.7) | 2.021 (1.233-3.312) | 0.005 |
Abbreviation: AFP, alpha fetoprotein; HR, hazard ratio; CI, confidence interval; Ref. Referent.
Multivariate analyses of factors associated with OS and RFS of HCC (n=119)
| variables | OS | RFS | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95%CI | P-value | |
| AFP level (<20ng/mL vs. ≥20ng/mL) | 1.615 | 0.925-2.820 | 0.092 | 1.742 | 1.051-2.888 | 0.031 |
| Tumor number (single vs. multiple) | 1.555 | 0.860-2.813 | 0.144 | 2.177 | 1.246-3.802 | 0.006 |
| Tumor size (<5cm vs. ≥5cm) | 2.004 | 1.114-3.605 | 0.02 | 1.998 | 1.174-3.399 | 0.011 |
| Tumor thrombi (no vs. yes) | 1.802 | 1.002-3.244 | 0.049 | 1.843 | 1.075-3.160 | 0.026 |
| TNM stage (I+II vs. III+IV) | 1.434 | 0.712-2.888 | 0.313 | 1.188 | 0.630-2.239 | 0.595 |
| Tumor differentiation (I+II vs. III+IV) | 1.341 | 0.781-2.305 | 0.287 | 1.321 | 0.807-2.162 | 0.268 |
| rs9926344 (GG vs. AA+AG) | 1.621 | 0.917-2.866 | 0.097 | 1.787 | 1.042-3.064 | 0.035 |
Abbreviation: AFP, alpha fetoprotein; HR, hazard ratio; CI, confidence interval.
WWOX expression in HCC patients' samples
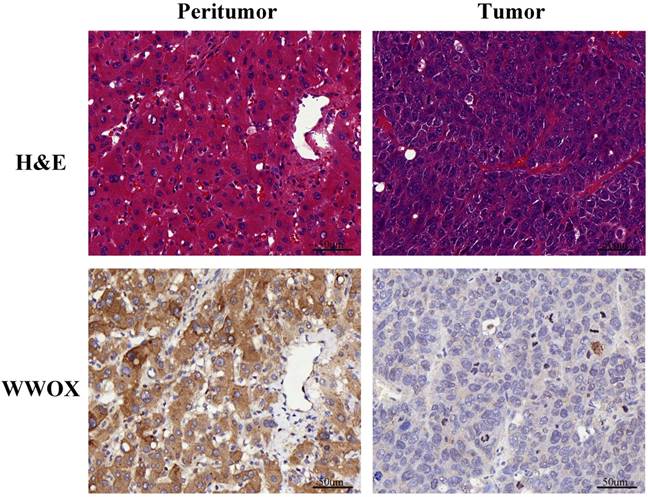
Haematoxylin and eosin (H&E) staining results showed that the malignant cells were relatively homogeneous in the tumor tissue. Then, we performed immunohistochemistry (IHC) analysis of WWOX expression in 100 HCC patients, which were selected randomly among those patients above. First, we compared the difference of WWOX expression between the adjacent normal liver tissues (the peritumor tissue) and the tumor tissues. As shown in Fig. 3, the staining pattern of WWOX was mainly cytoplasmic in both the tumor and the corresponding peritumor tissues and IHC staining revealed that the WWOX expression was significantly lower in tumor tissues compared to the peritumor tissues. Thereafter, we compared the difference of WWOX expression between the AA group or AG group and the GG group and the results showed that the WWOX down-regulation is more frequent in patients carrying rs9926344 AG genotype than those carrying GG genotype, which is 75.0% (12/16) to 43.9% (36/82), with the difference being statistically significance (P = 0.023). And this result suggested the poor prognostic value of A allele. No significance was found between the AG and AA genotypes (75.0% vs. 0%, P = 0.098) or the AA and GG genotypes (0% vs. 43.9%, P = 0.504).
Discussion
Increasing evidences have indicated that single nucleotide polymorphisms (SNPs) within oncogenes or tumor suppressors are widely viewed as the key factors in the carcinogenesis and progression of human tumors [17-20]. Our previous studies focused on the gene polymorphisms on chromosome 8p and their relationship to HCC metastasis and we found genetic variations such as single nucleotide polymorphisms on chromosome 8p may contribute to the modification of the promoter transcriptional activity and gene expression, which may influence HCC metastatic potential and clinical prognosis [21-24]. In the present study, using Genome-Wide Human SNP Array 6.0 and time-of-flight mass spectrometry, we detected the genomic SNPs within WWOX from HCC patients and identified the significant SNPs genotype. We analyzed the difference between different groups and investigated their values in predicting tumor recurrence and prognosis in HCC patients. We found that the AA or AG genotypes of rs9926344 within tumor suppressor gene WWOX indicate higher risk of HCC recurrence and, therefore, can be used as an independent predictor of HCC recurrence. Immunohistochemical expression analysis showed that the expression of WWOX is significantly down regulated in HCC tissue, when comparing to the pericancerous tissues. Next, in order to further investigate the possible correlation between the rs9926344 polymorphism and WWOX protein expression, the protein expression levels in tumorous samples between different genotypes were also analyzed. And the result indicated that the WWOX protein down-regulation is more frequent in patients with AG genotype compared to the GG genotype, suggesting the poor prognostic significance of A allele. However, the WWOX expression level in patients with AA genotype is not significantly different when compared to those in AG or GG group. This phenomenon may contribute to small number of patients with the AA genotype (only 2 samples) and large standard error. If the sample enlarged, some differences among these groups might be found. Therefore, further investigation is required to rigorously verify this conjecture.
Representative pictures of immunohistochemical analyses for WWOX in the liver tissue specimens and corresponding HE staining results. Scale bar, 50 μm; original magnifications, ×400.
WWOX gene's official name is WW domain containing oxidoreductase, also known as WOX1 or FOR. This gene spans the common fragile site FRA16D, located in 16q23.3-24.1 locus. The 1.3Mb open reading frame (ORF) of WWOX, containing 9 exons, encodes two WW binding domains at its N terminal region, a nuclear localization sequence (NLS) and a short-chain alcohol dehydrogenases/reductases (SDR) domain at C terminal [25]. Evidence shows that WWOX possess apoptosis properties by interacting with proteins in different signaling pathways. When cells affected by stress signals or tumor necrosis factor (TNF), WWOX regulates the apoptosis by down regulating apoptosis inhibitors Bcl-2 and Bcl-xL, binding and stabilizing the activating form of p53 protein, translocating into the nucleus and mitochondria to induce apoptosis synergistically [26-29]. Another study points out that WWOX enhances TNF cytotoxicity in L929 fibroblasts and is regarded as a downstream effector in TNF signaling pathway [27]. A zinc finger-like protein Zfra acts in a dual role in regulating cytotoxic effects of TNF, and physically interacts with the N-terminal first WW domain and C-terminal SDR domain of WWOX [30]. Research shows that staurosporine- induced apoptosis results in WWOX and Zfra cotranslocting to mitochondria, and Zfra suppressing the expression of Bcl-2 and invoking apoptosis via mitochondrial pathway [31]. However, another research indicates that TNF can increase the binding of Zfra and WWOX, and the over expressed Zfra sequester WWOX in the cytoplasm. The binding abolishes both apoptotic function of WWOX and Zfra [30], indicating that Zfra is a negative regulator of WWOX.
Moreover, it is considered that the LOH and homozygous deletions in the WWOX gene are widely detected and have an intimate connection with human disease, including cancer[32-34]. However, the next generation sequencing made more point mutations of WWOX been noticed. The c.358 C>T (Arg120Trp) missense polymorphism is found in colorectal tumors [16], non- small-cell lung cancers (NSCLC) [35], and esophageal cancers[36]. For instance, the c.358 C>T (Arg120Trp) and the c.760 C>T (Arg254Cys) are in exon 4 and 7, and they are considered to be related to lung carcinogenesis[35]. While the c.844 C>G (Pro282Ala) is a risk factor for differentiated thyroid carcinoma[37] and is associated with gastric cardia adenocarcinoma (GCA) susceptibility in Northern Chinese[38]. To our knowledge, only few studies has been investigating on the relationship between different polymorphisms of WWOX gene and tumor occurrence and progression in patients with HCC. After genotyping 5 single-nucleotide polymorphisms (SNPs) of the WWOX gene in 354 HCC patients and 708 normal controls using an ABI StepOne TM Real-Time PCR System, Lee HL and colleagues[17] found that a polymorphism within WWOX (rs12918952) can be used as a significant and reliable biomarker for predicting both early HCC occurrence and disease progression. In brief, WWOX is involved in so many pathways that affect apoptosis and metastasis, manifesting that there are potential effects of WWOX in regulating tumor activities to be uncovered. Based on those valuable results, we further proposed a significant difference between HCC patients with and without tumor recurrence in genetic variations of WWOX gene. To test this hypothesis and identify potential SNPs, genotype data from 8 HCC patients with tumor recurrence and 8 patients without recurrence were obtained using Affymetrix SNP 6.0 Array. The difference between recurrence group and non-recurrence group was calculated and the candidate SNPs were screened and further verified by TOF-MS. Intriguingly, our present results were consistent with previous studies reconfirming that there is correlation between the recurrence and WWOX polymorphisms, that provides insights for further efforts that investigate WWOX tumor suppression gene.
To summarize, we found that a polymorphism in WWOX gene (rs9926344) has an obvious impact on tumor recurrence in HCC patients after curative operation. This interesting finding has a potential clinical significance in assessing tumor progression and predicting postoperative tumor recurrence in patients with hepatocellular carcinoma.
Abbreviations
AFP: alpha fetoprotein; CCFS: the common chromosomal fragile site; CI: confidence interval; GCA: gastric cardia adenocarcinoma; gDNA: genomic DNA; HBV: hepatitis B virus; HCC: hepatocellular carcinoma; HR: hazard ratio; IHC: immunohistochemistry; LOH: loss of heterozygosity; MRI: magnetic resonance imaging; NLS: nuclear localization sequence; ORF: open reading frame; OS: overall survival; RFS: recurrence-free survival; SDR: short chain dehydrogenase/reductase domain; SNP: single nucleotide polymorphism; TNF: tumor necrosis factor; TOF-MS: time-of-flight mass spectrometry; WWOX: WW domain-containing oxidoreductase.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81472672, 81572301, 81502487) and the Shanghai Rising Star of Young Scientist Project (Grant No. 13QH1401000).
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010;7(8):449-58
2. El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142(6):1264-73
3. Torre LA, Bray F, Siegel RL. et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87-108
4. Gluer AM, Cocco N, Laurence JM. Systematic review of actual 10-year survival following resection for hepatocellular carcinoma. HPB (Oxford). 2012;14:285-290
5. Bednarek AK, Laflin KJ, Daniel RL. et al. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60(8):2140-2145
6. Ried K. et al. Common chromosomal fragile site FRA16D sequence: Identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum Mol Genet. 2000;9(11):1651-1663
7. Aqeilan RI, Abu-Remaileh M, Abu-Odeh M. The common fragile site FRA16D gene product WWOX: roles in tumor suppression and genomic stability. Cell Mol Life Sci. 2014;1(23):4589-4599
8. Schrock MS, Huebner K. WWOX: a fragile tumor suppressor. Exp Biol Med (Maywood). 2015;240(3):296-304
9. Abu-Odeh M, Salah Z, Herbel C. et al. WWOX, the common fragile site FRA16D gene Product, regulates ATM activation and the DNA damage response. Proc Natl Acad Sci USA. 2014;111(44E):4716-25
10. Abu-Remaileh M, Joy-Dodson E, Schueler-Furman O. et al. Pleiotropic Functions of Tumor Suppressor WWOX in Normal and Cancer Cells. J Biol Chem. 2015;290(52):30728-35
11. Choo A, O'Keefe LV, Lee CS. et al. Tumor suppressor WWOX moderates the mitochondrial respiratory complex. Genes Chromosomes Cancer. 2015;54(12):745-61
12. Li G, Sun L, Mu Z. et al. Ectopic WWOX Expression Inhibits Growth of 5637 Bladder Cancer Cell In Vitro and In Vivo. Cell Biochem Biophys. 2015;73(2):417-425
13. Sun W, Dou J, Zhang L. et al. Expression of CD133, E-cadherin and WWOX in colorectal cancer and related analysis. Pak J Med Sci. 2017;33(2):425-429
14. Wen J, Xu Z, Li J. et al. Decreased WWOX expression promotes angiogenesis in osteosarcoma. Oncotarget. 2017 doi: 10.18632/oncotarget.17126
15. Aderca I, Moser CD, Veerasamy M. et al. The JNK inhibitor SP600129 enhances apoptosis of HCC cells induced by the tumor suppressor WWOX. J Hepatol. 2008;49(3):373-83
16. Paige AJ, Taylor KJ, Taylor C. et al. WWOX: a candidate tumor suppressor gene involved in multiple tumor types. Proc Natl Acad Sci USA. 2001;98(20):11417-22
17. Lee HL, Cheng HL, Liu YF. et al. Functional genetic variant of WW domain-containing oxidoreductase (WWOX) gene is associated with hepatocellular carcinoma risk. PLoS One. 2017;12(4e):0176141
18. Lin WR, Hsu CW, Yeh CS. et al. Combinations of single nucleotide polymorphisms WWOX- rs13338697, GALNT14-rs9679162 and rs6025211 effectively stratify outcomes of chemotherapy in advanced hepatocellular carcinoma. Asia Pac J Clin Oncol. 2017 doi: 10.1111/ajco. 12745
19. Lien MY, Lin CW, Tsai HC. et al. Impact of CCL4 gene polymorphisms and environmental factors on oral cancer development and clinical characteristics. Oncotarget. 2017;8(19):31424-31434
20. Liu W, Tian T, Liu L. et al. A functional SNP rs1892901 in FOSL1 is associated with gastric cancer in Chinese population. Sci Rep. 2017;7:41737
21. Qin LX, Tang ZY, Ye SL. et al. Chromosome 8p deletion is associated with metastasis of human hepatocellular carcinoma when high and low metastatic models are compared. J Cancer Res Clin Oncol. 2001;127(8):482-8
22. Wu X, Jia HL, Wang YF. et al. HTPAP gene on chromosome 8p is a candidate metastasis suppressor for human hepatocellular carcinoma. Oncogene. 2006;25(12):1832-40
23. Ren N, Wu JC, Dong QZ. et al. Association of specific genotypes in metastatic suppressor HTPAP with tumor metastasis and clinical prognosis in hepatocellular carcinoma. Cancer Res. 2011;71(9):3278-86
24. Wu JC, Jia HL, Li ZR. et al. Genomic aberrations in the HTPAP promoter affect tumor metastasis and clinical prognosis ofhepatocellular carcinoma. PLoS One. 2014;9(3e):90528
25. Bednarek AK, Laflin KJ, Daniel RL. et al. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, aregion frequently affected in breast cancer. Cancer Res. 2000;60(8):2140-5
26. Lo JY, Chou YT, Lai FJ. et al. Regulation of cell signaling and apoptosis by tumor suppressor WWOX. Exp Biol Med (Maywood). 2015;240(3):383-91
27. Chang NS, Pratt N, Heath J. et al. Hyaluronidase induction of a WW domain-containing oxidoreductase that enhances tumornecrosis factor cytotoxicity. J Biol Chem. 2001;276(5):3361-70
28. Chang NS, Doherty J, Ensign A. et al. WOX1 is essential for tumor necrosis factor-, UV light-, staurosporine-, and p53-mediated celldeath, and its tyrosine 33-phosphorylated form binds and stabilizes serine 46-phosphorylatedp53. J Biol Chem. 2005;280(52):43100-8
29. Chen ST, Chuang JI, Cheng CL. et al. Light-induced retinal damage involves tyrosine 33 phosphorylation, mitochondrial and nucleartranslocation of WW domain-containing oxidoreductase in vivo. Neuroscience. 2005;130(2):397-407
30. Hong Q, Hsu LJ, Schultz L. et al. Zfra affects TNF-mediated cell death by interacting with death domain protein TRADD andnegatively regulates the activation of NF-kappaB, JNK1, p53 and WOX1 during stress response. BMC Mol Biol. 2007;8:50
31. Hsu LJ, Hong Q, Schultz L. et al. Zfra is an inhibitor of Bcl-2 expression and cytochrome c release from the mitochondria. Cell Signal. 2008;20(7):1303-12
32. Driouch K, Prydz H, Monese R. et al. Alternative transcripts of the candidate tumor suppressor gene, WWOX, are expressed at highlevels in human breast tumors. Oncogene. 2002;21(12):1832-40
33. Ruan X, Liu H, Boardman L. et al. Genome-wide analysis of loss of heterozygosity in breast infiltrating ductal carcinoma distant normal tissue highlights arm specific enrichment and expansion across tumor stages. PLoS One. 2014;9(4e):95783
34. Ben-Salem S, Al-Shamsi AM, John A. et al. A novel whole exon deletion in WWOX gene causes early epilepsy, intellectual disability and optic atrophy. J Mol Neurosci. 2015;56(1):17-23
35. Baykara O, Demirkaya A, Kaynak K. et al. WWOX gene may contribute to progression of non-small-cell lung cancer (NSCLC). Tumour Biol. 2010;31(4):315-20
36. Gonzalez-Garay ML, McGuire AL, Pereira S. et al. Personalized genomic disease risk of volunteers. Proc Natl Acad Sci USA. 2013;110(42):16957-62
37. Cancemi L, Romei C, Bertocchi S. et al. Evidences that the polymorphism Pro-282-Ala within the tumor suppressor gene WWOX is a newrisk factor for differentiated thyroid carcinoma. Int J Cancer. 2011;129(12):2816-24
38. Guo W, Dong Z, Dong Y. et al. Genetic and epigenetic alterations of WWOX in the development of gastric cardiaadenocarcinoma. Environ Mol Mutagen. 2013;54(2):112-23
Author contact
![]() Corresponding author: Dr. Ning Ren (e-mail: ren.ningsh.cn, Tel: +86-21-64037181) and Dr. Qinghai Ye (e-mail: ye.qinghaish.cn, Tel: +86-21-64037181), Department of Liver Surgery, Zhongshan Hospital, Fudan University, No.180 Fenglin Road, Shanghai 200032, China.
Corresponding author: Dr. Ning Ren (e-mail: ren.ningsh.cn, Tel: +86-21-64037181) and Dr. Qinghai Ye (e-mail: ye.qinghaish.cn, Tel: +86-21-64037181), Department of Liver Surgery, Zhongshan Hospital, Fudan University, No.180 Fenglin Road, Shanghai 200032, China.

 Global reach, higher impact
Global reach, higher impact